Abstract
Marseilleviruses are a group of double-stranded DNA viruses infecting Acanthamoeba within the phylum Nucleocytoviricota and are ubiquitous in water and soil globally. Here, we report six strains of marseilleviruses isolated from environmental samples in the Yangtze River Delta and the Pearl River Delta, China. Viral particles exhibited icosahedral shaped capsids measuring about 220 ~ 240 nm in diameter. Based on stability assays, viral particles were halotolerant and acid-tolerant, but sensitive to chloroform and high temperature. Genomics and phylogenetic analyses showed that these strains were highly conserved compared with other reported marseilleviruses. Diverse members of the small multidrug resistance (SMR) family of transporter, which is a type of antibiotics resistance gene (ARG) and contribute to the feature of antibiotic resistance in bacteria, to our best knowledge, are firstly described in Marseilleviridae. The alignments of primary structures and in-silico tertiary structures reveal structural and potential functional similarity between giant viral and bacterial SMR, suggesting a possible role in viruses’ interaction with antibiotics. The biological properties of marseillevirus and the discovery of viral SMR provide insight in the external and intracellular environment fitness of these large amoeba-infecting viruses.
Similar content being viewed by others
Introduction
The first marseillevirus was isolated from a water sample collected in a cooling tower in Paris, France1. It is the founder of a new viral family, officially recognized and named Marseilleviridae by the International Committee of Taxonomy of Viruses2. Since 2009, it has been shown that marseilleviruses are ubiquitous globally in water, soil, sediment, biospecimens and so on3. Based on the molecular phylogenetic analysis of the major capsid protein, the isolated members of Marseilleviridae are classically organized into five phylogenetic lineages (A, B, C, D and E)3.
The morphogenesis of marseillevirus particles slightly varies in different lineages. Marseillevirus massiliense strain T19, a member of lineage A, has an icosahedral virion with a 250 nm diameter with fibrils on the surface1. Meanwhile, for members of lineage B, negative staining of transmission electron microscopy (TEM) showed that Losannavirus lausannense strain 7715 has an icosahedral virion with a 220 nm diameter without fibrils4 and cryo-electron microscopy showed virion of Jyvaskylavirus strain fiAc85 with a 250 nm diameter without fibrils5. M. massiliense is able to form giant vesicles containing various numbers of viral particles. Giant vesicles have a higher efficacy of infection and more resistance to high temperature than naked particles, indicating that giant vesicles confer a physical advantage to marseillevirus6. However, as far as we know, the study of biological properties of giant viral particles under different environments remains limited.
Amoeba is a predator of microbes and the host of most isolated giant viruses. It constitutes a gene melting pot allowing intracellular microbial pathogens to evolve by gene acquisition7. Thus, it was hypothesized that the genome repertoire of marseillevirus is composed of genes derived from several distinct sources3. In addition, Yi et al. suggested that viruses within the phylum Nucleocytoviricota served as a potential vehicle for the transmission of ARG in the biome8. It has been proven that the dihydrofolate reductase (DHFR) encoded by L. lausannense, which is a type of ARG, is found to confer resistance to trimethoprim and pyrimethamine while expressed in Saccharomyces cerevisiae and Escherichia coli8,9. What’s more, ribosomal protection protein (RPP) encoded by Marseilleviridae, another type of ARG, is predicted to confer resistance to tetracycline8.
In this study, we present six strains of marseillevirus, namely Shanghaivirus strain J2016JW, Quyangvirus strain J2018QY, Dashavirus strain R2023E, Dashavirus strain R2023DC, Dashavirus strain R2023PK, and Dashavirus strain R2023JZZ. They were isolated from different water samples collected from the Yangtze River Delta and the Pearl River Delta, China. These viruses exhibit high conservation with other described marseilleviruses in morphogenesis of viral particles and genomes. The biological properties of viral particles and giant vesicles reveal viruses’ fitness of extracellular environment. Among marseillevirus genes without significantly similar sequences in databases and function unassigned, we discovered diverse novel sequences that shared structural similarities with bacterial SMR. These SMR-like genes indicate the potential interaction between giant viruses and chemical components and shed insights into viruses’ fitness in intracellular environment.
Results
Isolation of marseilleviruses from different water samples
During the survey period from 2016 to 2024, we collected a total of 4 and 14 different water samples in the Yangtze River Delta and the Pearl River Delta, respectively (Table S1). After six passages of amoebal co-culture with these water samples, eleven of the eighteen inocula showed stable lytic activity against Acanthamoeba castellanii. Subsequent virus cloning, followed by transmission electron microscopy (TEM) and PCR verification using primers targeting the DNA polymerase delta catalytic subunit (DNApol), D5-like primase-helicase (D5), and major capsid protein (MCP) gene, we identified six marseilleviruses that caused A. castellanii cell death (Fig. 1, S1).
