Abstract
The use of nanoparticles as an alternative to traditional fertilizers, aiming at a more efficient use of nutrients, is a recently developed concept that requires a thorough understanding of the processes occurring in the soil-plant system. A crucial aspect in this framework is to decipher the plant responses to the unique characteristics of these materials. In this work, we aim at decoding the transcriptional responses of cucumber and maize roots to FePO4 nanoparticles applied as P and Fe sources, respectively. The results demonstrate that P and Fe supplied as nanoscale salts support plant nutrition with an efficiency comparable to that of ionic forms of the nutrients. This supposition is confirmed by transcriptomic profiles that show no significant upregulation of transcripts typically induced by deficiencies in P and Fe in cucumber and maize plants in which these nutrients were provided by FePO4 nanoparticles. The analysis further revealed that nanoparticles alter the expression of genes involved in root development and stress responses, effect that appeared to be independent on the nutritional status of the plants. Our data further underline the challenge to identify generalizable elements of the impact of nanomaterials on plant species, as responses are intimately linked to the type of nanomaterials and differ among plant species.
Similar content being viewed by others
Introduction
Driven by global population growth and changing dietary patterns, global food demand is projected to increase by 70% by 20501. Additionally, the intensive use of pesticides and fertilizers had significant environmental impacts, including pollution, soil salinization, and the deterioration of soil biological health2. Therefore, a challenge of modern agriculture is to develop new technologies that enhance food supply while mitigating adverse environmental effects and addressing climate change3. Nano-scaled particles, ranging from 1 to 100 nm in at least one dimension4, possess unique physical and chemical properties that are being rapidly exploited across various scientific fields including agriculture5. When used to support plant nutrition, these particles are known as nanofertilizers6. The peculiar properties of nanomaterials enable them to release nutrients in a controlled manner under specific environmental conditions, significantly enhancing nutrient use efficiency7,8,9. A critical aspect of crop fertilization is to minimize nutrient losses and to synchronize nutrient release with their interception by plant roots, thereby reducing chemical inputs and environmental impact4,6,10.
Phosphorus (P), an element playing crucial structural, energetic, and post-translational regulatory roles in plants11, is one of the most limiting nutrients in soils and the one with the lowest nutrient use efficiency, accounting for only 10–15%12. In response to P deficiency, plants activate a wide range of morpho-physiological acclimations including a modified root architecture and reduced shoot growth, resulting in an enhanced root/shoot biomass ratio, reduced leaf expansion, and, consequently, an increase in chlorophyll content13,14. Furthermore, P-deficient roots induce the expression of high-affinity Pi transporters and enhance the extrusion of organic acids, protons, and phosphatases to solubilize immobile P forms, thereby increasing their acquisition by crops15.
Among the micronutrients, iron (Fe) is required in the largest amount by plants16. However, its solubility in soil is extremely low, particularly in calcareous soils, which account for about 30% of the world’s cultivated soils17. Iron plays a pivotal role in essential metabolic processes such as respiration, photosynthesis, and chlorophyll biosynthesis16,18. Leaf chlorosis is the most evident symptom of Fe deficiency16. Plants have evolved two strategies to face Fe shortage in the environment: Strategy I, employed by non-grasses, is based on acidification of the rhizosphere and reduction of insoluble ferric Fe species18 and Strategy II, adopted by grasses, which is based on the chelation of such forms of Fe18.
Most transcriptional investigations on the effects of nanoparticle (NP) application in plants have primarily focused on the phytotoxicity of metal-based and carbon-based nanomaterials19. The mechanisms of phytotoxicity were found to be highly dependent on the type of nanomaterials, although some studies highlight a common response of genes related to both abiotic and biotic stress in plants exposed to various types of NPs20,21,22,23,24. A much smaller number of studies have evaluated gene expression changes when NPs are applied as a source of mineral nutrients25,26,27. The rationale for the present study derived from previous findings of our research group. Specifically, we developed a method for synthesizing citrate-capped FePO₄ NPs28, which were shown to be an efficient source of both P and Fe, particularly when compared to their bulk, non-nano counterparts in hydroponic systems. It has been suggested that, due to their sub-micron size, NPs can reach the root surface in greater amounts29. Moreover, their high surface-to-volume ratio, combined with root activity, is expected to enhance their dissolution rate compared to bulk FePO₄, leading to species-specific plant responses29. Moreover, the NPs demonstrated the ability to sustain P nutrition in pot-grown cucumber plants as triple superphosphate7 without significantly impacting soil biology29.
Here, we attempted to catalogue the early transcriptomic responses (after 24 h) of hydroponically-grown cucumber and maize roots when FePO₄ nanoparticles were applied as sources of P and Fe, respectively. We compared the transcriptional profiles of roots which acquired P or Fe from NPs with those of plants treated with the corresponding ionic forms of the nutrients. We hypothesized that, in addition to their nutritional effects, NPs might induce additional root responses due to their nano-sized solid nature.
Results and discussion
Plant growth and physiological analyses
Previous research by Sega et al.29 demonstrated that FePO4 NPs are an efficient source of P and Fe for cucumber and maize plants, respectively. The experiments presented in the current manuscript corroborate these findings as evidenced by the plant growth parameters and SPAD index (Figs. 1 and 2, Figs. S1, S2, S3 and S4). Specifically, the application of FePO4 NPs as a P source (PNP) to cucumber plants resulted in shoot weights that were similar to control plants, no significant differences were observed in root weights between the two conditions (Figs. 1 and 3). Furthermore, the application of FePO₄ NPs as an Fe source allowed maize plants to attain shoot and root dry weights comparable to those of control plants (Fig. 2). These observations suggest that FePO4 NPs can effectively supply essential nutrients, promoting healthy plant growth under nutrient-deficient conditions. However, we observed significant differences in root morphology between FeNP-treated and control maize plants (Fig. 4), with FeNP-treated plants showing lower total and lateral root lengths. This indicates that FePO4 NPs can adequately supply Fe but may negatively impact root morphology in maize.
