Abstract
Since the detection of the Asian mosquito Anopheles stephensi in Dijbouti in 2012, it has spread throughout the Horn of Africa. This invasive vector continues to expand across the continent and is a significant threat to malaria control programs. Vector control methods, including insecticide-treated nets and indoor residual spraying, have substantially reduced the malaria burden. However, the increasing prevalence of mosquitoes resistant to insecticides, including An. stephensi populations, undermines ongoing malaria elimination efforts. Understanding population structure, gene flow between populations, and the distribution of insecticide resistance mutations is essential for guiding effective malaria control strategies. Here, we generated whole genome sequencing data for An. stephensi sourced from Awash Sebat Kilo, Ethiopia (n = 27) and compared with South Asian populations (n = 45; India and Pakistan) to assess genomic diversity, population structure, and uncovering insecticide resistance mutations. Population structure analysis using genome-wide single nucleotide polymorphisms (n = 15,533,476) revealed Ethiopian isolates clustering as a distinct ancestral group, separate from South Asian isolates. Three insecticide resistance-associated SNPs (gaba gene: A296S and V327I; vgsc L1014F) were detected. Evidence of ongoing selection was found in several loci, including genes previously associated with neonicotinoids, ivermectin, DDT, and pyrethroid resistance. This study represents the first whole genome population genetics study of invasive An. stephensi, revealing genomic differences from South Asian populations, which can be used for future assessments of vector population dispersal and detection of insecticide resistance mechanisms.
Similar content being viewed by others
Introduction
The Asian malaria vector, Anopheles (An.) stephensi, was first detected in Africa in Djibouti in 20121. Since then, this mosquito species has spread throughout the Horn of Africa (HOA), including Somalia, Ethiopia, Eritrea, South Sudan, Kenya, and with recent reports in Nigeria and Ghana2,3,4,5,6,7. Historically, An. stephensi was distributed across South Asia, the Persian Gulf, and the Arabian Peninsula. It is a primary vector of human malaria in India and Pakistan, proficiently transmitting both Plasmodium falciparum and P. vivax8,9. Unlike many traditional malaria vectors, An. stephensi is well-adapted to urban environments. Its spread into the HOA has coincided with a surge in malaria cases and urban outbreaks. In Djibouti, An. stephensi’s emergence has been epidemiologically linked to a dramatic resurgence in malaria cases, which increased 35-fold between the years 2013 and 202110,11. In Ethiopia, the role of An. stephensi in regional urban malaria transmission was confirmed in a recent outbreak of P. falciparum in Dire Diwa12.
This species is filling a currently empty ecological niche in urban areas in Africa, breeding in both small artificial water sources and natural aquatic habitats near human dwellings13,14,15. This adaptability allows An. stephensi to thrive in both rural and urban environments, unlike the primary malaria vectors in the HOA - An. arabiensis, An. gambiae s.s, and An. funestus s.s,- which typically occupy rural areas or regions of high agricultural activity. The ongoing urbanization across Africa and the expanding geographical range of An. stephensi is estimated to put a further 120 million Africans at risk from malaria6,14,16. With the World Health Organization’s (WHO’s) target to reduce the global malaria burden by 90% by 2030, the need to control this vector has never been more crucial17.
Enhancing our understanding of how An. stephensi became so well-established in the HOA is essential for predicting its potential spread to new regions. Population genetics has been widely applied to vector species to gain insights into population structure, ongoing selection, and gene flow18,19. For An. stephensi, studying population structure and genomic architecture can help elucidate its invasion routes and any adaptive evolution that may have occurred since its introduction. Alongside gene flow, selection is of particular interest, as it can reveal alleles associated with insecticide resistance and illustrate how these alleles are spreading through populations20.
In Africa, malaria vector control relies on long-lasting insecticide-treated nets (LLINs) and indoor residual spraying (IRS). There are five main classes of insecticides in use: pyrethroids, organochlorides, organophosphates, carbamates, and pyrroles, each with varying modes of action. However, resistance to all major adulticide classes has been reported across many Anopheles species, including An. stephensi across the HOA, India, Pakistan, Sri Lanka, and the WHO Eastern Mediterranean region3,21,22,23. Resistance to insecticides can arise through multiple mechanisms: target site resistance, overexpression of metabolic enzymes, behavioural changes, microbiome alterations, and thickening of the insect cuticle24,25,26. Predominantly, resistance results from metabolic or target site alterations. Target-site resistance arises from single nucleotide polymorphisms (SNPs) that alter the target protein’s amino acid sequence and result in conformational changes that prevent the insecticide from binding properly25. The target site mutation A296S in the gaba receptor has been reported in An. stephensi from Ethiopia, and is associated with resistance to dieldrin (rdl), an organochlorine banned in the 1990s due to concerns about its environmental persistence and potential impact on human health27,28,29. The L1014 knockdown resistance (kdr) mutation in the voltage-gated sodium channel (vgsc) is linked to cross-resistance to Dichlorodiphenyltrichloroethane (DDT) and pyrethroids and has been reported in An. stephensi from Ethiopia, Afghanistan, and India28,30,31,32,33. However, there have been examples of pyrethroid resistance in An. stephensi in the absence of this mutation, implying other mechanisms may contribute to the development of resistance23,34. These alternative mechanisms could include metabolically mediated resistance, characterized by increased detoxification of insecticides through the overexpression of glutathione s-transferases or cytochrome P450s, thereby reducing insecticide efficacy24,25. The genomic changes leading to metabolic resistance are generally more complex than those associated with target-site resistance. In some cases, missense SNPs increase insecticide metabolism, such as the L119F mutation in the GSTe2 gene, which causes cross-resistance to pyrethroids and DDT in An. funestus35. Other, more complex mechanisms include structural variants or copy number variations, like the 6.5 kb insertion that mediates pyrethroid resistance, also observed in An. funestus6. These examples underscore the importance of looking beyond simple SNPs and small insertions or deletions (INDELs) to fully understand the molecular mechanisms underlying insecticide resistance.
To date, the detection of insecticide resistance markers and population genetics of the invasive An. stephensi species has been limited to the examination of a few candidate genes. This species emergence in Africa is not fully understood, but one hypothesis is that An. stephensi was transported through either human mediated travel or cattle transportation18,36,37. Other theories include long-distance wind-borne migration38. The Ethiopian isolates used in this study were collected in Awash Sebat Kilo, a town 200 km east of Addis Ababa and a main transportation corridor from Addis Ababa to Djibouti. Awash Sebat Kilo is an area of high malaria transmission and with large populations of An. stephensi with reports of insecticide resistance, making it an important site for investigating the population genetics of An. stephensi8,18,21,40.
