Abstract
Abelmoschus Manihot is an important medicinal and edible plant known for its functional secondary metabolites. However, little is known about the key genes involved in production of secondary metabolites in A. manihot. This is largely due to the lack of effective gene expression detection systems for A. manihot, making the screening of real-time PCR reference genes a prerequisite. In this study, 11 candidate reference genes were screened and cloned from A. manihot, and their expression stability was evaluated in different tissues under different flowering stages using four algorithms: geNorm, NormFinder, BestKeeper, and RefFinder. The expression stability of eIF and PP2A1 was the highest, while that of tubulin alpha (TUA) was the lowest. The combined use of the two most stable reference genes, eIF and PP2A1, met the experimental requirements for normalizing gene expression in A. manihot. Furthermore, the gene expression of transcription factors bHLH147 and bHLH148 was further validated by data normalization. This study identified potential reference genes in different A. manihot tissues, paving the way for functional gene analysis and dissecting metabolite regulation mechanisms in A. manihot.
Similar content being viewed by others
Introduction
Abelmoschus Manihot is an annual or perennial herbal plant of the Malvaceae family and is widely distributed in China, India, Sri Lanka Card, and Australia1. Abelmoschus Manihot has high nutritional and medicinal value and is a popular green leafy vegetable in the South Pacific Islands2. Moreover, this plant is listed in the ‘Chinese Pharmacopoeia’ because its flowers are traditionally used for medicinal purposes due to their physiological capacities to reduce swelling, detoxify, and clear heat and dampness. Consequently, this plant is also consumed as functional foods and floral teas. It has been demonstrated that A. manihot contains various bioactive compounds, such as flavonoids, polysaccharides, phenolic compounds, vitamin E, organic acids, nucleosides, and amino acids3, exhibiting pharmacological activities, such as antitumor4 and neuroprotection activities5. It is extensively cultivated as an important medicinal plant for treating inflammation, primary glomerulonephritis6, and type 2 diabetic nephropathy7.
Recent research on A. manihot was mainly focused on the isolation, characterization, and pharmacological examination of its bioactive components8,9,10. However, due to a lack of relevant systems and platforms, molecular biology studies on A. manihot are scarce, and the regulatory mechanisms of nutritional component synthesis are not well understood. Gene expression analysis is one primary method for characterizing regulatory mechanisms in plants, and thus establishing a gene expression detection system is of great significance to elucidate the molecular mechanisms underlying the biosynthesis of these natural compounds.
Quantitative real-time PCR (qPCR) is one main molecular technique for gene expression analysis and is known for its high precision, accuracy, and repeatability11,12,13,14. Reference gene selection is essential for reliable qRT-PCR results as a suitable reference gene is needed to normalize the expression level of the gene of interest. The ideal reference gene should demonstrate consistent expression levels across diverse plant tissues and developmental stages15. Housekeeping genes are frequently used as reference genes. However, a growing body of evidence has shown that the transcripts of these reference genes are not always equally abundant under different experimental conditions16,17,18. Unstably expressed reference genes are incapable of accurately calibrating the expression level of the target gene and may precipitate erroneous interpretations of experimental results19. To date, no reference genes are available for transcript normalization in A. manihot.
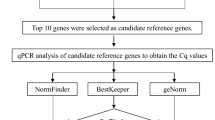
This study was performed to identify the most suitable reference genes in A. manihot for use with different tissues and developmental stages. Eleven candidate reference genes (eIF, GTP, PP2A1, EIF4α, PP2A2, ACT2, PP2A3, vps, PHD, YLS, and TUA) were screened from an A. manihot transcriptome20. The basis for the screening was to select candidate gene families based on previously reviewed literature on the same genus and to select genes with stable changes in fragments per kilobase of exon model per million mapped fragments (FPKM) values between different tissues; these were compared with housekeeping genes using blast. Four statistical algorithms, namely, geNorm, NormFinder, BestKeeper, and RefFinder, were used to evaluate the accuracy and stability of these candidate internal reference genes for qPCR analysis. These four algorithms have been widely used in several internal reference gene screening studies in recent years, and their accuracy has been verified. Expression levels of two key genes, bHLH147 and bHLH148, were examined to verify the applicability and feasibility of the selected reference genes. The bHLH transcription factor family comprises MBW complexes21 involved in the transcriptional regulation of flavonoid metabolism in plants. bHLH147 and bHLH148 are key regulatory genes in the flavonoid biosynthetic pathway, and they were identified in a previous joint metabolomics and transcriptomics analysis of A. manihot20. The results of this study may serve as a foundation for further research on key genes of flavonoid synthesis and related metabolic regulation mechanisms in A. manihot.