Phenotypic parameters of FeNP-treated cucumber plants. a Shoot dry weight; b Root dry weight; c Leaf SPAD index after 13 d of growth in hydroponics. Control plants (C); plants grown without P (-P); plants grown with NPs as a source of P (PNP). Data are means ± S.E. of three independent experiments with three plants each (one-way ANOVA with Tukey’s test, n = 9, three plants for each independent growth experiment, p < 0.05).
Phenotypic parameters of FeNP-treated maize plants. a Shoot dry weight; b Root dry weight; c Leaf SPAD index of maize plants after 10 d of growth in hydroponics. Control plants (C); plants grown without Fe (-Fe); plants grown with NPs as a source of Fe (FeNP). Data are means ± S.E. of three independent experiments with three plants each (one-way ANOVA with Tukey’s test, n = 9, three plants for each independent growth experiment, p < 0.05).
Total, main, and lateral root length cucumber plants after 13 days of growth in hydroponics. Control plants (C); plants grown without P (-P); plants grown with NPs as a source of P (PNP). Data are means ± S.E. of three independent experiments with three plants each (one-way ANOVA with Tukey’s test, n = 9, three plants for each independent growth experiment, p < 0.05).
Total, main, and lateral root length maize plants after 10 days of growth in hydroponics. Control plants (C); plants grown without Fe (-Fe); plants grown with NPs as a source of Fe (FeNP). Data are means ± S.E. of three independent experiments with three plants each (one-way ANOVA with Tukey’s test, n = 9, three plants for each independent growth experiment, p < 0.05).
Root transcriptional changes in response to NP treatment
To understand the action of FePO4 NPs on nutritional processes related to the acquisition of P (in cucumber) and Fe (in maize), we studied the early transcriptomic responses in roots of these two species to short-term exposure to FePO4 NPs. Specifically, we identified transcripts that were differentially expressed in roots among plants treated for 24 h with FePO4 NPs, plants grown in complete nutrient solution (C), and plants grown in the absence of P (-P, for cucumber) or Fe (-Fe, for maize). The number of differentially expressed transcripts (DETs) identified in the two root transcriptome comparisons carried out for each species is reported in Table 1. The differential expression of a selected set of transcripts was also analysed by RT-qPCR experiments (Table S1). The highest number of DETs was observed in maize roots when the FePO4 NP treatment was compared to control plants (Table 1). The results of a GO enrichment analysis of the DETs are reported in Figs. S5 and S6.
Transcriptional differences between NP-treated and P-sufficient cucumber roots
To determine whether the role of FePO4 NPs was solely nutritional (i.e., by providing P intercepted by the roots) without affecting other physiological or developmental processes, we first compared transcripts that were differentially expressed between NP-treated and P-sufficient plants (PNP vs. C, Additional file 1). In this comparison, no transcripts related to P uptake, P metabolism, or cell wall metabolism linked to root developmental processes were identified after 24 h, highlighting the role of FePO4 NPs as an efficient source of the macronutrient for cucumber29. Interestingly, FePO4 NPs seem to enhance the stress responses in the roots of cucumber plants. We recorded upregulation of two transcripts (Cucsa.197540.10 and Cucsa.241740.1, Table 2), homologous to AtNHL10 and AtICS2, respectively, that are putatively involved in biotic stress responses. AtNHL10 is a pattern-triggered immunity-responsive gene30, while AtICS2 encodes isochorismate synthase 2, which plays a role in salicylic acid synthesis and is involved in the enhancement of plant immunity at low temperatures31. Another transcript induced by FePO4 NPs (Cucsa.326110.1, Table 2) shows homology to AtPRX52, which is responsive to the fungal toxin fumonisin B1 and may play a role in modulating H2O2 levels during the oxidative burst in response to microbial pathogen attack32. Only one transcript linked to biotic stress response was found to be downregulated in FePO4 NP-treated plants (Cucsa.254210.1, Table 2). This gene is homologous to AtSTP13, encoding a sugar transporter involved in resistance to Botrytis cinerea in Arabidopsis33. Our data on biotic stress responses contrast with previous findings in Arabidopsis, where NPs composed of different materials (TiO₂-NPs, Ag-NPs, and multi-walled carbon nanotubes) suppressed pathogen-responsive genes and those involved in salicylic acid-mediated pathways after 12 h. This response was linked to increased bacterial colonization in infection experiments19. It thus appears that data obtained with different experimental systems (e.g., a certain combination of NPs and plants) can lead to different results, rendering a prediction of the outcome of such experimental difficult. FePO4 NPs may be perceived on the root surface as xenobiotic compounds, as indicated by the upregulation of transcripts involved in detoxifying these exogenous substances (Cucsa.256030.1 and Cucsa.058370.1, Table 2). These two transcripts encode proteins homologous to AtGSTU7 and AtHYR1, respectively. AtGSTU7 is putatively involved in herbicide detoxification in roots34, while AtHYR1 is an UDP glucosyl transferase that mediates resistance to hypostatin, an inhibitor of cell expansion35.