The application of whole genome sequencing (WGS) provides a holistic view of the genomic landscape, allowing for insights into mosquito ancestry, ongoing genetic selection, and the identification of insecticide resistance markers. The availability of increasingly affordable and high throughput platforms for WGS is resulting in a growing number of Anopheles species with available genomic data19,40,41. Previous population genetics studies of An. stephensi from Ethiopia have either investigated limited gene numbers or used double digest restriction-site associated DNA sequencing (ddRADseq). While these approaches yielded valuable insights into insecticide resistance and population structure, they provided only a partial view of the genome18,21,40. Here, we generate the first whole genome sequence data from An. stephensi collected in Africa, specifically from Awash Sebat Kilo, Ethiopia. We conduct a population genetic analysis of these Ethiopian An. stephensi WGS data (n = 27), alongside publicly available data sourced from South Asia (India and Pakistan, n = 45) to gain a deeper understanding of the genomic architecture of this vector, to gain insight into its ancestry, phylogenetics, and uncover loci under selective pressure.
Results
Whole genome sequence data and nucleotide diversity
Whole genome data was generated from 27 mosquitoes sourced from Awash Sebat Kilo, Ethiopia, in 2019. The number of raw sequenced paired reads across the 27 samples ranged between 26,841,458 and 85,763,451. After mapping to An. stephensi’s three chromosomes, this resulted in an average coverage of 33.7-fold (standard deviation 7.5-fold). A further 45 isolates with publicly available WGS data from India (Bangalore and Mangalore, N = 21; insectary colonies, N = 16) and Pakistan (insectary colony, N = 8) were also mapped, and across the combined dataset (N = 72), 15,533,476 SNPs passed filtering criteria (details in Methods). Sliding window analysis (size 100kbp) revealed low nucleotide diversity with π averaging < 0.03 across An. stephensi’s three chromosomes (Supplementary Fig. S1 and Supplementary Table S1).
Population differentiation and ancestral analysis reveals distinct geographic groups
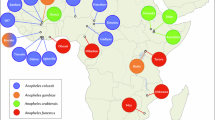
Using the 15,533,476 SNPs, both principal component and phylogenetic analysis revealed that the Ethiopian, Pakistan colony, and Indian field An. stephensi isolates formed distinct genetic clusters (Fig. 1). The Indian field isolates also clustered with a subset of Indian colony mosquitoes, whilst the remaining (n = 5) Indian colony isolates appear genetically closer to the Pakistani SDA500 colony isolates. This separation is based on their origin, the five clustering with Pakistani colony samples are the Walter Reed colony strain, and those appearing with the Indian field isolates are a different laboratory-reared colony strain. Application of the pairwise FST population differentiation metric, which quantifies differences in allele frequencies, confirmed that the greatest genetic distinctness was between Ethiopian field and Pakistan insectary colony samples (70,898 SNPs with FST > 0.8; 3,472 SNPs with FST = 1, average FST = 0.133) (Table 1). Indian field and colony samples were the most genetically similar (2,411 SNPs with FST > 0.8, 354 SNPs with FST =1, average FST = 0.073) (Supplementary Table S2).
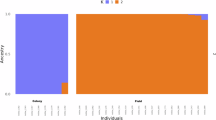
Admixture analysis was performed to identify any potential ancestral relationships between the isolates and revealed five ancestral populations (labelled, k1-k5): Ethiopian, Indian field, Pakistani colony, and the separation of Indian colony isolates, as observed in the phylogenetic tree. The Ethiopian isolates consisted of one sub-population and the same ancestral population was also observed in a sample from Pakistan colony and Indian field isolates (Admixture plots K = 2, K = 3, K = 4 in Supplementary Fig. S2, and K = 5 in Fig. 2). In the Indian field isolates, the k4 ancestral sub-population was dominant, and no clear distinction could be seen between Bangalore and Mangalore isolates (Fig. 2). The k3 ancestral sub-population appeared as the only ancestry present in a subset of Indian colony samples. In the remaining Indian colony samples, k5 was dominant, although two isolates shared the k2 ancestral sub-population that was predominant in colony samples from Pakistan. The single Pakistani colony sample sharing ancestry with Ethiopia also had k3 and k5 ancestral types seen in Indian field and colony samples(Fig. 2).
Genome wide admixture analysis of An. stephensi isolates. Each isolate is represented by a column. Five ancestral populations were identified using ADMIXTURE (K = 5 ancestral populations, labelled k1-k5), across the four isolate groups (Ethiopia (n = 27), Indian colony (n = 21), Indian field (n = 16), and Pakistan colony (n = 8)) analyses.
Selection analysis reveals genes involved with insecticide resistance
Genome-wide selection scans were performed to identify signals of recent positive selection using the Cross-Population Extended Haplotype Homozygosity (XP-EHH) method between populations and integrated Haplotype Score (iHS) within each population. Using XP-EHH, 275 SNPs were identified as having ongoing directional selection (XP-EHH > 4.0), with 148 SNPs within coding sequences (CDS) of 81 genes, and 127 SNPs within introns (Supplementary Table S3). One gene was identified as a candidate potentially involved in insecticide resistance (acetylcholine receptor (nAChR) subunit beta-like 2) (two SNPs, XP-EHH values > 5.3) in a comparison of Indian and Ethiopian field samples (Table 2). In single population (iHS) analysis, a total of 997 loci were identified as having significant iHS scores (iHS > 4.0), 439 were within non-coding regions, and the remaining 559 occurred within CDS across 325 different genes (Supplementary Table S4). Of these, five were candidate genes that had loci with iHS scores indicative of selection (iHS > 4.0), including the nAChR subunit beta-like 2 in the Indian field isolates. The remaining four genes were nAChR alpha-like (Ethiopia and Indian field samples), glutamate-gated chloride channel (Pakistani Colony), cytochrome P450 307a1-like (CYP307a1) (Indian field isolates), and gaba subunit beta (India field samples and Pakistan colony).