Results
Primer specificity and amplification efficiency of reference genes in A. manihot
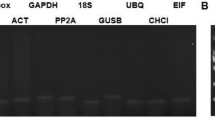
A total of 11 candidate genes were tested to examine whether they could be used as reference genes. Gene-specific primers were designed for these candidates (Table 1). The specificity and application efficiency of the primers were verified using agarose gel electrophoresis (1.8% w/v) and melt curve analysis. An amplification product with the expected molecular weight (92–234 bp) in each primer-specific lane (Fig. 1). Melt curve analysis demonstrated that all primers exhibited specific amplification, with only one unique peak on the melting curve (Figure S1). Additionally, standard curve analysis showed that the amplification efficiencies (E) of the 11 candidate genes were greater than 90% (92.4–101.6%), with a regression coefficient (R2) spanning from 0.983 (eIF) to 0.999 (ACT2 and PP2A3) that met the general criteria for reference genes.
Expression profile of reference genes in A. manihot
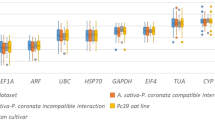
The threshold cycle (Ct) value directly reflects the approximate expression of the reference gene. The average Ct values of the 11 candidate genes ranged from 19.59 (TUA) to 28.14 (PP2A3), with most values falling between 15 and 35 (Fig. 2). Ct values were negatively correlated with gene expression, where lower Ct values indicated higher gene expression levels. The expression profiles of 11 genes varied significantly among the samples. Among these genes, the average Ct values for PP2A1, PP2A2, PHD, and GTP were notably less variable, exhibiting a tendency to cluster around 25. The average Ct values for ACT2, EIF4α, and eIF were lower (20.0–23.6), indicating higher expression levels. Furthermore, TUA displayed the most considerable variation in average Ct values among the tissue samples, with a range from 20.5 in stems to 28.8 in flowers. Among the genes under consideration, ACT2 exhibited the lowest average Ct, indicative of the greatest expression level, whereas PP2A3 displayed the highest average Ct, indicative of the lowest expression in A. manihot.
Stability analysis by geNorm
geNorm software was used to analyze the expression stability of candidate genes of A. manihot by calculating the average expression stability (M) value. Reference genes with an M value below 1.5 can be employed for expression analysis, with lower M values indicating higher expression stability. The average M values of the 11 reference genes in the composite sample are shown in Fig. 3. The rank of reference genes by geNorm was not consistent among different tissues (Tables S1 and S2). In the root, PP2A1 and vps showed the greatest stability (M = 0.07), while eIF exhibited the least stability (M = 0.48). In the stem, ACT2 and YLS exhibited the highest stability (M = 0.08), whereas PP2A3 exhibited the lowest (M = 0.75). In the leaves, ACT2 and TUA (M = 0.03) showed the highest stability, while YLS (M = 0.79) displayed the lowest. In the flower, eIF and PHD (M = 0.11) had the highest stability, and YLS (M: 1.01) showed the lowest. In the fruit samples, PP2A2 and vps (M = 0.10) were the most stable, while TUA (M = 1.16) was the least stable. Furthermore, in the flower bud samples, ACT2 and EIF4α (M = 0.09) displayed the greatest stability, while PP2A3 (M: 0.87) exhibited the lowest. In general, PP2A1 and PP2A2 (M = 0.58) exhibited the greatest stability, while TUA (M = 1.56) showed the lowest.
Pairwise variation (V) obtained from geNorm analysis (Vn/Vn + 1) was employed to evaluate the minimum number of reference genes to obtain reliable normalization in gene expression analysis. In general, a Vn/Vn + 1 value below 0.15 indicates that the use of n reference genes is sufficient to normalize relative expression levels obtained by qRT-PCR data. As shown in Fig. 4, the V2/V3 values observed among tissue samples were below 0.15, indicating that the combined use of the two most stable candidate genes in each tissue represents an optimal practice for gene expression analysis. However, when considering mixed samples, the paired variance value was above the threshold of 0.15 for V2/V3 (0.21) and below it for V3/V4 (0.14), suggesting that the use of three reference genes for normalization is recommended in mixed tissue samples.