Transcriptional differences between NP-treated and P-deficient cucumber roots
To investigate early effects of FePO4 NPs on the nutritional status of cucumber plants, we analysed the transcriptomic profiles of plants supplemented with NPs and plants grown without P (-P) (Additional file 1). The identified DETs indicate that FePO4 NPs may influence root development and metal homeostasis of the plants. A putative effect of NPs on root development is evidenced by the downregulation of an expansin transcript (Cucsa.355990.1, Table 2) and increased abundance of a transcript encoding a pectin lyase (Cucsa.249050.1). Expansins are known to facilitate cell wall loosening, a process that is crucial for cell growth and the emergence of lateral roots36. However, lateral root development also hinges on the activities of pectate lyases and the de-esterification of pectins by pectin methyl esterases36. Notably, FePO4 NPs caused downregulation of a transcript encoding a plant invertase/pectin methylesterase inhibitor (Cucsa.286280.1, Table 2). This protein family was shown to promote lateral root development and overall root growth37. The supposition that FePO4 NPs promote root growth is further supported by their ability to downregulate the expression of glucan synthase-like 4 (Cucsa.058230.1, Table 2), a homolog of AtGSL4 involved in callose deposition in plasmodesmata. Callose deposition can inhibit root growth, particularly under stress conditions38. Conspicuously, these findings appear to be congruent with our phenological analyses, revealing that plants treated with FePO4 NPs develop a root system akin to control plants supplied with ionic forms of P (Fig. 3). Further evidence supporting the efficiency of these NPs as a P source is provided by the regulation of transcripts involved in root hair elongation. Generally, root hair development is responsive to the P nutritional status of the plant39. In Arabidopsis, P starvation stimulates root hair elongation through induction of the bHLH proteins AtRSL2 and AtRSL440. Adequate P supply suppresses root hair elongation41. NP-treated plants showed reduced transcript abundance of the AtRSL2 homolog Cucsa.352750.1 relative to P-starved plants (Table 2), indicative of an adequate P supply.
The presence of FePO4 NPs appears to alter metal homeostasis in roots. We observeddifferential expression of a heavy metal transport/detoxification superfamily protein (HIPP/HPP) transcript (Cucsa.257230.1, Table 2), possibly associated with metal transport42. This transcriptional behaviour aligns with the observed efficiency of FePO4 NPs as a P source for cucumber plants and the previously reported accumulation of excess Fe in the apoplast29. A role of FePO4 NPs in metal homeostasis is further supported by the downregulation of two transcripts (Cucsa.090660.1 and Cucsa.092390.1, Table 2) homologous to Arabidopsis AtNAS4. AtNAS4 is critical for the biosynthesis of the Fe chelator nicotianamine and known to be induced in roots under Fe deficiency43,44.
Transcriptional differences between treated NP-treated and Fe-sufficient maize roots
Maize is a Strategy II species for Fe acquisition and has been shown to efficiently utilize Fe delivered by FePO4 NPs29. Similar to our analysis on cucumber, we aimed to determine whether the role of FePO4 NPs in maize was purely nutritional or if the supplied nanoparticles also impacted other processes. To validate such a role of FePO4 NPs, we compared the root transcriptional profiles of plants 24 h after supplying FeNP as an Fe source with those grown in the presence of a soluble Fe form (FeEDTA) as a control (Additional file 2). No genes involved in Fe acquisition or cellular Fe homeostasis were differentially expressed between treated and non-treated plants, suggesting that NP-treated plants were adequately supplied with the micronutrient29. However, several DETs encode proteins involved in lateral root formation and stress response.
Plants supplied with FePO4 NPs showed a reduction in the expression of two class III peroxidase transcripts (GRMZM2G138450_T01 and GRMZM2G015280_T01, Table 2), possibly pointing to a less developed root apparatus. Class III peroxidases is known to play a role in lateral root emergence in Arabidopsis45. This assumption is further supported by the induction of a transcript that shows homology to the HOMEOBOX transcription factor AtHB22 of Arabidopsis in FeNP-treated roots (GRMZM2G417229_T01, Table 2). AtHB22 and its close homolog AtHB25 are involved in the feedback regulation of auxin-signalling-related SMALL AUXIN-UP RNA (SAUR) through deposition of the histone variant H2A.Z, thereby reducing IAA-induced lateral root formation46. The hypothesized repression of lateral root formation due to FePO4 NPs supply is also supported by the upregulation of a DET (GRMZM2G346095_T01, Table 2) encoding a chalcone synthase homologous to AtTT4. AtTT4 negatively controls lateral root development by synthesizing flavonoids that scavenge superoxide ions, a driver of lateral root emergence47. Additionally, FeNP supply induced the expression of GRMZM2G090381_T01 (Table 2), encoding a protein homologous to AtCNGC, which is involved in Ca2+ uptake and primary root growth48. Primary roots grow faster in the Arabidopsis cngc1 mutant48, suggesting that higher transcript levels of this gene in FeNP-treated plants exerts a negative control on root growth. Taken together, these results suggest that FePO4 NPs predominantly repress molecular processes involved in lateral root development. Iron deficiency can enhance ectopic lateral root branching in maize49, an observation that confirms that plants supplied with FePO4 NPs were in a sufficient Fe nutritional status29. This assumption is further supported by the upregulation of an HIPP transcript (AC217910.3_FGT001, Table 2), potentially involved in metal homeostasis42.