Identification of three insecticide resistance associated SNPs
A total of 25 missense SNPs were identified across four genes previously associated with insecticide resistance in Anopheles mosquitoes (ace-1, gaba (rdl), GSTe2, and vgsc) (Table 3). Three of these are SNPs known to be associated with target-site mediated resistance (vgsc L1014F; gaba A296S and V327I). The vgsc L1014F SNP mediates resistance to pyrethroids and was identified exclusively in the Ethiopian population. It was only identified as a heterozygous genotype in 2 samples (allelic frequency of 7.4% (2/27 samples)). The gaba A296S mutation was found in all four populations (allelic frequency of 18.8%; 27/72 samples), with all samples being heterozygous at this position, no homozygotes were found for this mutation. The V327I mutation was only present in the Indian field and colony populations (5 samples). This SNP was also present as the heterozygous genotype and only in samples that also had the gaba A296S mutation. A further 22 missense mutations were identified across the four key insecticide resistance-associated genes (ace-1 (n = 5), gaba (n = 9), GSTe2 (n = 9), vgsc (n = 2)), with allelic frequencies ranging from 1 to 83% (Table 3). No nonsense or nonstop mutations were detected in these genes.
Detection of structural variants: copy number variants and indels
For coverage-based analysis, only one significant deviation from the median genome coverage for the Ethiopian population was observed (Supplementary Figure S3). A possible copy number variant (CNV) was detected on chromosome two in a region containing a cluster of cytochrome P450 genes (CYP). The CYP6a cluster on chromosome two (NC_050202.1) was observed to have elevated coverage in comparison to both the median coverage and other populations.
From analysis with Delly, a structural variant prediction tool, 57,815 deletions were identified after quality control filtering (specified in the method section). Of these, 36 were identified across 18 genes previously identified as involved in insecticide resistance or as belonging to a gene family possibly involved in insecticide metabolism41. These included two deletions in cytochrome P450 genes: a 494 bp intronic deletion in CYP307a1 was detected in an Ethiopian sample and a frameshift-causing deletion in CYP6a1, detected in a single Indian sample (SRR1529388). This deletion was one of two detected in the CYP6a1 gene, where the other was a 3’ UTR variant found in both Indian colony and field samples. There were 2901 duplications originally found across 44 genes and 25 intergenic regions. Of these, 351 (12.1%) resulted in a frameshift variant. An 181 bp duplication event was identified in the CYP9f2 candidate gene in one Indian colony sample (SRR1529388, see above) but was annotated as a non-coding transcript. Delly did not detect any Copy Number Variants (CNVs) in the chromosome 2 CYP6a cluster with elevated coverage identified in Supplementary Figure S3.
Discussion
The invasion of An. stephensi into the HOA is a significant threat to malaria control and elimination efforts in sub-Saharan Africa. Understanding the origins of this invasive species and ongoing gene flow can provide greater insight into its emergence in this region and improve predictions of future spread. Here we explore whole genome sequence data of An. stephensi and demonstrate, using population structure analysis, that our Ethiopian field isolates from Awash Sebat Kilo, are genetically distinct from Indian and Pakistani insectary colony and field populations. Ancestry analysis indicated that each field population constituted a distinct ancestral population. The Indian field isolates, despite the large geographical distance between the regions (Bengaluru and Mangaluru, around 350 km), shared an ancestral population (K4) in the presence of several minor ones. The Indian colony samples were divided into two groups based on their origin (Walter Reid or laboratory-reared strains). All samples from Ethiopia inherited the k1 ancestry, which was present at a minor proportion in some field Indian samples and a single Pakistan SDA500 colony sample (intermediate form). This observation suggests a possible South Asian origin of the Ethiopian samples, as previously reported, however further comparison with genomic data from other geographical regions is necessary to enhance the accuracy of origin inference2. Previous studies using candidate gene analysis have identified high levels of genetic diversity across Ethiopian populations, implying either one large invasion incident or multiple smaller colonisation events18,37. The samples analysed here were collected in the same market town, which could have resulted from a population expansion in this area, resulting in the homogenous ancestry observed. The ancestry observed in Ethiopia might have been dominant in other areas in South Asia at the time of the introduction or introductions events. A similar relationship is observed between the Indian insectary-reared and field isolates, where the former was collected in 2016 in Chennai, India and has only k5 ancestral subpopulation. This is shared with 14 Indian field samples with the k1 Ethiopian ancestral subpopulation, suggestive of shared ancestry between the Indian populations, despite the large geographical distance between Chennai, Bengaluru, and Mangaluru. Other studies have indicated that Anopheles spp. population structure remains stable over time, and physical distance is a larger driver of genetic variation42,43. However, there are also cases where species dynamics change relatively quickly, leading to genomic changes, such as in response to increased insecticide exposure, as well as examples where physical distance and genetic distance deviate, indicative of processes like consecutive founder events44,45,46,47. It is clear that to better estimate the origins and dispersion of An. stephensi, a more comprehensive WGS dataset is needed from these and other native and recently invaded geographical regions, including India, Pakistan, Afghanistan, Saudi Arabia, Iran, as well as elsewhere in the Arabian Peninsula and HOA. One of this study’s key limitations is the abundance of colony samples analysed, which may introduce bias into the results of selection and genetic diversity tests. Colonies such as the Pakistani SDA500 strains have been laboratory reared for decades, and in these conditions, limited gene flow results in reduced genetic diversity, inbreeding, and increased linkage disequilibrium. The local field populations from the regions where the colony samples originated would likely exhibit far greater genetic diversity, which is not represented in the samples used here.
The kdr L1014F insecticide resistance mutation was detected at a low frequency in this Ethiopian population and was absent in the Indian field samples. This kdr mutation confers resistance to pyrethroids and DDT which has been found previously in Indian populations of An. stephensi collected in 2016, and in Afghan populations, collected in 201831,32. The kdr L1014F mutation is absent in the Mangalore and Bangalore populations analysed here, despite evidence of extensive pyrethroid resistance in both these cities3,48. The low proportion of Ethiopian isolates with this SNP, along with its presence as a heterozygous genotype, implies it has recently arisen in this population. This is supported by a previous work in Awash, where no evidence of the kdr mutation was found in samples collected a year prior to those examined here18. We have previously identified this kdr L1014F mutation at low frequency in a set of samples from the same regions/year when using an amplicon assay28. The low allelic frequency of the resistance marker, in the context of phenotypic resistance in the sampling location, indicates mechanisms other than target site modifications are responsible for this observed phenotype.