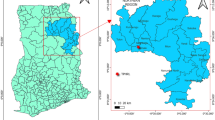
Geographical sources of marseilleviruses. (A) The location of the origin of the samples and TEM results of marseilleviruses in giant vesicles in cytoplasmic of A. castellanii cells at 12 h p.i. The map was generated using Ocean Data View v5.7.2 (https://odv.awi.de/). (B) Agarose gel electrophoresis analysis showed PCR products amplified from different co-cultures of water sample with amoeba using a pair of primers targeting DNApol. Lane 1 to 18 (location as described in Table S1): (1) Jiangwan Campus, (2) Quyang Sewage Treatment Plant, (3) Dongqu Sewage Treatment Plant, (4) North Guoquan Road, (5) Futian River, (6) Futian Mangrove Reserves, (7) Futian Mangrove Reserves, (8) Dasha River, (9) Seven Star Bay, (10) Tungchung Tourist Area, (11) Shenzhen Talent Park, (12) Xiwan Mangrove Park, (13) Xiaoao Mountain, (14) Jiazi Town, (15) Laichang Ferry, (16) Former Australian Port, (17) Red Bay, and (18) Yonghe River.
Biological properties of marseillevirus
The morphology of the marseillevirus particles observed in cytoplasmic of A. castellanii cells using TEM showed that they were icosahedral. The diameter of Shanghaivirus virion was 243 ± 17 nm, and that of Quyangvirus and Dashavirus were 216 ± 14 nm and 210 ± 12 nm, respectively. Giant vesicles containing dozens of viral particles were observed in the cytoplasm at 12 h post-infection (p.i.) before cell lysis.
The one-step growth curve showed that their replication cycles were completed within approximately 16 h (Fig. 2A). Following treatment with lysis buffer (see Methods), the increase in the total number of viral particles varied from 150% for Shanghaivirus to 283% for Dashavirus, indicating that the number of particles in the giant vesicles varied among different marseilleviruses (Fig. 2B). The burst size is the total number of infectious particles released, which were treated with lysis buffer, at the end of one cycle of replication/number of infected amoebae. The burst sizes varied from 2,563 ± 822 particles/cell for Shanghaivirus to 4,759 ± 788 particles/cell for Dashavirus. Both giant vesicles and viral particles of all marseilleviruses were highly sensitive to chloroform treatment (Fig. 2C, S2A). The inactivation of viral particles indicated that lipid components were important for marseillevirus infection. However, no morphological change was detected between chloroform-treated and untreated viral particles (Fig. 2D–F) because chloroform sensitivity alone did not show the presence of lipids on the surface of viral particles.
Biological properties of marseilleviruses. (A) The viral replication logarithmic curve of six strains of marseillevirus. Histogram of the percentage of infectious particles after treatment with (B) lysis buffer, or (C) chloroform. TEM negative staining of (D) a giant vesicle, (E) a viral particle, and (F) a chloroform-treated viral particle. Histogram of the percentage of infectious viruses after treatment with (G) 5 M of NaCl, (H) PBS with pH at 2, (I) PBS with pH at 12, and (J) PBS at 60 °C. Data points represent the mean values and standard deviations from triplicate experiments.
Considering the fact that marseilleviruses were isolated from different water samples, we tested the impacts of salinity, pH, and temperature on the stability of giant vesicles and viral particles. NaCl incubation experiments showed that both giant vesicles and viral particles were halotolerant, with 5 M NaCl treatment having no effect on their infectivity (Fig. 2G, S2B). The pH incubation experiments showed that giant vesicles displayed acid and alkaline tolerance with infectivity maintained above 80% at pH 2 and 12, respectively, whereas the infectivity of viral particles remained below 30% (Fig. 2H, I, S2C). The thermal stability tests were consistent with the previous finding that giant vesicles had higher resistance to thermal stress than viral particles6 (Fig. 2J, S2D). These results indicated that the difference in acid/alkaline and thermal tolerance between giant vesicles and viral particles highlighted the importance of the membrane, which wraps viral particles to form giant vesicles6, in enhancing virus fitness in diverse external environments.
Genomic content and phylogenetic analysis
Illumina sequencing and sequence assembly yielded a single contig for each new marseillevirus genome, with sizes ranging from 360,239 to 381,001 bp (Table S2), which was in line with the genome sizes of reported marseilleviruses3. The GC contents of the six marseilleviruses ranged from 42.88 to 44.70%, which were concordant with reported marseilleviruses. The numbers of predicted protein genes varied between 472 for Shanghaivirus and 547 for Dashavirus (Table S2). The average distance between consecutive genes varied between 48-nt for Dashavirus and 56-nt for Shanghaivirus, resulting in an average coding density of 93.18%. We found no evidence for the presence of tRNA and aminoacyl tRNA synthetase genes. A total of 3,096 proteins, ranging from 29 to 2,065 amino acids, with an average length of 221, were predicted. All these protein genes were under purifying selection, as the average values of omega (dN/dS ratio, which was the ratio of non-synonymous mutations to synonymous mutations) of Shanghaivirus versus M. massiliense and Dashavirus strain 2023E versus Noumeavirus, which was isolated from New Caledonia11, were 0.205 ± 0.347 (median = 0.110) and 0.165 ± 0.260 (median = 0.096), respectively. This supported that most viral protein genes contribute to virus fitness10.