Regarding DETs related to stress responses, we observed increased abundance of a transcript involved in the detoxification of xenobiotics50, the Tau GST GRMZM2G480439_T01 (Table 2) in FePO4 NP-treated roots relative to control plants (FeNP vs. C), suggesting that, similar to what has been observed in cucumber (see above), FeNP can activate biochemical pathways involved in xenobiotic detoxification in maize. We also observed an increase in the expression of a transcript corresponding to a protein homologous to SUGAR-DEPENDENT1 (SDP1) TAG lipase (AtSDP1) (GRMZM2G004276_T01, Table 2), which may be involved in lipid remodelling during the cold response51. Downregulation of two transcripts homologous to AtAOX1a (GRMZM2G125669_T01 and GRMZM2G074761_T01, Table 2) in FeNP-treated roots compared to controls (FeNP vs. C) suggests that roots of plants supplied with FePO4 NPs as a source of Fe did not experience oxidative stress. AtAOX1a is typically upregulated in response to various stress conditions52.
Transcriptional differences between NP-treated and Fe-deficient maize roots
We also analysed the effects of FePO4 NPs as a Fe source by comparing the root transcriptional profiles of FeNP-treated and Fe-deficient plants (FeNP vs. -Fe, Additional file 2). We observed significant changes in the abundance of transcripts potentially involved in root developmental processes and responses associated with Fe availability. Considering cell wall metabolism associated with root developmental processes, we recorded an induction of transcripts encoding a homolog of the P-type H+-ATPase AtAHA8 (GRMZM2G148374_T01), a pectin lyase (GRMZM2G177940_T01), an extensin (GRMZM5G888797_T01), and two peroxidases (GRMZM2G361475_T01 and GRMZM2G061776_T01, Table 2), indicative of active cell wall processes specific to root elongation and differentiation zones36. These changes in gene expression corroborate the observation that the root systems of FeNP-treated plants were better developed compared to those grown without Fe (Fig. 4).
NP-induced transcriptional changes that were independent of the nutritional status
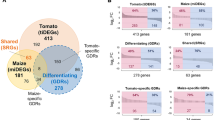
Venn diagram analysis identified DETs that were commonly modulated in both cucumber comparisons (PNP vs. C and PNP vs. -P) and maize comparisons (FeNP vs. C and FeNP vs. -Fe). These DETs, totalling two in cucumber and 26 in maize (Fig. 5, Additional files 1 and 2), represent transcripts specifically affected by FePO4 NPs and seem to be independent of the nutritional status of the plants. Notably, expression of these transcripts followed the same pattern when FePO4 NP-treated roots were compared to both control and nutrient-deficient conditions (-P for cucumber and -Fe for maize). Thus, these transcriptome changes could be attributed to the inherent properties of the NPs. In cucumber, the two DETs were repressed by FePO4 NPs and could play roles in root development. One of these DETs (Cucsa.322910.1, Table 3) encodes a protein homologous to AtDVL17, a member of a class of small polypeptides putatively involved in coordinating differentiation, development, and growth53. The other DET encodes a heavy chain-like protein (Cucsa.048300.5, Table 3).
Maize roots appear to be more responsive than cucumber to the FePO4 NPs, a total of 26 DETs were common in the two comparisons. Within this group, we observed the upregulation of a transcript (GRMZM2G086401_T01) encoding a protein similar to AtSPK1. This protein is involved in the SPK1-ROP6-RIC1 pathway that is activated by auxin and inhibits the internalization of PIN2, an auxin efflux transporter crucial for the polar transport of the hormone54. This pathway controls the longitudinal spacing of lateral roots55. Notably, both the spk1 null mutant and pin2 knockout plants exhibited an increase in lateral root density54. Moreover, NP-inducted expression of a KINASE-ASSOCIATED PROTEIN PHOSPHATASE (KAPP, GRMZM2G150608_T01), encoding a protein with high similarity to AtRAG1, further indicates reduced lateral root development, as the rag1-1 mutant of Arabidopsis exhibited enhanced lateral root formation under salt stress56. On the contrary, we recorded induction of a pectin lyase transcript (GRMZM2G177940_T01) with a putative role in cell wall metabolism in the zone of lateral root emergence36,57. All these observations are consistent with the phenotype shown by maize plants treated with FePO4 NPs, which, after 10 d of treatment, exhibited a less developed root system compared to control plants (Fig. 4).
Other effects of the FePO4 NP treatment are related to hormone signalling. Application of NPs induced a GUANINE NUCLEOTIDE EXCHANGE FACTOR (GEF) transcript (GRMZM2G442148_T01) with a putative role in signal transduction58. GEF proteins activate RHO OF PLANTS (ROPs), which in turn regulate cell growth and shape, subcellular localization of proteins, and responses to stresses59. ROPs are also involved in the regulation of ABA signalling as well as in the signalling and transport of auxin59. In relation to ABA, we also observed an upregulation of a transcript (GRMZM2G336766_T01) encoding a protein similar to the ABF/AREB transcription factor DPBF3, which is involved in the response to drought stress in Arabidopsis by remodelling the actin cytoskeleton and promoting stomatal closure60. In addition, FePO4 NPs caused induction of two transcripts (GRMZM2G158117_T01 and GRMZM2G114503_T01) encoding ABA-inducible MYB transcription factors homologous to AtRMS2 and AtRMS361. Also in line with the modulation of ABA responses caused by FePO4 NPs is the upregulation of a transcript (GRMZM2G000052_T01) for a protein similar to the polyamine oxidase AtPAO2. In Arabidopsis, AtPAO2 appears to act antagonistically to ABA inhibition of primary and secondary root growth62.
Brassinosteroids (BRs) are hormones involved in growth, development, and stress responses63. Induction of a UDP-glucosyltransferase (GRMZM2G341771_T01) with homology to AtUGT73C5 may hint to a role of FePO4 NPs in governing BR activity. In Arabidopsis, AtUGT73C5 controls the level of active BRs through inactivation by conjugation64. Notably, it has been reported that BR and ABA pathways antagonize each another63. Additionally, BRs are known to promote the growth of lateral roots by enhancing the acropetal transport of auxin65, and a reduction in active BR levels could be associated with decreased lateral root development. In summary, FePO4 NPs could affect hormone signalling, regulate root development, and promote plant tolerance to abiotic stresses. This is consistent with the effects of FePO4 NPs treatments of various materials applied to different plant species, including maize66.