The other known target site mutation identified in the Ethiopian isolates was gaba A296S, which confers resistance to dieldrin27,29. This substitution was also identified in the Indian field populations, highlighting its prevalence, despite the insecticide being banned since the 1990s. The presence of this mutation corroborates the historical shared ancestry of the isolates analysed here. The gaba V327I mutation was also identified, and has a strong association with the A296S alteration, with five Indian field and colony samples carrying both SNPs49,50. A further 22 putatively novel SNPs were identified in genes associated with insecticide resistance. Six of these SNPs appeared in two Pakistani colony samples (Ste32 and Ste58) known to be insecticide susceptible. Two of these SNPs occurred in the ace-1 gene (N177D and V59A), and four in GSTe2 (F196Y, C146S, H97A, and G66A). The remaining 16 missense SNPs occur in populations where the resistance status is unknown, so it is not possible to infer any impact on insecticide susceptibility. None of these missense SNPs, including those previously associated with insecticide resistance, had any evidence of ongoing selection using either the iHS or XP-EHH statistics.
Of the genes identified as having ongoing directional selection, five could be potentially involved in insecticide resistance. The nAChR receptor subunit beta was found under selection by both within (iHS) and between (XP-EHH) population analysis. The nAChR receptor subunit alpha was also identified using the iHS metric as being under ongoing selection in Indian and Ethiopian populations. These two subunits of the receptor are the result of splicing of the nAChR gene; the presence (alpha) or absence (beta) of two cysteines determines their type51. Mutations within nAChR have been reported to result in resistance to neonicotinoids, which are pesticides that mediate synaptic transmission via nAChR, resulting in insect mortality52,53. This type of pesticide usage has been reported to result in neonicotinoid resistance in other Anopheles species54. Worryingly, neonicotinoids are considered an alternative to pyrethroids, for vectors with high levels of resistance to pyrethroids3,55,56,57. In Ethiopia, the President’s Malaria Initiative have been using SumiShield, which contains a neonicotinoid, since 2021. However, this is after the collection of these samples, so it is unknown whether they had been exposed to the chemical58. In addition, significant directional selection was detected in glutamate-gated channel genes in the Pakistan colony isolates. Mutations in these loci have been associated with ivermectin resistance in Drosophila59,60,61. Ivermectin is an anti-parasitic/endectocide, often used in mass drug administrations, which kills both the Plasmodium parasite, and Anopheles spp. mosquito when they ingest the blood of a treated host62,63,64. Ivermectin has been trialled as a vector control method using mass drug administration to help reduce malaria cases65.
Distinct signals of selection in a gaba gene were identified in India field and Pakistan colony samples. Fipronil, an insecticide that targets gaba receptors, induces neurotoxicity, which may be linked to these selection patterns66. This phenylpyrazole has also been proposed as part of a One Health approach to vector control67. Similarly to ivermectin, fipronil can be used in mass drug administration, particularly in livestock to target zoophilic vectors like An. stephensi. This approach has been successfully trialled in several studies68,69,70. Resistance to fipronil has been reported in Iranian An. stephensi isolates, with both kdr and rdl mutations hypothesised to be involved71. Mutations within gaba receptors have been associated with reduced insecticide efficacy of fipronil, although this has not been observed in Anopheles spp72,73,74. Further surveillance of this gene could provide valuable insights into its potential involvement in fipronil resistance.
Another notable gene exhibiting strong directional selection by iHS was CYP307a1, identified in the Indian field populations. A 494 bp deletion was detected in the intronic region of this locus in one Ethiopian isolate, and may impact gene expression or result in alternative splicing75. CYP307a1 is a member of the cytochrome P450 gene family and has previously been linked to resistance to both DDT and pyrethroids in An. funestus76,77. Similarly, in other insect species (Cydia pomonella), the upregulation of CYP307a1 has been associated with deltamethrin (pyrethroid) resistance78. Typically, CYP307a1 is involved in ecdysteroid hormone biosynthesis, these hormones control mosquito behaviour, nervous system development, and reproduction79. This gene has not been confirmed to directly result in insecticide resistance but warrants further investigation.
Other structural variants were detected that resulted in amino acid alterations, including a 67 bp deletion in the CYP6a1 gene, found in a single Indian colony sample. Additionally, a 3’ UTR variant in the same gene was detected in both Indian colony samples and field samples. Altered expression levels of CYP6a1 have previously been associated with deltamethrin resistance in D. melanogaster and C. pipiens80,81. With the absence of kdr mutations in these Indian field samples, but phenotypic pyrethroid resistance reported near the collection sites, it is likely that other mechanisms contribute to resistance; such as metabolic resistance3,23,34,82. This CYP6a1 gene identified here is not in the CYP6a cluster that was observed to have elevated coverage proportional to the genome median in Ethiopian isolates. To further investigate this gene cluster, read orientations and breakpoints would need to be identified to confirm whether this increased coverage was due to a copy number variation in the population.
A limitation of this study was the lack of phenotype data on insecticide response. Further investigations, using WGS or targeted amplicon sequencing, in tandem with susceptibility bioassays, are needed to investigate the impact of these mutations on insecticide response. The novel missense SNPs potentially linked with resistance should be used as targets in high-throughput molecular assays, to support surveillance and assist functional work to understand and validate underlying mechanisms associated with resistant phenotypes.
In conclusion, this study gives greater insight into the population genetics of cross-continental An. stephensi. Applications of WGS analysis to larger An. stephensi sample cohorts, across different geographical regions, will be key to understanding gene flow and identifying insecticide resistance markers. Such insights will enable public health authorities to make informed choices about vector surveillance and insecticide usage.
Methods
Mosquito collection and identification
An. stephensi mosquitoes were sourced from an LSHTM colony (Sind Kasur strain, SDA500, originally from Pakistan in 1982). Field samples were collected in Awash Sebat Kilo, Ethiopia (GPS coordinates 9.003009, 40.167630), between April and September 2019. The study protocol was conducted in accordance with relevant ethical guidelines and regulations, as described in the “Ethical Approval and Consent” section. The samples were collected in one of three ways: CDC mini light traps, aspiration from cattle shelters, and human landing collection. All mosquitoes were identified morphologically as An. stephensi before a multiplex qPCR with ITS2 and cox-1 genes was used for molecular confirmation8,83,84.
DNA extraction
Mosquitoes were individually suspended in 1X phosphate buffered saline (PBS), before being mechanically lysed with a Tissue Ruptor II (Qiagen, Hilden, Germany) for 30 s, or until all body parts were no longer visible. DNA was extracted using the Qiagen Blood and Tissue kits, according to manufacturer’s instructions. The study protocol was conducted in accordance with relevant ethical guidelines and regulations, more information on this can be found in the “Ethical Approval and Consent” section. DNA concentrations for each sample were quantified using the Qubit 2.0 fluorimeter HS DNA kit (ThermoFisher). DNA was then stored at − 20 °C.