Expression stability analysis by NormFinder
The NormFinder algorithm was used to calculate intra- and inter-group variations and to obtain stability values (SV) for the reference genes in different tissue samples. The candidate gene with the smallest SV was likely to have the most stable expression. Unlike the geNorm analyses, the stability rankings of PP2A1 and EIF4α were both in the top three, indicating their high stability, while YLS and TUA were at the bottom, indicating their poor stability.
Expression stability analysis by BestKeeper
BestKeeper analyzes the expression stability of candidate genes by calculating the standard deviation (SD) and coefficient of variation (CV). Gene expression stability is negatively correlated with SD and CV values. In general, genes with an SD value less than 1 can be used as reference genes. The CV value indicates the degree of dispersion of the data. In circumstances where the SD values are relatively similar, a lower CV value indicates more reliable data. As shown in Table 2, for the mixed sample group, PP2A1 had an SD of 0.42, indicating that its expression was the most stable one, while YLS and TUA, with SD values exceeding 1, were deemed unstable and therefore subsequently excluded. Consistent with the NormFinder results, PP2A1, eIF, and EIF4α demonstrated relatively good stability, while YLS and TUA had relatively poor stability.
Expression stability analysis by RefFinder
Due to the inherent differences between the algorithms described above, the sorting results were not entirely consistent. In most cases, several highly stable genes were at the top but their rankings differed. Therefore, we used RefFinder for further analysis. This tool combines several of the algorithms mentioned above to provide a more comprehensive analysis and a more credible stability ranking (Table 2, S1, and S2). RefFinder analysis indicated that in the mixed sample group, eIF and PP2A1 were ranked at the top, suggesting that they the two most suitable reference genes, while YLS and TUA were the least stable reference genes for mixed tissues. Based on the fact that the top-ranked genes in terms of stability varied among tissues, and the most appropriate combination of reference genes should be selected based on a comprehensive analysis to ensure experimental accuracy.
Stability verification
The feasibility of using these reference genes was verified based on geNorm, NormFinder, BestKeeper, and RefFinder analyses. We achieved this by examining the expression levels of two target genes (bHLH147 and bHLH148) in A. manihot at different flower development stages. Given that both eIF and PP2A1 ranked high in the mixed sample rankings of each of the aforementioned software and are ranked second for stability in the comprehensive analysis (RefFinder), these two genes were considered to be genes with the highest versatility and the best stability in A. manihot. Therefore these two reference genes, as well as their combination (eIF + PP2A1), were the optimal choice for use as the normalization factor. This enabled the detection of the relative expression patterns of bHLH147 and bHLH148 at different flower development stages. As shown in Fig. 5, qRT-PCR and transcriptome analysis showed consistent expression patterns for bHLH147 and bHLH148 in A. manihot flowers at four developmental stages (Fig. 6) after normalization with eIF, PP2A1, and eIF + PP2A1. The expression levels of bHLH147 and bHLH148 were lowest at the bud stage and continued to increase from the bud to the bloom stage, followed by a decreasing trend at the withering stage. Statistical analysis (Tukey’s post hoc test) for normalization of the two target genes’ relative expression with different reference genes or combinations is shown in Table S3 and Table S4. In addition, correlation analysis was also performed on the expression levels of the two target genes at each stage, together with their FPKM values (Fig. 7). The analysis demonstrated strong correlations when using the stable reference genes eIF, PP2A1, and eIF + PP2A1 as normalization factors (bHLH147: 0.79–0.96; bHLH148: 0.71–0.87), while normalization with the least stable reference gene, TUA, resulted in a poor correlation (bHLH147: 0.31; bHLH148: 0.35), which did not exceed 0.5. These results affirmed the stability of reference genes evaluated and recommended by algorithms such as RefFinder.
qRT-PCR results and transcriptome data (FPKM) of bHLH147 (A) and bHLH148 (B) at four developmental stages in Abelmoschus Manihot flowers. Columns indicate the FPKM values of the target genes at different stages of flower development, and different colored folds indicate the relative expression of the target genes normalized by different reference genes.