Conclusion
In conclusion, the data presented here confirm that Fe and P-containing FePO4 NPs are good nutritional sources for cucumber and maize plants. Transcriptional profiling of NP-treated plants did not reveal indications for nutrient deficiencies, corroborating the ability of FePO4-NPs to provide Fe and P that can be readily absorbed by the roots. As depicted in Fig. 6, alongside their nutritional function, these nanomaterials exert species-specific molecular actions on the root systems already 24 h post-treatment. It is notable that some of these actions are entirely independent of nutritional aspects and appear to be related to the nanoparticulate nature of the material used for the treatment. Our findings further confirm that the impact of nanomaterials on plants depends not only on the characteristics of the nanoparticles but also on the physiological and metabolic specificity of the plant species under investigation, making it challenging to derive generalizable plant responses.
Schematic diagram depicting the effects of NPs. a Effects of NPs as P- source for cucumber b Effects of NPs as Fe sources for maize. The root transcriptional changes (24 h) and shoot and root parameters (13 d for cucumber and 10 d for maize) are reported; the symbols + and − indicate processes that were enhanced or repressed by NPs, respectively.
Materials and methods
FePO4 NPs synthesis
The FePO₄ nanoparticles were citrate-capped and synthesized using a batch method as described by Sega et al.28 where extensive characterization, including TEM and FT-IR analyses, was performed. The FePO₄ nanoparticles used in this study originated from the same batch characterized in a previous study7. These NPs were spheroidal and tended to aggregate, with a size peak at 91 nm7. The Fe and P concentrations were 0.15 M and 0.12 M, respectively, with a Fe/P ratio of 1.257.
Plant growth and physiological analyses
Purchased seeds of Cucumis sativus Viridis F1 hybrid (Franchi Sementi S.p.A.) and of Zea mays L. inbred line PR33T56 (Pioneer Hybrid Italia S.p.A.) were germinated on paper soaked with 1 mM CaSO4 in dark at 25 °C. Six seedlings were transferred in one 2-L pot filled with 1.8 L of continuously aerated nutrient solution. One pot was set up for each treatment. The control nutrient solution had the following composition: 0.7 mM K2SO4, 2 mM Ca(NO3)2, 0.5 mM MgSO4, 0.1 mM KH2PO4, 0.1 mM KCl, 100 µM FeNaEDTA, 10 µM H3BO3, 0.5 µM MnSO4, 0.5 µM ZnSO4, 0.2 µM CuSO4, and 0.01 µM (NH4)6Mo7O24. In addition to the control, conditions were set up as follows. For cucumber, plants were grown in NS without KH2PO4 (-P) and in NS without KH2PO4 and supplied with FePO4 NPs as P source (PNP). In the case of maize, plants were grown in NS without FeNaEDTA (-Fe) and in NS without FeNaEDTA and supplied with FePO4 NPs as Fe source (FeNP). FePO4 NPs were used in a concentration of P and Fe equivalent to 100 µM. The nutrient solution was changed twice per week. Plants were grown in a 16/8 day/light photoperiod at 25 °C and 200 µmol m− 2 sec− 1 PPFD (Photosynthetic Photon Flux Density). Roots of three out of six seedlings per pot (treatment) were sampled after 24 h and pooled to obtain samples for RNA extraction and further transcriptomic analyses. The remaining three cucumber and maize seedlings were grown for 13 and 10 days, respectively, in accordance with the occurrence of nutrient deficiency symptoms. For these seedlings, the SPAD index was calculated as average of all leaves of each plant. For each leaf, the average value of five measurements performed using a SPAD-502 Plus Chlorophyll meter® (Konica Minolta) was calculated. In addition, root parameters were determined from images of the roots of each seedling, acquired with an EpsonV700 perfection scanner. Images were then analysed with the WinRHIZO™ software 2015a Pro version (Regent Instruments Inc.) using the “root morphology” mode to determine the volume, length, and surface area of the roots. Three independent experiments (biological replicates) were performed.