Whole genome sequencing and bioinformatic analysis
The DNA of 27 An. stephensi isolates were sequenced on the Illumina MiSeq using 2 × 250 bp paired end configuration. Twenty-five of these isolates were collected in Awash Sebat Kilo, Ethiopia, and the remaining two were colony mosquitoes. Publicly available An. stephensi WGS data was included in the population analyses, this includes 21 field Indian samples (11 from Bangalore, and 10 from Mangalore)85. A total of 24 publicly colony samples were included in the analysis, 16 of which were Indian colony samples, and eight Pakistan colony samples. The raw WGS sequence data was first trimmed using trimmomatic software (version 0.39), before being aligned to the UCI_ANSTEP_V1.0 (An. stephensi) reference genome, using bwa-mem (version 0.7.17) software (with default parameters)86,87,88. Coverage statistics from the resulting bam files were calculated using samtools (version 1.9)89. Variants (SNPs and INDELs) were called and validated using GATK software (version 3.8) with the HaplotypeCaller90. Once individual VCF files had been generated, a multi-sample VCF was created using GATK’s GenomicsDBImport and GenotypeGVCFs function. The multi-sample VCF was then filtered to contain only chromosomal variants using bcftools (version 1.9), this package was also used to sort and normalise multi-allelic variant sites. Further filtering was conducted using vcftools (version 0.1.16), removing variants with a depth < 5 and more than 50% missingness91. Missingness refers to the percentage of missing data for each SNP site; each SNP must have at least 50% of samples with sufficient depth for a reliable SNP call. A total of 16,580,599 SNPs were initially identified and reduced to 15,533,476 after filtering.
Identification of insecticide resistance associated SNPs
A bed file (Supplementary Table S6) containing genes associated with insecticide resistance was created based on a literature search. This search included the para, gaba, and ace-1 genes associated with target site resistance, along with cytochrome P450’s, and esterase’s linked to metabolic-based resistance. The bed file included the 500 bp before the gene start codon, and 500 bp after the stop codon to identify any variants within the promoter or terminator regions. The bed file was then applied to the filtered multi-sample VCF using bcftools. The package snpEff was then used to annotate these variants, using a custom-built database from the UCI_ANSTEP_V1.0 gene feature file in gff format gff86,92.
Population genetic analysis
A pairwise-genetic distance matrix was generated from the filtered multi-sample VCF file, using an in-house script93. This distance matrix was used as the basis for the generation of a neighbourhood joining tree, and principal component analyses generated in R using ape and qqman packages94,95. The resulting neighbourhood joining (NJ) tree was visualised and annotated using iTOL96. ADMIXTURE software (v1.3) was used to conduct admixture analysis97. PLINK package (version 1.90b6.21) was first used to convert the VCF file to a bed file for these analyses98. The optimum K value (estimated number of ancestral populations) was calculated by cross-validation of 1–10 dimensions of eigenvalue decay (k = 5). This value along with the bed file was used by ADMIXTURE software (version 1.3.0) to identify shared ancestral populations. The output was then visualised in R. To investigate genetic differentiation in An. stephensi, Fst was calculated using the Weir and Cockerham estimator between Ethiopian and Indian field populations, using vcftools and visualised in the R statistical tool. Nucleotide diversity was also calculated across the genome, using 100 kb windows. Genomic regions under directional selection, were detected with the R-based package rehh99. The Integrated haplotype statistic (iHS) was used to find selection within populations, a positive iHS value indicates selection for the ancestral (reference) allele, whilst a negative iHS score suggests selection for the derived (alternate) allele. Extended haplotype homozygosity (XP-EHH) was used to identify selection ongoing between populations, with a positive score suggesting selection in population A and a negative score indicating population B is under selection. In this instance, a score > 4.0 or <-4.0 is considered significant.
Identification of structural variants
Two methods were utilised to detect copy number variants for this data set. First, a coverage-based method was used focussing on clusters of CYP genes identified in the genome (Supplementary Table S5). Sample coverage was averaged by collection location resulting in four populations: Ethiopia, Indian field, Indian colony, and Pakistani colony. Coverage for each population was normalised using Kernel-smoothing, and then plotted against the median genome coverage depth for that population.
Secondly, Delly software (version 0.7.6) was used to identify structural variants (SVs) > 60bp100. Delly uses multiple methods to identify these variants: read-pair analysis, split-read analysis, and de novo assembly, integrating the information from these three methodologies to validate variants and reduce false positives. For individual samples that had insertions and deletions, sample BCF format files were merged and filtered based on sample missingness (< 50%), followed by the removal of heterozygous calls. As described above, the population differentiation statistic FST was calculated using vcftools, to identify SVs unique to populations. To confirm any SVs occurring in genes associated with insecticide resistance, the bcftools software was used for visual inspection.
Data availability
All raw fastq files generated in this work for Ethiopian An. stephensi is publicly available (see PRJEB66077 for accession numbers, https://www.ncbi.nlm.nih.gov/bioproject/PRJEB66077/). Accession numbers for publicly available raw used in this study can be found in Supplementary Table S7. Analysis scripts are available at https://github.com/LSHTMPathogenSeqLab.
References
Faulde, M. K., Rueda, L. M. & Khaireh, B. A. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti, Horn of Africa. Acta Trop. 139, 39–43 (2014).
Carter, T. E. et al. First detection of Anopheles stephensi Liston, 1901 (Diptera: culicidae) in Ethiopia using molecular and morphological approaches. Acta Trop. 188, 180–186 (2018).
WHO. Global Malaria Programme: Malaria Threats Map. https://www.who.int/teams/global-malaria-programme/surveillance/malaria-threats-map
Ahmed, A., Khogali, R., Elnour, M. A. B., Nakao, R. & Salim, B. Emergence of the invasive malaria vector Anopheles stephensi in Khartoum State, central Sudan. Parasit. Vectors. 14, 511 (2021).
Ali, S., Samake, J. N., Spear, J. & Carter, T. E. Morphological identification and genetic characterization of Anopheles stephensi in Somaliland. Parasit. Vectors. 15, 247 (2022).
Sinka, M. E. et al. A new malaria vector in Africa: Predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc. Natl. Acad. Sci. 117, 24900–24908 (2020).
Organization, W. H. Partners Convening: A Regional Response To the Invasion of Anopheles Stephensi in Africa: Meeting Report, 8–10 March 2023 (World Health Organization, 2023).
Tadesse, F. G. et al. Anopheles stephensi mosquitoes as vectors of plasmodium Vivax and falciparum, Horn of Africa, 2019. Emerg. Infect. Dis. 27, 603–607 (2021).