In addition, the average nucleotide identity (ANI) analysis of the whole genome was performed to compare the nucleotide sequence divergence of marseilleviruses. The ANI of Shanghaivirus compared with M. massiliense, which was isolated from France1, was 97.42% with an alignment percentage of 93.33%. Meanwhile, the ANIs of Quyangvirus and four Dashaviruses compared with Noumeavirus, varied from 90.87% (85.81%) to 96.10% (87.63%) (Fig. 3). Furthermore, to estimate the phylogenetic position of six marseilleviruses in Marseilleviridae, molecular phylogenetic analysis was performed based on five core genes of Nucleocytoviricota12. Phylogenetic analysis revealed that Quyangvirus and four Dashaviruses formed a close evolutionary relationship with Noumeavirus, a representative of lineage B. Similarly, Shanghaivirus exhibited a closer evolutionary relationship with M. massiliense, representing lineage A. Concatenated phylogenetic analysis of five core genes, combined with ANI results, confirmed that Shanghaivirus is a conserved member of lineage A, while Quyangvirus and four Dashaviruses are conserved members of lineage B (Fig. 3). These results indicated extreme sequence conservation among marseilleviruses isolated from very distant locations.
Molecular phylogenetic analysis based on the amino acid sequences of the concatenated NCLDV core genes. Numbers on each branch represent the percentage of trees containing that branch. Background colors of marseillevirus names represent different lineages. Solid symbols adjacent to virus names represent the environmental source for isolation. The middle panel of the heat map shows the average nucleotide identity of whole genomes with percentage of alignment. The right panel shows the presence (violet) or absence (pink) of antibiotic resistance genes. DHFR dihydrofolate reductase, RPP ribosomal protection protein, SMR small multidrug resistance family of transporter.
Putative antibiotic resistance genes
Considering a high proportion of marseillevirus genes corresponding to genes without significantly similar sequences in databases3, we applied protein structure prediction analyses on conserved proteins, which were identified in more than half of the Marseilleviridae members (12/23 strains analyzed in this study). We found three new SMR genes with high confidence (Fig. S3. Table S3-S5). SMR genes were under purifying selection among all members of Marseilleviridae, as the average values of omega were 0.198 ± 0.216 (median = 0.078), 0.064 ± 0.035 (median = 0.045) and 0.015 ± 0.009 (median = 0.018), suggesting they were important for virus fitness. Therefore, they are highly conserved and ubiquitous across all isolated marseilleviruses (Fig. 3). For instance, in Dashavirus strain R2023E, which is a member of lineage B, the tertiary structures of MarDSR188, MarDSR189 and MarDSR284 were composed of four transmembrane (TM) helices, showing similarity to that of EmrE, which is the prototypical member of the bacterial SMR family13,14. Similar SMRs were observed for MarSH128, MarSH129 and MarSH228 in Shanghaivirus, which is a member of lineage A. The primary structures of three Dashaviral SMRs were diverse, since only 25% of amino acid residues of MarDSR188 (25/101) were identically aligned to that of MarDSR189, and no identical residue was found in MarDSR284 compared with the previous two viral SMRs. Therefore, viral SMRs formed divergent and distant branches in the result of molecular phylogenetic analysis (Fig. 4, S4), we named the types of marseilleviral SMR as pimav1, pimav2, and pimav3, represented by MarDSR188, MarDSR189 and MarDSR284, respectively. Similar to EmrE15, conserved amino acid residues were found in transmembrane (TM) helices, while amino acid residues in loops were divergent (Fig. 5, S5). Since EmrE functions as a homodimer of a small four-TM protein13, the homodimers of viral SMRs were predicted with high confidence (Fig. S3). The in-silico tertiary structure of viral SMR homodimer revealed the antiparallel arrangement of two monomers and suggested the function similarity between viral SMRs and EmrE (Fig. 6, S6). Notably, similar highly conserved residues, which were reported in EmrE to be important and implicated in substrate binding and proton-dependent transport, especially Glu1413,16, were found in the members of the pimav1 type of SMR (Fig. 6, S6). Overall, the in-silico tertiary structures of viral SMRs and their homodimers suggest potential functional similarities to those seen in antibiotic-resistant bacteria.