RNA extraction, cDNA synthesis, and microarray expression analysis
Total RNA was extracted from three biological replicates (e.g. three independent growth), each consisting of pooled roots from three seedlings collected 24 h after treatment, for both cucumber and maize. Total RNA extractions were performed using 80 mg of maize and cucumber root tissue homogenized in liquid nitrogen and the Spectrum™ Plant Total RNA kit (Sigma-Aldrich). Total RNA quantity was determined using a NanoDropTM 1000 (Thermo Scientific) spectrophotometer, RNA quality was analysed with Bioanalyzer 2100 using a Bioanalyzer Chip RNA 6000 Nano kit (Agilent). The cRNA was synthesized and labelled using 200 ng of total RNA from each sample, the Low Input Quick Amp Labeling Kit, and Cyanine 3 (Cy3)-CTP fluorescent dye following the instructions of the Agilent technical manual (http://www.agilent.com). For each sample, 1.65 µg of Cy3-labeled cRNA was used for the hybridization reactions. The Cy3-labeled cRNA samples of maize and cumber were hybridized on two different 4 × 44k Agilent arrays according to the manufacturer’s manual for 17 h at 65° C and scanned on an Agilent G2565CA Microarray Scanner System (Agilent). For cucumber, microarray analyses were carried out using a chip that allows to analyse the expression of 30,364 transcripts predicted by the cucumber (Gy14) v1 genome annotation (http://cucurbit-genomics.org/organism/7). Probe design was performed using Agilent eArray (http://www.genomics.agilent.com). A complete description of the chip is available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entry (GPL34058). For maize, we used the chip described by Santi et al.67, which allowed for analysing the expression of 39,372 maize transcripts predicted from the B73 reference genome whose description is available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; GPL22578). Feature intensities were extracted using Agilent’s Feature Extraction Software 12.0 (www.agilent.com). Hybridization data of all samples were normalized using the value of the 75th percentile and Log2 transformed. Hierarchical clustering analyses were carried out with MeV software. After, differentially expressed transcripts (DETs) were identified through t-tests carried out using the MeV software (http://mev.tm4.org/#/welcome) for the comparisons PNP vs. -P and PNP vs. C for cucumber, and for the comparisons FeNP vs. -Fe and FeNP vs. C for maize. T-tests were performed, setting a p-value based on permutation with a critical value of 0.05. DETs were then filtered based on fold change values (|FC|≥2). Data are available at the GEO data repository (http://www.ncbi.nlm.nih.gov/geo) with the series entries GSE252553 and GSE252443. The complete lists of inferentially expressed transcripts identified in the different transcriptional comparisons are reported in Additional file 1 for cucumber and Additional file 2 for maize, respectively. For each maize and cumber transcript, the annotation of Phytozome for Cucumis sativus v1.0 (Csativus_122_annotation_info.txt) and the B73 maize reference genome (Zmays_284_5b+.annotation_info.txt) was used. GO enrichment analysis of the differentially expressed transcripts was carried out on the platform PlantRegMap68, the results were visualized with tools of the SRplot platform69.
Quantitative reverse transcription PCR (RT-qPCR) analysis
DNase I treatment was carried out for each sample using 1000 ng of total RNA and the RQ1 RNase-Free DNase (Promega, Madison, WI, USA) according to the manufacturer’s procedure. These treated samples were used to produce cDNA using the ImProm-II Reverse Transcription System (Promega). RT-qPCR analyses were performed with a final volume of 10 µL, a primer concentration of 350 nM, and 1 µL of an 1:3 solution of cDNA sample using the FastSYBR® Green Master Mix (ThermoFisher Scientific). The reaction was carried out with StepOnePlus™ (ThermoFisher Scientific) using the following reaction conditions: 20 s at 95 °C for initial denaturation, then 3 s at 95 °C, and 30 s at 60 °C for 40 cycles. The sequences of primers are reported in Table S2 and Table S3. Mean Normalized Expression (MNE)70 was calculated for each sample. Two housekeeping transcripts were used both for cucumber (Cucsa.313280.1 and Cucsa.238470.1) and maize (GRMZM2G047204_T01 and GRMZM2G149286_T01) analysis. The PCR efficiency of each primer couple was calculated using the LinRegPCR software71 based on fluorescence raw data. Mean normalized expression (MNE) was determined for each transcript and each sample using the two housekeeping transcripts70. Then, the geometric mean of the two MNE values obtained for each transcript and each sample was calculated72. Fold change (FC) values were then calculated in the MNE of samples in each considered comparison.
Data availability
Microarray data are available at the GEO (http://www.ncbi.nlm.nih.gov/geo) with the series entries GSE252553 and GSE252443. The data of the current study are available from the corresponding author on reasonable request.
References
Bindraban, P. S., Dimkpa, C. O., Angle, S. & Rabbinge, R. Unlocking the multiple public good services from balanced fertilizers. Food Secur. 10, 273–285 (2018).
Tyczewska, A., Woźniak, E., Gracz, J., Kuczyński, J. & Twardowski, T. Towards food security: current state and future prospects of agrobiotechnology. Trends Biotechnol. 36, 1219–1229 (2018).
Fincheira, P. et al. Nanotechnology advances for sustainable agriculture: current knowledge and prospects in plant growth modulation and nutrition. Planta 254, 1–25 (2021).
Liu, R. & Lal, R. Potentials of engineered nanoparticles as fertilizers for increasing agronomic productions. Sci. Tot. Environ. 514, 131–139 (2015).
Duhan, J. S. et al. Nanotechnology: the new perspective in precision agriculture. Biotechnol. Rep. 15, 11–23 (2017).
Derosa, M. C., Monreal, C., Schnitzer, M., Walsh, R. & Sultan, Y. Nanotechnology in fertilizers. Nat. Nanotechnol. 5, 91 (2010).
Ciurli, A. et al. A novel FePO4 nanosized fertilizer is as effective as triple superphosphate in sustaining the growth of cucumber plants. Pedosphere https://doi.org/10.1016/j.pedsph.2023.12.005 (2023).
Monreal, C. M., Derosa, M., Mallubhotla, S. C., Bindraban, P. S. & Dimkpa, C. Nanotechnologies for increasing the crop use efficiency of fertilizer-micronutrients. Biol. Fertil. Soils. 52, 423–437 (2016).
Shen, M., Liu, S., Jiang, C., Zhang, T. & Chen, W. Recent advances in stimuli-response mechanisms of nano-enabled controlled-release fertilizers and pesticides. Eco Environ. Health. 2, 161–175 (2023).
Marchiol, L., Iafisco, M., Fellet, G. & Adamiano, A. Chapter Two—Nanotechnology support the next agricultural revolution: perspectives to enhancement of nutrient use efficiency. in (ed. Sparks, D. L. B. T.-A. In A.) vol. 161, 27–116 (Academic, 2020).