The dominant Anopheles. vectors of human malaria in the Asia-Pacific region: Occurrence data, distribution maps and bionomic précis | Parasites & Vectors | Full Text. https://parasitesandvectors.biomedcentral.com/articles/https://doi.org/10.1186/1756-3305-4-89
Seyfarth, M., Khaireh, B. A., Abdi, A. A., Bouh, S. M. & Faulde, M. K. Five years following first detection of Anopheles stephensi (Diptera: Culicidae) in Djibouti, Horn of Africa: Populations established—malaria emerging. Parasitol. Res. 118, 725–732 (2019).
Moussa, R. A. et al. Molecular investigation of malaria-infected patients in Djibouti City (2018–2021). Malar. J. 22, 147 (2023).
Emiru, T. et al. Evidence for a role of Anopheles stephensi in the spread of drug- and diagnosis-resistant malaria in Africa. Nat. Med. 29, 3203–3211 (2023).
Al-Eryani, S. M. et al. Public health impact of the spread of Anopheles stephensi in the WHO Eastern mediterranean region countries in Horn of Africa and Yemen: Need for integrated vector surveillance and control. Malar. J. 22, 187 (2023).
Takken, W. & Lindsay, S. Increased threat of urban malaria from Anopheles stephensi mosquitoes, Africa. Emerg. Infect. Dis. 25, 1431–1433 (2019).
Surendran, S. N. et al. Anthropogenic factors driving recent range expansion of the malaria vector Anopheles stephensi. Frontiers Public. Health 7 (2019).
WHO. WHO Initiative to Stop the Spread of Anopheles Stephensi in Africa. (2022). https://www.who.int/publications/i/item/WHO-UCN-GMP-2022.06
Global Malaria Programme. WHO Global. WHO World Malaria Report 2022. WHO.
Samake, J. N. et al. Population genomic analyses reveal population structure and major hubs of invasive Anopheles stephensi in the Horn of Africa. Mol. Ecol. 32, 5695–5708 (2023).
Consortium, A. et al. Genetic diversity of the African malaria vector Anopheles Gambiae. Nature 552, 96–100 (2017).
Dennis, T. P. W. et al. Signatures of adaptation at key insecticide resistance loci in Anopheles Gambiae in Southern Ghana revealed by reduced-coverage WGS. Sci. Rep. 14, 8650 (2024).
Balkew, M. et al. An update on the distribution, bionomics, and insecticide susceptibility of Anopheles stephensi in Ethiopia, 2018–2020. Malar. J. 20, 263 (2021).
Yared, S. et al. Insecticide resistance in Anopheles stephensi in Somali region, Eastern Ethiopia. Malar. J. 19, 180 (2020).
Enayati, A., Hanafi-Bojd, A. A., Sedaghat, M. M., Zaim, M. & Hemingway, J. Evolution of insecticide resistance and its mechanisms in Anopheles stephensi in the WHO Eastern mediterranean region. Malar. J. 19, 258 (2020).
World Health Organisation. Global Report on Insecticide Resistance in Malaria Vectors: 2010–2016 Global Malaria Programme.
Liu, N. Insecticide resistance in mosquitoes: Impact, mechanisms, and research directions. Annu. Rev. Entomol. 60, 537–559 (2015).
Dada, N., Sheth, M., Liebman, K., Pinto, J. & Lenhart, A. Whole metagenome sequencing reveals links between mosquito microbiota and insecticide resistance in malaria vectors. Sci. Rep. 8, 2084 (2018).
ffrench-Constant, R. H., Anthony, N., Aronstein, K., Rocheleau, T. & Stilwell, G. Cyclodiene insecticide resistance: From molecular to population genetics. Annu. Rev. Entomol. 45, 449–466 (2000).
Acford-Palmer, H. et al. Identification of two insecticide resistance markers in Ethiopian Anopheles stephensi mosquitoes using a multiplex amplicon sequencing assay. Sci. Rep. 13, 5612 (2023).
Independent mutations in the Rdl locus confer dieldrin resistance to Anopheles gambiae and An. arabiensis - Du – 2005 - Insect Molecular Biology - Wiley Online Library. https://resjournals.onlinelibrary.wiley.com/doi/https://doi.org/10.1111/j.1365-2583.2005.00544.x
Samake, J. N. et al. Detection and population genetic analysis of Kdr L1014F variant in Eastern Ethiopian Anopheles stephensi. Infect. Genet. Evol. 99, 105235 (2022).
Dykes, C. L. et al. Knockdown resistance (kdr) mutations in Indian Anopheles stephensi (Diptera: Culicidae) populations. J. Med. Entomol. 53, 315–320 (2016).
Safi, N. H. Z. et al. Status of insecticide resistance and its biochemical and molecular mechanisms in Anopheles stephensi (Diptera: Culicidae) from Afghanistan. Malar. J. 18, 249 (2019).
Martinez-Torres, D. et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles Gambiae S.s. Insect Mol. Biol. 7, 179–184 (1998).
Safi, N. H. Z. et al. Evidence of metabolic mechanisms playing a role in multiple insecticides resistance in Anopheles stephensi populations from Afghanistan. Malar. J. 16, 100 (2017).
Riveron, J. M. et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 15, R27 (2014).
Ahn, J., Sinka, M., Irish, S. & Zohdy, S. Modeling marine cargo traffic to identify countries in Africa with greatest risk of invasion by Anopheles stephensi. Sci. Rep. 13, 876 (2023).
Carter, T. E. et al. Genetic diversity of Anopheles stephensi in Ethiopia provides insight into patterns of spread. Parasit. Vectors. 14, 602 (2021).
Lehmann, T. et al. Urban malaria may be spreading via the wind—here’s why that’s important. Proc. Natl. Acad. Sci. 120, e2301666120 (2023).
Teshome, A. et al. Resistance of Anopheles stephensi to selected insecticides used for indoor residual spraying and long-lasting insecticidal Nets in Ethiopia. Malar. J. 22, 218 (2023).
Ingham, V. A. et al. Integration of whole genome sequencing and transcriptomics reveals a complex picture of the reestablishment of insecticide resistance in the major malaria vector Anopheles coluzzii. PLoS Genet. 17, e1009970 (2021).
Lucas, E. R. et al. Whole-genome sequencing reveals high complexity of copy number variation at insecticide resistance loci in malaria mosquitoes. Genome Res. 29, 1250–1261 (2019).