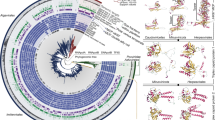
Molecular phylogenetic analysis based on the amino acid sequences of SMR. Color blocks of leave name represent different species taxonomic groups and color categories of branches represent different types of viral SMR. The imiterv type was found in mimivirus and klosneuvirus, the pimav type was found in marseillevirus, and the algav type was found in alga virus. Circles on each branch represent the percentage of trees containing that branch. The tertiary structures of bacterial and viral SMR are attached to the leaf names and the crystal structures are highlighted with grey surface covered. NCBI accession numbers of all SMR protein sequences and their species names are shown in Supplementary Fig. 3.
Primary and secondary structure of bacterial and viral SMRs. (A) Multiple amino acid sequence alignments of bacterial and viral SMRs using MAFFT alignment. High consensus residues are highlighted in red. Secondary structures of TM helices are represented above and below the multiple sequence alignments, respectively. RCSB accession numbers for E. coli SMRs: 7MH6.A and 2I68.A. NCBI accession numbers for viral SMRs: Shanghaivirus strain J2016JW (MarSH128, AVR52833.1), Dashavirus strain R2023E (MarDSR188, WNL50227.1), Tunisfontainevirus strain 484 (TNSORF421, AHC55139.1), Brazilianmarseillevirus strain BH2014 (ORFR113, YP_009238618.1), and Goldenmarseillevirus strain Golden (GMAR39, YP_009310156.1). (B) Secondary structures of Dashavirus SMR (MarDSR188, left) and E. coli SMR (2I68.A, right). Highly conserved residues between different species are in violet circles and conserved residues within marseilleviruses are in medium turquoise circles. Residues of E. coli SMR reported to be implicated in substrate binding and transport13 are highlighted with red frame. The four TM helices are indicated in bold and the membrane shown by the light orange box.
In-silico tertiary structure of a viral SMR homodimer. (A) Side view of the homodimer of MarDSR188 encoded by Dashavirus strain R2023E. The four TM helices, N-terminus and C-terminus of a monomer are label. (B) Top view of the viral SMR structures, with red spheres indicating the positions of residues are similar to that of which were reported to be implicated in substrate binding and transport in E. coli SMR13. One monomer is rendered in color gradient with label in bold and the other one is shown in gray.
Additionally, two viral ARGs, DHFR and RPP, which had been described in literature8,9, were found in all our marseilleviruses (Fig. 3). DHFRs and RPPs were under purifying selection, as the average values of omega were 0.081 ± 0.063 (median = 0.064) and 0.054 ± 0.016 (median = 0.055), respectively, indicating their importance for virus fitness in intracellular environments.
Discussion
Since the discovery of Mimivirus bradfordmassiliense, it has been found that giant viruses, like their amoeba hosts, are distributed worldwide17,18,19. Marseilleviruses and mimiviruses were isolated from the same laboratory acanthamoeba host and grouped into the same class, Megaviricetes1,2. The structural integrity of mimivirus particles exhibited distinct responses to different environmental conditions. Under an alkaline condition (pH 11), as well as when exposed to organic solvents or enzymatic treatments, the particles maintained their stability, with no observable disruption of virions. In contrast, exposure to acidic conditions (pH 3), high temperatures (100 °C), or high salt concentrations (4 M NaCl) led to significant morphological alterations, resulting in the disruption of the virions20. Herein, the stability assays showed that the biological properties of marseillevirus was differed from that of mimivirus. Marseillevirus particles were halotolerant and acid-tolerant, but sensitive to chloroform and alkaline. Since marseillevirus particles were wrapped in membranes to form giant vesicles6, the stability assays further revealed giant vesicles exhibited better thermal stability and a broader pH stability range than viral particles, indicating the importance of host membranes for virus fitness under different external environments.
Transporters of membrane are essential for microbial survival in dynamic environments. They bridge the interior of the cell with the external environment and permit the translocation of ions, nutrients, metabolic byproducts, and toxins across the membrane barrier. The SMRs are ubiquitous in bacteria and archaea15. They are small proteins with four TM helices, initially gaining recognition for their role in antibiotic and antiseptic resistance, as well as their unique dual topology architecture. EmrE of E. coli is one of the best-studied transporters, which couples the efflux of drugs to the inward movement of protons across the cell membrane to defend against a variety of xenobiotics21. Although viral SMR-like genes have been mentioned in algae-infecting phycodnavirus and Vermamoeba-infecting Fadolivirus22,23, their diversity and function remain largely unknown. In this study, we reported three new SMRs encoded by Marseilleviridae showing high sequence conservation under purifying selection, which indicated the importance of SMR for marseillevirus infection. The tertiary structures of three SMRs were predicted to fold in a similar way of EmrE, and especially, one type of viral SMR showed conserved residues required for substrate binding and proton-dependent transport with EmrE. Tetracycline is reported to inhibit the proliferation of Acanthamoeba by mitochondrial dysfunction and inhibition of protein biosynthesis24,25. Thus, we hypothesize that viral SMRs may play a role in antibiotic transport, potentially maintaining host cellular metabolism and mitochondrial function to facilitate virus replication. As a limitation of this study, biochemical experimental validations are needed to test our hypothesis in future works. Besides, the wide use of antibiotic cocktails in the isolation experiments of giant viruses18,19,25 probably introduces a strong bias towards detecting giant viruses that harbor drug-resistance proteins. This bias may explain why marseilleviruses are readily isolated from environment samples.