Discussion
As a medicinal and edible plant with a long history of cultivation in China, A. manihot is rich in nutrients and popular both in China and internationally. In particular, the abundant flavonoids and polysaccharides in A. manihot have been widely used in the treatment of chronic kidney disease22, and they also have antioxidant23 and anti-inflammatory applications24. In recent years, the discovery of its anti-aging and skincare effects has extended its industrial chain. The flavonoids in A. manihot have been widely studied in terms of their extraction process25 and pharmacological mechanisms26. In recent years, there is a trending demand for molecular studies on A. manihot. For example, Hou et al.27 analyzed the flavonoid synthesis mechanism in flowers through transcriptome and metabolome analysis, while Zhou et al.28 explored the changes and biosynthetic mechanisms of flavonoids in different plant organs in the Malvaceae family. Gene expression analysis is one key step for advancing research on the regulation of plant flavonoids. According to previous studies, flavonoids in Malvaceae plants function as nutritional products. Additionally, flavonoids are involved in the regulation of plant growth and development, resistance to pests and diseases, and reproductive growth. Some suggest that flavonoids are important in the growth cycle of mallow plants and are therefore targets for exploration.
The accuracy and reliability of qRT-PCR, the most widely used technique for gene expression analysis, is greatly influenced by the selected reference gene. However, the expression levels of the same reference genes vary under different experimental conditions and in different plant species29,30. To our knowledge, no systematic studies of qRT-PCR reference genes for A. manihot have been performed to date.
Here we ranked the stability of 11 A. manihot candidate genes (eIF, GTP, PP2A1, EIF4α, PP2A2, ACT2, PP2A3, vps, PHD, YLS, and TUA) using the geNorm, NormFinder, BestKeeper, and RefFinder algorithms. We found slight differences in the results obtained by these programs. The difference in results is likely because these programs used different algorithmic formulas and produced distinct results, which can reflect the stability of the reference genes. Also, because of the mutual independence of the algorithms, it was difficult to determine the obvious advantages and disadvantages of the algorithms. Therefore, when we analyzed the results, we attempted to summarize a reasonable and accurate explanation for reference. Based on these considerations, we used RefFinder, integrating other calculation algorithms to offset biases, and obtained a more reliable, comprehensive ranking of gene expression stability.
Considering all these results, we concluded that eIF and PP2A1 exhibited the highest degree of stability and can be sued as reference genes for A. manihot. eIF is stably expressed in different tissues of okra under various abiotic stressors31. PP2A1, a protein phosphatase 2 A, also exhibited good stability in the mixed and different tissue samples of A. manihot. Previous studies have shown that PP2A can be stably expressed in some species, such as tobacco32, Siberian wild rye33, Suaeda glauca34, and Oenanthe javanica (BI.), under abiotic stress35. In contrast, although TUA exhibited highly stable expression in different soybean tissues36 and YLS exhibited decent expression in Aegilops tauschii (Coss.) under cold stress37, they did not perform well in A. manihot samples.
Based on the screened reference genes, there are many possibilities for future molecular research in A. manihot. We can use the screened internal reference genes for qPCR experiments to further clarify the synthesis mechanism of various metabolite pathways in A. manihot, such as polysaccharides, polyphenols, and flavonoids, which are rich in A. manihot. Flavonoids and polysaccharide compounds are extensively used in medicine and in more healthcare products. The development in the beauty industry in recent years has demonstrated that A. manihot has great potential in the KangYang (health + old age) market. The current problem of large differences in metabolite content under different growing conditions in A. manihot can also be addressed by further mapping its mechanism through molecular studies. In addition, based on the phenomenon of the short flowering time of Malvaceae plants, molecular mechanisms can also be studied to enhance the ornamental value of marshmallow as an ornamental flower and further improve its social benefits. However, although in some parts of the world A. manihot is consumed daily, in China, its daily application and popularity are very low. The promotion of okra is an example of vegetable factorization of A. manihot, and the understanding of its synthesis mechanism of various types of nutrients will also help in its promotion. In China, A. manihot is primarily being cultivated in the north. In southern China, water and fertilizer management can hypothetically achieve flowering three to four times or more per year. Thus, A. manihot can be combined with low temperature, drought stress, and other related experiments for research, which will help factory nursery development.