Population genetic structure of the malaria mosquito Anopheles. arabiensis across Nigeria suggests range expansion - Onyabe – 2001 - Molecular Ecology - Wiley Online Library. (2001). https://onlinelibrary.wiley.com/doi/full/10.1046/j.0962-1083.01387.x?sid=nlm%3Apubmed.
Kent, R. J., Mharakurwa, S. & Norris, D. E. Spatial and Temporal genetic structure of Anopheles arabiensis in Southern Zambia over consecutive wet and drought years. Am. J. Trop. Med. Hyg. 77, 316–323 (2007).
Campos, M. et al. The origin of Island populations of the African malaria mosquito, Anopheles coluzzii. Commun. Biol. 4, 630 (2021).
Schmidt, H. et al. Transcontinental dispersal of Anopheles Gambiae occurred from West African origin via serial founder events. Commun. Biol. 2, 473 (2019).
Norris, L. C. et al. Adaptive introgression in an African malaria mosquito coincident with the increased usage of insecticide-treated bed nets. Proc. Natl. Acad. Sci. U. S. A. 112, 815–820 (2015).
Main, B. J. et al. Complex genome evolution in Anopheles coluzzii associated with increased insecticide usage in Mali. Mol. Ecol. 24, 5145–5157 (2015).
Tiwari, S., Ghosh, S. K., Ojha, V. P., Dash, A. P. & Raghavendra, K. Reduced susceptibility to selected synthetic pyrethroids in urban malaria vector Anopheles stephensi: a case study in Mangalore City, South India. Malar. J. 9, 179 (2010).
Yang, C., Huang, Z., Li, M., Feng, X. & Qiu, X. RDL mutations predict multiple insecticide resistance in Anopheles sinensis in Guangxi, China. Malar. J. 16, 482 (2017).
Liu, N., Feng, X. & Qiu, X. RDL mutations in Guangxi Anopheles sinensis populations along the China–Vietnam border: distribution frequency and evolutionary origin of A296S resistance allele. Malar. J. 19, 23 (2020).
Jones, A. K., Buckingham, S. D., Brown, L. A. & Sattelle, D. B. Alternative splicing of the Anopheles Gambiae nicotinic acetylcholine receptor, Agamαβ9, generates both alpha and beta subunits. Invert. Neurosci. 9, 77 (2009).
Shimada, S. et al. The mechanism of loop C-neonicotinoid interactions at insect nicotinic acetylcholine receptor Α1 subunit predicts resistance emergence in pests. Sci. Rep. 10, 7529 (2020).
Bass, C. et al. Mutation of a nicotinic acetylcholine receptor Β subunit is associated with resistance to neonicotinoid insecticides in the aphid Myzus persicae. BMC Neurosci. 12, 51 (2011).
Mouhamadou, C. S. et al. Evidence of insecticide resistance selection in wild Anopheles coluzzii mosquitoes due to agricultural pesticide use. Infect. Dis. Poverty. 8, 64 (2019).
Zoh, M. G. et al. Experimental evolution supports the potential of neonicotinoid-pyrethroid combination for managing insecticide resistance in malaria vectors. Sci. Rep. 11, 19501 (2021).
Oxborough, R. M. et al. Susceptibility testing of Anopheles malaria vectors with the neonicotinoid insecticide Clothianidin; results from 16 African countries, in Preparation for indoor residual spraying with new insecticide formulations. Malar. J. 18, 264 (2019).
Tchouakui, M. et al. Comparative study of the effect of solvents on the efficacy of neonicotinoid insecticides against malaria vector populations across Africa. Infect. Dis. Poverty. 11, 35 (2022).
U.S. President’s Malaria Initiative: Ethiopia Malaria Operational Plan FY 2023.
Kane, N. et al. (ed, S.) Drug-resistant drosophila indicate glutamate-gated chloride channels are targets for the antiparasitics nodulisporic acid and Ivermectin. Proc. Natl. Acad. Sci. 97 13949–13954 (2000).
McCavera, S., Rogers, A. T., Yates, D. M., Woods, D. J. & Wolstenholme, A. J. An Ivermectin-Sensitive Glutamate-Gated chloride channel from the parasitic nematode Haemonchus contortus. Mol. Pharmacol. 75, 1347–1355 (2009).
Atif, M. et al. GluClR-mediated inhibitory postsynaptic currents reveal targets for Ivermectin and potential mechanisms of Ivermectin resistance. PLoS Pathog. 15, e1007570 (2019).
Chaccour, C., Lines, J. & Whitty, C. J. M. Effect of Ivermectin on Anopheles Gambiae mosquitoes fed on humans: The potential of oral insecticides in malaria control. J. Infect. Dis. 202, 113–116 (2010).
Kobylinski, K. C., Foy, B. D. & Richardson, J. H. Ivermectin inhibits the sporogony of plasmodium falciparum in Anopheles Gambiae. Malar. J. 11, 381 (2012).
Azevedo, R., Mendes, A. M. & Prudêncio, M. Inhibition of plasmodium sporogonic stages by Ivermectin and other avermectins. Parasit. Vectors. 12, 549 (2019).
Marathe, A. et al. Potential impact of 5 years of Ivermectin mass drug administration on malaria outcomes in high burden countries. BMJ Glob Health. 6, e006424 (2021).
Ikeda, T., Nagata, K., Kono, Y., Yeh, J. Z. & Narahashi, T. Fipronil modulation of GABAA receptor single-channel currents. Pest Manag. Sci. 60, 487–492 (2004).
Ruiz-Castillo, P., Rist, C., Rabinovich, R. & Chaccour, C. Insecticide-treated livestock: A potential one health approach to malaria control in Africa. Trends Parasitol. 38, 112–123 (2022).
Makhanthisa, T. I., Braack, L. & Lutermann, H. The effect of cattle-administered Ivermectin and fipronil on the mortality and fecundity of Anopheles arabiensis Patton. Parasit. Vectors. 14, 349 (2021).
Dreyer, S. M. et al. Fipronil and Ivermectin treatment of cattle reduced the survival and ovarian development of field-collected Anopheles albimanus in a pilot trial conducted in Northern Belize. Malar. J. 18, 296 (2019).
Poché, R. M. et al. Preliminary efficacy investigations of oral fipronil against Anopheles arabiensis when administered to Zebu cattle (Bos indicus) under field conditions. Acta Trop. 176, 126–133 (2017).
Davari, B. et al. Selection of Anopheles stephensi with DDT and dieldrin and cross-resistance spectrum to pyrethroids and fipronil. Pestic Biochem. Physiol. 89, 97–103 (2007).