Overall, we reveal the biological properties of conserved marseilleviruses isolated from different environmental samples from the Yangtze River Delta and the Pearl River Delta, China. We also identify the diverse SMRs from marseillevirus genes with function unassigned and indicate antibiotic-resistance feature of giant viruses. Our results broaden our horizon about the fitness of giant viruses against extracellular and intracellular environments.
Materials and methods
Marseillevirus isolation and purification
A. castellanii strain Neff (ATCC30010™) were cultivated in peptone-yeast extract with glucose medium (PYG) as described in the literature at 27 °C for 2 d25 and then the medium was replaced with Page’s amoeba saline (PAS) containing antibiotic cocktail26.
About 200 ml surface water sample (0.1–0.5 m in depth) was collected at each sampling site in the Yangtze River Delta and the Pearl River Delta in Shanghai and Guangdong province, China, respectively (detailed locations are listed in Table S1). The collected water sample was filtered through a 0.22-µm filter membrane (Merck Millipore, Darmstadt, Germany). The filter membrane was washed with 5 mL PAS at room temperature, and the elution was collected. The 500 µL elution of filter membrane was mixed with 500 µL amoeba (approximately 1 × 106 cells) in a 6-well plate. After 3 d, under an optical microscope, most of the vegetative form of amoeba became round and floating and cell lysis eventually, implying the cytopathic effects (CPE) and the existence of amoeba virus.
The protocols of marseillevirus isolation, cloning, and titration, as well as recipes of solutions, have been described in the literature25,26. In brief, cell lysate was filtered through a 1.2-µm filter (Merck Millipore) to remove amoeba debris, and the elution was layered onto a 24% sucrose cushion in an ultracentrifuge tube. The tube was centrifuged at 35,250 × g, for 30 min at 4 °C in an ultracentrifuge to collect the pellet containing marseillevirus. End-point dilution assays were performed to determine the viral titer of marseilleviruses, which involved serial dilutions of the viral solution and inoculation with A. castellanii previously plotted on 96-well plates. After incubation, CPE was observed under an optical microscope, and the titer (TCID50) was calculated as described by Reed and Muench (1938). Amoeba cells were seeded into a well in a 24-well culture plate with 250 µL of PYG, with an additional 250 µL of the isolated virus solution added. After 1 h inoculation, excess viruses were removed, and the cells were washed three times with 500 µL of PYG and then harvested by scraping. Serial dilutions were performed in the next 23 wells by mixing 100 µL of the previous well with 100 µL of fresh PYG. A total of 400 µL fresh amoeba cells were added to each well and were cultured for 5 d. The last well showing CPE was collected to obtain the viral clones, which were then amplified and stored for later use.
Infectivity assay
The infectivity assays were performed by inoculating amoeba cells with the marseillevirus at a multiplicity of infection (MOI) of 10. At 10 min p.i., the PYG medium was removed, and the amoeba cells were washed with PAS three times. The infected cells were collected between 20 and 1,080 min p.i. using a cell scraper into 1.5-ml sterile centrifuge tubes and then frozen and thawed to release viruses trapped in the cells. The cells were centrifuged at 356 × g for 5 min at room temperature to remove the cell debris, and then the supernatants were titrated using the end-point dilution method26. To evaluate the burst size of marseillevirus, the number of amoeba cells in each well was counted using a counting chamber (Sangon, Shanghai, China). The total viral particles were obtained from amoeba cells infected (MOI = 10, 18 h p.i.).
Purification of giant vesicles and viral particles
To release viral particles from vesicles, the protocol and recipes of solutions were adopted from the literature6. The supernatant obtained from amoeba cells infected with marseillevirus (MOI = 10, 18 h p.i.) was centrifugated at 20,000 × g for 5 min. The vesicle-containing pellet was collected, while the supernatant containing only naked particles was discarded. The pellet was resuspended in phosphate-buffered saline (PBS). To collected particles released from vesicles, the supernatant was mixed with a lysis buffer containing 1% NP-40 and 1% sodium deoxycholate (v/v%) at a ratio of 1:1 for 10 min at room temperature, and was centrifuged at 35,250 × g for 1 h to collect pellets and resuspended in PBS.