Conclusions
In summary, this study screened 11 candidate reference genes for A. manihot and normalized the gene relative expression data in different tissues at different flowering stages using qRT-PCR. The normalization results were analyzed using different algorithms, namely geNorm, NormFinder, BestKeeper, and RefFinder, to identify the most appropriate reference genes and combinations. We found that using eIF or PP2A1 alone or a combination of these two stable reference genes, depending on the sample, improved the accuracy of qRT-PCR gene expression analysis. These findings will facilitate the quantitative expression analysis of key genes involved in flavonoid biosynthesis in A. manihot, providing a foundation for elucidating the regulatory mechanisms of the flavonoid biosynthesis pathway in plants.
Materials and methods
Plant materials
Healthy mature A. manihot seeds were sown in the experimental field of South China Agricultural University (113°37’E, 23°16’N) in Guangzhou, China. The seedlings were grown to maturity during the summer season. Different tissue samples, namely, roots, stems, and leaves, were collected from 4-week-old plants, and the flower buds and flowers were collected at three developmental stages (pre-bloom, bloom, and withering) (Fig. 6) from 13–16-week-old plants. All experimental samples were collected from three or more biological replicates. Tissue samples were frozen in liquid nitrogen and stored at - 80 ℃ immediately after collection.
RNA extraction and cDNA synthesis
Total RNA was extracted from each tissue sample using a Fast Universal Plant RNA Extraction Kit (Huayueyang, Beijing, China) according to the manufacturer’s instructions. RNA samples were treated with RNase-free DNase I (Takara Bio Inc., Kyoto, Japan) to remove residual genomic DNA (gDNA). The quality and concentration of the extracted RNA were confirmed using a nanophotometer (Implen, Munich, Germany). Integrity was assessed by electrophoresis on 1% (w/v) agarose gel. First-strand cDNA was synthesized using the HiScript® III RT SuperMix for qPCR Kit (Vazyme, Nanjing, China) following the manufacturer’s instructions. Subsequently, the synthesized cDNA was diluted (1:5) for subsequent qRT-PCR analysis.
Reference gene selection and primer design
Eleven reference genes were selected using the coefficient of variation (CV) method38. Gene expression levels were quantified as transcripts per million (TPM), with biological replic ates averaged for analysis. Genes with low expression (log2(TPM) ≤ 5) were excluded to minimize quantification bias. The CV was calculated as CV = standard deviation (SD) of log2(TPM) / average log2(TPM) (mean)39, where SD and mean values were computed in Microsoft Excel. Candidate genes were required to meet stability thresholds (SD < 1, CV ≤ 0.2) defined as the cutoff for consistency of expression across tissues. Based on these criteria, and in combination with a homology-based screening strategy for reference gene families within the same genus as reported in the literature, 11 candidate reference genes (eIF, GTP, PP2A1, EIF4α, PP2A2, ACT2, PP2A3, vps, PHD, YLS, and TUA) were identified from the transcriptome data. The selection process involved evaluating the stability of FPKM (fragments per kilobase of exon model per million mapped reads) values across different tissues and validating the conservation of candidate genes against housekeeping genes using BLAST31. All specific primers of the candidate genes (Table 1) for qRT-PCR were designed using Primer Premier 5.0 software, in accordance with conventional qRT-PCR primer parameters40.
qRT‑PCR analysis
qRT-PCR was performed using the LightCycler® 480 Real-Time PCR System (Roche, Los Angeles, CA, USA). According to the manufacturer’s instructions, the reaction mixture was composed of 1 µL of 5-times diluted cDNA, 10 µL of 2×ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China), 0.4 µL of each 10 µM forward and reverse primers, and 8.2 µL of ddH2O. The PCR reaction procedure was as follows: 95 ℃ for 30 s; 40 cycles of 95 ℃ for 3 s, 60 ℃ for 10 s; a final step at 95 ℃ for 15 s; and melting curve analysis. Three biological and three technical replicates were carried out for each qRT-PCR reaction. Primer specificity was analyzed using the melting curves and agarose gel electrophoresis. The amplification efficiency and correlation coefficient were determined through standard curve calculations41.
Analysis of the expression stability of candidate reference genes
Four programs, namely geNorm (https://genorm.cmgg.be/)42, NormFinder (https://moma.dk/normfinder-software)43, BestKeeper (https://www.gene-quantification.de/bestkeeper.html)44, and RefFinder (https://blooge.cn/RefFinder/? type = reference)45, we-re used to evaluate the expression stability of candidate reference genes in dif-ferent A. manihot tissue samples.