Zhang, Y. et al. Synergistic and compensatory effects of two point mutations conferring target-site resistance to fipronil in the insect GABA receptor RDL. Sci. Rep. 6, 32335 (2016).
Nakao, T. Mechanisms of resistance to insecticides targeting RDL GABA receptors in planthoppers. NeuroToxicology 60, 293–298 (2017).
Tian, Y., Gao, Y., Chen, Y., Liu, G. & Ju, X. Identification of the fipronil resistance associated mutations in Nilaparvata lugens GABA receptors by molecular modeling. Molecules 24, 4116 (2019).
How introns enhance gene expression - ScienceDirect. https://www.sciencedirect.com/science/article/pii/S1357272517301541?via%3Dihub
Tchigossou, G. et al. Molecular basis of permethrin and DDT resistance in an Anopheles funestus population from Benin. Parasit. Vectors. 11, 602 (2018).
Sandeu, M. M., Mulamba, C., Weedall, G. D. & Wondji, C. S. A differential expression of pyrethroid resistance genes in the malaria vector Anopheles funestus across Uganda is associated with patterns of gene flow. PLoS ONE. 15, e0240743 (2020).
Insects | Free Full-Text. | Genome-Wide Selective Signature Analysis Revealed Insecticide Resistance Mechanisms in Cydia pomonella. https://www.mdpi.com/2075-4450/13/1/2
Pondeville, E. et al. Microarray and RNAi analysis of P450s in Anopheles Gambiae male and female steroidogenic tissues: CYP307A1 is required for ecdysteroid synthesis. PLoS ONE. 8, e79861 (2013).
Sun, Y. et al. Functional characterization of an arrestin gene on insecticide resistance of culex pipiens pallens. Parasit. Vectors. 5, 134 (2012).
Cariño, F. A., Koener, J. F., Plapp, F. W. & Feyereisen, R. Constitutive overexpression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect Biochem. Mol. Biol. 24, 411–418 (1994).
Shetty, V., Sanil, D. & Shetty, N. J. Insecticide susceptibility status in three medically important species of mosquitoes, Anopheles stephensi, Aedes aegypti and culex quinquefasciatus, from Bruhat Bengaluru Mahanagara Palike, Karnataka, India. Pest Manag. Sci. 69, 257–267 (2013).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Glick, J. I. Illustrated key to the female Anopheles of Southwestern Asia and Egypt (Diptera: Culicidae). Mosq. Syst. 24, 125–153 (1992).
Thakare, A. et al. The genome trilogy of Anopheles stephensi, an urban malaria vector, reveals structure of a locus associated with adaptation to environmental heterogeneity. Sci. Rep. 12, 3610 (2022).
Chakraborty, M. et al. Hidden genomic features of an invasive malaria vector, Anopheles stephensi, revealed by a chromosome-level genome assembly. BMC Biol. 19, 28 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinforma Oxf. Engl. 30, 2114–2120 (2014).
H.Li. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arxiv.org n/a, n/a (2013).
Danecek, P. et al. Twelve years of samtools and BCFtools. GigaScience 10, 1–4 (2021).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. 201178 Preprint at (2018). https://doi.org/10.1101/201178
Danecek, P. et al. The variant call format and vcftools. Bioinformatics 27, 2156–2158 (2011).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin). 6, 80–92 (2012).
fastq2matrix/scripts. at master · LSHTMPathogenSeqLab/fastq2matrix. GitHub https://github.com/LSHTMPathogenSeqLab/fastq2matrix/tree/master/scripts
Turner, S. D. Qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open. Source Softw. 3, 731 (2018).
Paradis, E. & Schliep, K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Alexander, D. H. & Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry Estimation. BMC Bioinform. 12, 246 (2011).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
rehh 2. 0: a reimplementation of the R package Rehh to detect positive selection from haplotype structure - Gautier – 2017 - Molecular Ecology Resources - Wiley Online Library. https://doi.org/10.1111/1755-0998.12634
Rausch, T. et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012).
Funding
H.A.P. is funded by a LIDo-DTP PhD studentship. J.P. was funded by a Newton Institutional Links Grant (British Council, no. 261868591). T.G.C. is funded by the MRC UK (Grant no. MR/M01360X/1, MR/N010469/1, MR/R025576/1, and MR/R020973/1). S.C. was funded by MRC UK grants (ref. MR/M01360X/1, MR/R025576/1, and MR/R020973/1). T.W. was funded by a Wellcome Trust/Royal Society Sir Henry Dale Fellowship (101285/Z/13/Z). L.A.M. is jointly funded by the Foreign, Commonwealth & Development Office (FCDO), the Medical Research Council, Wellcome and Department of Health and Social Care (Grant Ref: MR/R006040/1). M.K. was funded by the Foreign, Commonwealth & Development Office (FCDO) RPC and a Wellcome Trust Biomedical Resources grant. F.G.T. was supported by the Bill and Melinda Gates Foundation (HAMMS; INV-048214) and Wellcome Trust Early Career Award (102348).
Author information
Authors and Affiliations
Contributions
T.G.C. and S.C. conceived and directed the project. T.B., L.A.M., and F.G.T. provided samples and performed species identification. L.A.M, M.K., T.W. undertook sample processing, DNA extraction and molecular identification. H.A-P., performed the bioinformatic analyses and results interpretation, with the assistance of E.M, J.E.P., M.H. and A.O., and under the supervision of T.G.C., S.C., J.P. and L.A.M. H.A-P. drafted the manuscript. All authors commented on manuscript drafts, and approved the final manuscript.
Corresponding author
Ethics declarations
Consent for publication
All authors have consented to the publication of this manuscript.
Competing interests
The authors declare no competing interests.
Ethics approval and consent
Study protocol was approved by the Institutional Ethical Review Board of the Aklilu Lemma Institute of Pathobiology of Addis Ababa University (ALIPB IRB/025/2011/2019), the Oromia Regional Health Bureau (BEFO/MBTFH/1331), and AHRI/ALERT Ethics Review Committee (AF-10-015.1, PO07/19). All participants or parents/legal guardians for participants < 18 years of age provided written informed consent. Persons who volunteered for human landing collection also provided written informed consent, were monitored for 3 weeks after collections, and if symptomatic and positive received treatment for Plasmodium according to the treatment guidelines of the country.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Acford-Palmer, H., Tadesse, F.G., Manko, E. et al. Genome wide population genetics and molecular surveillance of insecticide resistance in Anopheles stephensi mosquitoes from Awash Sebat Kilo in Ethiopia. Sci Rep 15, 16443 (2025). https://doi.org/10.1038/s41598-025-95814-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95814-0