TEM
The amoeba cells infected by viruses at MOI of 10 were harvested by centrifugation at 356 × g for 5 min, and washed with phosphate-buffered saline (PBS) twice. The cells were fixed with 2.5% glutaraldehyde and stained with 2% osmium tetroxide. The stained cells were dehydrated in a series of ethanol of increasing concentrations and embedded in PON-812 (SPI supplies, PA, USA). Embedded amoeba cells were sectioned at 70 nm thickness using an ultra-microtome (Leica, Wetzlar, German), and fished out onto the 150 meshes. The meshes were stained with 2% uranium acetate avoiding light for 8 min, and 2.6% lead citrate, avoiding CO2. After the meshes were dried at room temperature for 12 h, observations were performed using a HT7700 transmission electron microscope (Hitachi, Japan).
Chloroform sensitivity
The supernatant containing viral particles or giant vesicles was treated with chloroform resulting in different final concentrations (10%, 20%, 30%, 40% and 50% (v/v%)), and rotated at 60 rpm for 1 h using rotating mixer shaker (Joan Lab, Zhejiang, China) at room temperature. The chloroform was removed by centrifugation at 500 g for 5 min at room temperature and the aqueous phase was transferred to a new 1.5-ml sterile centrifuge tube and incubated for 12 h at 4 °C to remove any remaining chloroform.
pH stability assays
The purified viral particles or giant vesicles were resuspended in PBS, which was adjusted to the desired pH (1 to 13), and rotated at 60 rpm for 24 h using rotating mixer shaker at room temperature. The supernatant was centrifuged at 35,250 × g for 1 h to collect pellets and resuspended in PBS (pH 7) and allowed to equilibrate for 12 h. As a control, viral particles were also incubated in PBS (pH 7) for 37 h at room temperature.
Thermal stability assays
The marseillevirus supernatants containing viral particles or giant vesicles were incubated in a BioRad T100 thermal cycler at 40 °C, 50 °C, 60 °C, 70 °C and 80 °C, respectively, for 1 h and allowed to equilibrate at room temperature for 1 h. As a control, viral supernatants were also incubated for 2 h at room temperature.
High salty incubation
The purified viral particles or giant vesicles were resuspended in PBS, which was adjusted to the desired concentration of NaCl (1 M, 2 M, 3 M, 4 M and 5 M), and rotated at 60 rpm for 24 h using rotating mixer shaker at room temperature. The supernatant was centrifuged at 35,250 × g for 1 h to collect pellets and resuspended in PBS (CNaCl = 137 mM) and allowed to equilibrate for 12 h. As a control, viral particles were also incubated in PBS (137 mM) for 24 h at room temperature.
DNA Preparation
Genomic DNA of the cloned strain of marseillevirus was prepared from the supernatant of infected A. castellanii using PureLinkTM Genomic DNA Mini Kit (Invitrogen, Waltham, United States) following manufacturer’s protocol. The protocol of DNA extraction from gram-negative bacterial cells was used. Viral DNA was quantified with a NanodropTM 2000/2000c spectrophotometer (Thermo Fisher Scientific, Carlsbad, CA, USA), as well as by fluorometric quantitation with a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific).
Viral genome sequencing, assembly and annotation
High-quality genomic DNA (2 µg) was used for DNA Illumina sequencing, which was carried out by the BioMarker Technologies Inc. (Beijing, China). Viral DNA was fragmented into 400–500 bp using Covaris M220 ultrasonic instrument. The DNA library was prepared using TruSeqTM DNA Sample Prep Kit (Illumina, San Diego, CA, USA), and sequenced on an Illumina HiSeq Xten with paired-end 150 bp sequencing. Illumina raw reads were processed and the filtered Illumina paired-end reads were assembled de novo using SPAdes v3.15.227 to obtain the draft assembly.
PCR verification
Amplification reactions were performed using 2× Phanta Max Master Mix (Vazyme, Nanjing, China) in a GeneAmp 2400 thermal cycler (Perkin-Elmer-Cetus, Norwalk, CT, USA) with an initial enzyme activation step (95°C, 5 min) followed by 35 cycles of denaturation (95°C, 10 s), hybridization (55°C, 5 s) and elongation (60°C, 1 min).The amplification products were gel-purified using the HiPure Gel Pure DNA Mini Kit (Magen, China) and T/A-cloned using the 5 min TA/Blunt-Zero Cloning Kit (Vazyme). The forward primer (5’-CCAAACATYTTCCTYCTTGCGTT-3’) and the reverse primer (5’-TSTTCTCGAAAAAGAGRTACTAYACT-3’) were designed for the encoding region of the DNApol gene. The forward primer (5’-CYTGCATGCAAGAAGARACCAT-3’) and the reverse primer (5’-CGNGCRGAAAACTACGATGATT-3’) were designed for the encoding region of the D5 gene. The forward primer (5’-GTAACTGCTGCTTCCGGTTT-3’) and the reverse primer (5’-CTGYGAAAGGTTGATGGTCGTC-3’) were designed for the encoding region of the MCP gene. Abbreviations of mixed bases were as follows: S = G/C, Y = C/T, R = A/G, and N = A/T/G/C. Multiple recombinants were subjected to Sanger sequencing (GeneWiz Technologies Inc., Suzhou, China), and the sequencing results were compared to identify any nucleotide variations.