Hawkesford, M. et al. Chapter 6—Functions of Macronutrients. in (ed. Marschner, P. B. T.-M. M. N. of H. P. (Third E.) 135–189 (Academic Press, San Diego, 2012).
Syers, J. K., Johnston, A. E. & Curtin, D. Efficiency of soil and fertilizer phosphorus use. Food Agric. Organ. United Nations 9, 27–44 (2008).
Jiang, C., Gao, X., Liao, L., Harberd, N. P. & Fu, X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant. Physiol. 145, 1460–1470 (2007).
Carstensen, A. et al. The impacts of phosphorus deficiency on the photosynthetic electron transport chain. Plant. Physiol. 177, 271–284 (2018).
Chiou, T. J. & Lin, S. I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant. Biol. 62, 185–206 (2011).
Broadley, M., Brown, P., Cakmak, I., Rengel, Z. & Zhao, F. Chapter 7—Function of Nutrients: Micronutrients. in (ed. Marschner, P. B. T.-M. M. N. of H. P. (Third E.) 191–248 (Academic Press, San Diego, 2012).
Mori, S. Iron acquisition by plants. Curr. Opin. Plant. Biol. 2, 250–253 (1999).
Kobayashi, T. & Nishizawa, N. K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant. Biol. 63, 131–152 (2012).
García-Sánchez, S., Bernales, I. & Cristobal, S. Early response to nanoparticles in the Arabidopsis transcriptome compromises plant defence and root-hair development through Salicylic acid signalling. BMC Genom. 16, 341 (2015).
Van Aken, B. Gene expression changes in plants and microorganisms exposed to nanomaterials. Curr. Opin. Biotechnol. 33, 206–219 (2015).
Khodakovskaya, M. V. et al. Complex genetic, photothermal, and photoacoustic analysis of nanoparticle-plant interactions. Proc. Natl. Acad. Sci. USA 108, 1028–1033 (2011).
Chun, S. C. & Chandrasekaran, M. Chitosan and Chitosan nanoparticles induced expression of pathogenesis-related proteins genes enhances biotic stress tolerance in tomato. Int. J. Biol. Macromol. 125, 948–954 (2019).
Landa, P. et al. Nanoparticle-specific changes in Arabidopsis Thaliana gene expression after exposure to ZnO, TiO2, and fullerene soot. J. Hazard. Mater. 241–242, 55–62 (2012).
Kaveh, R. et al. Changes in Arabidopsis Thaliana gene expression in response to silver nanoparticles and silver ions. Environ. Sc I Technol. 47, 10637–10644 (2013).
Garza-Alonso, C. A. et al. ZnO nanoparticles as potential fertilizer and biostimulant for lettuce. Heliyon 9, e12787 (2023).
Sotoodehnia-Korani, S., Iranbakhsh, A., Ebadi, M., Majd, A. & Oraghi-Ardebili, Z. Efficacy of magnesium nanoparticles in modifying growth, antioxidant activity, nitrogen status, and expression of WRKY1 and bZIP transcription factors in pepper (Capsicum annuum); an in vitro biological assessment. R. J. Plant. Physiol. 70, 39 (2023).
Yang, M., Dong, C. & Shi, Y. Nano fertilizer synergist effects on nitrogen utilization and related gene expression in wheat. BMC Plant. Biol. 23, 26 (2023).
Sega, D. et al. FePO4 nanoparticles produced by an industrially scalable continuous-flow method are an available form of P and Fe for cucumber and maize plants. Sci. Rep. 9, 11252 (2019).
Sega, D., Baldan, B., Zamboni, A. & Varanini, Z. FePO4 NPs are an efficient nutritional source for plants: combination of nano-material properties and metabolic responses to nutritional deficiencies. Front. Plant. Sci. 11, 586470 (2020).
Po-Wen, C., Singh, P. & Zimmerli, L. Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant. Pathol. 14, 58–70 (2013).
Li, Z. et al. Low temperature enhances plant immunity via Salicylic acid pathway genes that are repressed by ethylene. Plant. Physiol. 182, 626–639 (2020).
Smith, S. J. et al. Isolation of Arabidopsis extracellular ATP binding proteins by affinity proteomics and identification of PHOSPHOLIPASE C-LIKE 1 as an extracellular protein essential for Fumonisin B1 toxicity. Plant. J. 106, 1387–1400 (2021).
Lemonnier, P. et al. Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant. Mol. Biol. 85, 473–484 (2014).
Pingarron-Cardenas, G. et al. Selective herbicide safening in dicot plants: a case study in Arabidopsis. Front. Plant. Sci. 14, 1335764 (2024).
Zhao, Y. et al. Chemical genetic interrogation of natural variation uncovers a molecule that is glycoactivated. Nat. Chem. Biol. 3, 716–721 (2007).
Somssich, M., Khan, G. A. & Persson, S. Cell wall heterogeneity in root development of Arabidopsis. Front. Plant. Sci. 7, 1242 (2016).
Coculo, D. & Lionetti, V. The plant invertase/pectin methylesterase inhibitor superfamily. Front. Plant. Sci. 13, 863892 (2022).
Liu, J., Liu, Y., Wang, S., Cui, Y. & Yan, D. Heat stress reduces root meristem size via induction of plasmodesmal Callose accumulation inhibiting phloem unloading in Arabidopsis. Int. J. Mol. Sci. 23, 2063 (2022).
Ma, Z., Bielenberg, D. G., Brown, K. M. & Lynch, J. P. Regulation of root hair density by phosphorus availability in Arabidopsis Thaliana. Plant. Cell. Environ. 24, 459–467 (2001).