Phylogenetic analysis
The nucleotide and amino acid sequences of viruses in the family Marseilleviridae including Marseillevirus massiliense strain T19 (GenBank accession number: NC_013756.1), Cajuinensisvirus strain I (OR991738.1), Cannes8virus strain Cannes8 (KF261120.1), Melbournevirus strain 1 (NC_025412.1), Tokyovirus strain A1 (NC_030230.1), Shenzhenvirus strain R2023E (OR157982.1), Futianvirus strain R2023F (OR343188.1), Kurlavirus strain BKC-1 (KY073338.1), Losannavirus lausannense strain 7715 (NC_015326.1), Noumeavirus strain NMV1 (NC_033775.1), Port-miou virus strain 1 (KT428292.1), Xiwanvirus strain R2023XWM (PQ010612.1), Xiwanvirus strain R2024YHH (PQ561596.1), Insectomimevirus strain V478 (KF527888.1), Tunisfontainevirus strain U484 (KF483846.1), Brazilianmarseillevirus strain BH2014 (NC_029692.1), Goldenmarseillevirus strain Golden (NC_031465.1) were retrieved from GenBank. Megavirus baoshanense strain SH (MH046811.2) was used as the outgroup. The italic virus names designated by the binomial nomenclature system can be searched on the International Committee on Taxonomy of Viruses (ICTV) taxonomy browser (2023 Release).
Five NCLDV core genes, including MCP, DNApol, D5, virus late transcription factor 3, and A32-like packaging ATPase were chosen for building of the phylogenic trees12. The amino acid sequences of EmrE were extracted from the literature15 and the UniProt database.
Orthologue sequences were aligned using MAFFT v3.8.155128. In IQ-TREE multicore v2.1.229, the best evolutionary models were calculated for each core gene with MFP program to be LG models with GAMMA distributed rates of 5 in all cases. Phylogenetic trees were generated using maximum-likelihood method with LG + G model and 1,000 bootstrap replicates. Phylogenetic trees were visualized using Chiplot v2.630.
Comparative genomics analysis
The average nucleotide identity (ANI) was calculated by JSpeciesWS v3.8.531 based on BLAST. The rate of nonsynonymous (dN) and synonymous (dS) substitutions and their ratio (dN/dS) were computed by PAML_X v1.3.1 using YN00 program with default options32.
Prediction of protein structures
The alignments of primary and secondary structures of SMR were deciphered and visualized using ESPript v3.033. The secondary structures of transmembrane were predicted and visualized using Protter v1.034. The amino acid sequences of viral proteins were submitted to HHpred webservers for functional prediction using PDB_mmCIF70_8_Mar35. The tertiary structures of proteins and their complexes were predicted using Alphafold2 and Alphafold3 webserver36, and the results were visualized in PyMol v2.5.5 (http://www.pymol.org/).
Data availability
The annotated genomes of Shanghaivirus strain J2016JW, Quyangvirus strain J2018QY, Dashavirus strain R2023E, Dashavirus strain R2023DC, Dashavirus strain R2023PK, and Dashavirus strain R2023JZZ were deposited in the GenBank database under accession numbers MG827395.1, MT663336.1, OR343189.1, PQ220252.1, PQ220253.1, PQ010614.1, respectively. The raw sequencing data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA)37 of China National GeneBank DataBase (CNGBdb) with accession number CNS1224376, CNS1224378, CNS1219734, CNS1219735, CNS1219736 and CNS1219739, respectively.
References
Boyer, M. et al. Giant marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U S A. 106, 21848–21853 (2009).
Walker, P. J. et al. Changes to virus taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2019). Arch. Virol. 164, 2417–2429 (2019).
Sahmi-Bounsiar, D. et al. Marseilleviruses: an update in 2021. Front. Microbiol. 12, 648731 (2021).
Thomas, V. et al. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 13, 1454–1466 (2011).
de Freitas Almeida, G. M. et al. Genomic and Structural Insights into Jyvaskylavirus, the First Giant Virus Isolated from Finland (eLife Sciences Publications, Ltd, 2025).
Arantes, T. S. et al. The large marseillevirus explores different entry pathways by forming giant infectious vesicles. J. Virol. 90, 5246–5255 (2016).
Moliner, C., Fournier, P. E. & Raoult, D. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34, 281–294 (2010).
Yi, X. et al. Giant viruses as reservoirs of antibiotic resistance genes. Nat. Commun. 15, 7536 (2024).
Mueller, L., Hauser, P. M., Gauye, F. & Greub, G. Lausannevirus encodes a functional dihydrofolate reductase susceptible to proguanil. Antimicrob. Agents Chemother. 61 (2017).
Doutre, G., Philippe, N., Abergel, C. & Claverie, J. M. Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to virus fitness. J. Virol. 88, 14340–14349 (2014).
Fabre, E. et al. Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 8, 15087 (2017).
Aylward, F. O., Moniruzzaman, M., Ha, A. D. & Koonin, E. V. A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol. 19, e3001430 (2021).
Chen, Y. J. et al. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. U. S. A. 104, 18999–19004 (2007).
Lucero, R. M., Demirer, K., Yeh, T. J. & Stockbridge, R. B. Transport of Metformin metabolites by guanidinium exporters of the small multidrug resistance family. J. Gen. Physiol. 156 (2024).
Bay, D. C. & Turner, R. J. Diversity and evolution of the small multidrug resistance protein family. BMC Evol. Biol. 9, 140 (2009).
Yerushalmi, H. & Schuldiner, S. An essential Glutamyl residue in EmrE, a multidrug antiporter from Escherichia coli. J. Biol. Chem. 275, 5264–5269 (2000).
La Scola, B. et al. A giant virus in amoebae. Science 299, 2033 (2003).
Morimoto, D., Tateishi, N., Takahashi, M. & Nagasaki, K. Isolation of viruses, including mollivirus, with the potential to infect Acanthamoeba from a Japanese warm temperate zone. PLoS One. 19, e0301185 (2024).
Machado, T. B. et al. A long-term prospecting study on giant viruses in terrestrial and marine Brazilian biomes. Virol. J. 21, 135 (2024).
Schrad, J. R., Abrahao, J. S., Cortines, J. R. & Parent, K. N. Structural and proteomic characterization of the initiation of giant virus infection. Cell 181, 1046–1061 (2020). e1046.
Chung, Y. J. & Saier, M. H. Jr. SMR-type multidrug resistance pumps. Curr. Opin. Drug Discov Devel. 4, 237–245 (2001).
Andreani, J. et al. Morphological and genomic features of the new Klosneuvirinae isolate fadolivirus IHUMI-VV54. Front. Microbiol. 12, 719703 (2021).
Zhang, W. et al. Four novel algal virus genomes discovered from Yellowstone lake metagenomes. Sci. Rep. 5, 15131 (2015).
Jha, B. K. et al. Tigecycline inhibits proliferation of Acanthamoeba castellanii. Parasitol. Res. 114, 1189–1195 (2015).
Pagnier, I. et al. A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba. Intervirology 56, 354–363 (2013).
Xia, Y. et al. Role of an FNIP repeat domain-containing protein encoded by megavirus Baoshan during viral infection. J. Virol. 96, e0081322 (2022).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Xie, J. et al. Tree visualization by one table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–w592 (2023).
Richter, M., Rossello-Mora, R., Glockner, O., Peplies, J. & F. & JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931 (2016).
Xu, B. & Yang, Z. PAMLX: a graphical user interface for PAML. Mol. Biol. Evol. 30, 2723–2724 (2013).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new endscript server. Nucleic Acids Res. 42, W320–324 (2014).
Omasits, U., Ahrens, C. H., Müller, S. & Wollscheid, B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886 (2014).
Gabler, F. et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinf. 72, e108 (2020).
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596, 583–589 (2021).
Guo, X. et al. CNSA: a data repository for archiving omics data. Database. (2020). (2020).
Acknowledgements
This research is supported by National Natural Science Foundation of China (No. 42406092), Guangdong Basic and Applied Basic Research Foundation (No. 2025A1515010802), Guangdong Major Project of Basic and Applied Basic Research Foundation (No. 2023B0303000017), Innovation Team Project of Universities in Guangdong Province (No. 2023KCXTD028), and Scientific Instrument Developing Project of Shenzhen University (Grant No. 2024YQ004). We thank Prof. Jiang Zhong and Prof. Zhexue Quan of Fudan University for kindly providing environmental samples from the Yangtze River Delta. We thank Dr. Bu Xu and Dr. Le Xie for collecting environmental samples from the Pearl River Delta. We are also grateful to Prof. Jintian Li of South China Normal University for his useful discussions.
Author information
Authors and Affiliations
Contributions
Y.X. and R.Z. designed research; Y.X., B.S., H.R. and F.L. performed research; Y.X., and B.S. analyzed data; Y.X., X.W., Y.W. and R.Z. wrote the paper. All authors contributed to manuscript revision, read, and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xia, Y., Su, B., Ren, H. et al. Conserved marseilleviruses harboring diverse antibiotic resistance genes isolated from the Yangtze river Delta and the Pearl river delta, China. Sci Rep 15, 10663 (2025). https://doi.org/10.1038/s41598-025-94967-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-94967-2