Bhosale, R. et al. A mechanistic framework for auxin dependent Arabidopsis root hair elongation to low external phosphate. Nat. Commun. 9, 1409 (2018).
Mangano, S., Denita-Juarez, S. P., Marzol, E., Borassi, C. & Estevez, J. M. High auxin and high phosphate impact on RSL2 expression and ROS-Homeostasis linked to root hair growth in Arabidopsis Thaliana. Front. Plant. Sci. 9, 1164 (2018).
Rono, J. K., Sun, D., Yang, Z. M. & Metallochaperones A critical regulator of metal homeostasis and beyond. Gene 822, 146352 (2022).
Klatte, M. et al. The analysis of Arabidopsis Nicotianamine synthase mutants reveals functions for Nicotianamine in seed iron loading and iron deficiency responses. Plant. Physiol. 150, 257–271 (2009).
Koen, E. et al. Arabidopsis Thaliana Nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant. Sci. 209, 1–11 (2013).
Manzano, C. et al. The emerging role of reactive oxygen species signaling during lateral root development. Plant. Physiol. 165, 1105–1119 (2014).
Sun, A. et al. Feedback regulation of auxin signaling through the transcription of H2A.Z and deposition of H2A.Z to SMALL AUXIN UP RNAs in Arabidopsis. New. Phytol. 236, 1721–1733 (2022).
Chapman, J. M. & Muday, G. K. Flavonols modulate lateral root emergence by scavenging reactive oxygen species in Arabidopsis Thaliana. J. Biol. Chem. 296, 100222 (2021).
Ma, W., Ali, R. & Berkowitz, G. A. Characterization of plant phenotypes associated with loss-of-function of AtCNGC1, a plant Cyclic nucleotide gated cation channel. Plant. Physiol. Biochem. 44, 494–505 (2006).
Ventouris, Y., Protopappa, S. T., Nikolopoulou, A. E., Bouranis, D. & Chorianopoulou, S. Ectopic lateral root branching in Fe-Deprived maize plants: searching for the genes underpinning the phenotype. Biol. Life Sci. Forum. 4, 7 (2021).
Dixon, D. P., Lapthorn, A. & Edwards, R. Plant glutathione transferases. Genome Biol. 3, reviews3004.1 (2002).
Klińska-Bąchor, S., Kędzierska, S., Demski, K. & Banaś, A. Phospholipid:diacylglycerol acyltransferase1-overexpression stimulates lipid turnover, oil production and fitness in cold-grown plants. BMC Plant. Biol. 23, 370 (2023).
Clifton, R. et al. Stress-induced co-expression of alternative respiratory chain components in Arabidopsis Thaliana. Plant. Mol. Biol. 58, 193–212 (2005).
Wen, J., Lease, K. A. & Walker, J. C. DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant. J. 37, 668–677 (2004).
Lin, D. et al. A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22, 1319–1325 (2012).
Laskowski, M. et al. Root system architecture from coupling cell shape to auxin transport. PLoS Biol. 6, e307 (2008).
Manabe, Y. et al. The Arabidopsis kinase-associated protein phosphatase regulates adaptation to Na+ stress. Plant. Physiol. 146, 323–324 (2008).
Sun, L. & van Nocker, S. Analysis of promoter activity of members of the pectate lyase-like (PLL) gene family in cell separation in Arabidopsis. BMC Plant. Biol. 10, 152 (2010).
Berken, A., Thomas, C. & Wittinghofer, A. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature 436, 1176–1180 (2005).
Feiguelman, G., Fu, Y. & Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant. Physiol. 176, 57–79 (2018).
Qian, D. et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 70, 435–446 (2019).
Yang, B. et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 14, e1007839 (2018).
Wimalasekera, R. et al. Polyamine oxidase2 of Arabidopsis contributes to ABA mediated plant developmental processes. Plant. Physiol. Biochem. 96, 231–240 (2015).
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E. & Yin, Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant. Cell. 32, 295–318 (2020).
Poppenberger, B. et al. The UGT73C5 of Arabidopsis Thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA. 102, 15253–15258 (2005).
Bao, F. et al. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant. Physiol. 134, 1624–1631 (2004).
Tripathi, D., Singh, M. & Pandey-Rai, S. Crosstalk of nanoparticles and phytohormones regulate plant growth and metabolism under abiotic and biotic stress. Plant. Stress. 6, 100107 (2022).
Santi, C., Zamboni, A., Varanini, Z. & Pandolfini, T. Growth stimulatory effects and genome-wide transcriptional changes produced by protein hydrolysates in maize seedlings. Front. Plant. Sci. 8, 433 (2017).
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J. & Gao, G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113 (2020).
Tang, D. et al. SRplot: a free online platform for data visualization and graphing. PLoS One. 18, e0294236 (2023).
Simon, P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440 (2003).
Ruijter, J. M. et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45 (2009).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Acknowledgements
The work was funded by Joint Project 2016 and Joint Project 2018 developed by the University of Verona and Fabbrica Cooperativa Perfosfati Cerea.
Author information
Authors and Affiliations
Contributions
A. Z. and Z.V.: conceived and designed the experiments. A.C.: acquired the data. A.C. and A.Z.: analysed the data. All the authors interpreted the data. A.C. and A.Z.: wrote the original draft. A. Z.: revised the draft. Z.V.: supervised the work and provided funding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ciurli, A., Zamboni, A. & Varanini, Z. Early transcriptomic changes in cucumber and maize roots in response to FePO4 nanoparticles as a source of P and Fe. Sci Rep 15, 11786 (2025). https://doi.org/10.1038/s41598-025-95989-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-95989-6