Both geNorm and NormFinder analysis were employed to evaluate the average Ct values and 2-ΔCt (ΔCt = Ct value of each sample − lowest Ct value) to assess the expression stability. In the geNorm algorithm, a lower M value signifies greater gene expression stability. In addition, geNorm provides the optimal number of reference genes for normalization by calculating the Vn/Vn + 1 value. NormFinder is based on analysis of variance and determines the SV of each reference gene by calculating the within- and between-group variance of the data. SV is negatively correlated with gene stability. BestKeeper evaluates gene stability by calculating the CV and SD, with genes with an SD of more than 1 considered unstable. RefFinder combines many of these algorithms with delta-Ct for comprehensive ranking analysis.
Validation of candidate reference genes by qRT-PCR analysis of target genes’ expression
bHLH147 and bHLH148 belong to the MYB-bHLH-WD40 (MBW) family46, which are crucial transcription factors that regulate flavonoid biosynthesis in plants. To determine the relative expression of bHLH147 and bHLH148, we performed qRT-PCR at four flower development stages in A. manihot. Data were normalized using the most and least stable reference genes identified by the four algorithms described above. The results were then analyzed using the 2−ΔΔCT method47 for relative quantification and one-way ANOVA followed by Tukey’s post-hoc tests (IBM SPSS Statistics 27.0.1) to assess the significance of the results48.
Data availability
The raw sequence data are deposited in the Genome Sequence Archive in the National Genomics Data Center, Chinese Academy of Sciences, under accession number CRA023777 which is publicly accessible at https://ngdc.cncb.ac.cn/gsa/s/GDdAx7sM. The datasets used and analysed during the current study available from the corresponding author on reasonable request.
References
Yang, Z. et al. Enrichment and purification of the bioactive flavonoids from flower of Abelmoschus Manihot (L.) Medic Using Macroporous Resins. Molecules 23 (2018).
Rubiang-Yalambing, L., Arcot, J., Greenfield, H. & Holford, P. Aibika (Abelmoschus manihot L.): genetic variation, morphology and relationships to micronutrient composition. Food Chem. 193, 62–68 (2016).
Du, L. et al. Comparative characterization of nucleotides, nucleosides and nucleobases in Abelmoschus manihot roots, stems, leaves and flowers during different growth periods by UPLC-TQ-MS/MS. J. Chromatogr. B. 1006, 130–137 (2015).
Hosseini, F. et al. Anti-Inflammatory and Anti-Tumor effects of Α-L-Guluronic acid (G2013) on cancer-Related inflammation in a murine breast cancer model. Biomed. Pharmacother. 98, 793–800 (2018).
Heitman, E. & Ingram, D. K. Cognitive and neuroprotective effects of chlorogenic acid. Nutr. Neurosci. 20, 32–39 (2017).
Yin, S. et al. Comparison of multiple bioactive constituents in the corolla and other parts of Abelmoschus manihot Molecules 26, (2021).
Chen, Y. Z. et al. Efficacy and safety of Flos Abelmoschus manihot (Malvaceae) on type 2 diabetic nephropathy: A systematic review. Chin. J. Integr. Med. 21, 464–472 (2015).
Wei, C. et al. The Pharmacological mechanism of Abelmoschus manihot in the treatment of chronic kidney disease. Heliyon 9, e22017 (2023).
Yuan, D. et al. In-Depth Insight Into the Mechanism of Incorporation of Abelmoschus Manihot Gum On the Enhancement of Gel Properties and in Vitro Digestibility of Frankfurters. Foods 12 (2023).
Guo, J. et al. Fast and automated characterization of major constituents in rat biofluid after oral administration of Abelmoschus manihot extract using Ultra-Performance liquid chromatography/quadrupole Time-of-Flight mass spectrometry and metabolynx. Rapid Commun. Mass. Spectrom. 24, 443–453 (2010).
Guo, W. et al. Selection and validation of reference genes for gene expression studies in Euonymus japonicus based on RNA sequencing. Genes 15, (2024).
Ma, Q., Zhuang, Z. M., Feng, W. R., Liu, S. F. & Tang, Q. S. Evaluation of reference genes for quantitative Real-Time PCR analysis of gene expression during early development processes of the tongue sole (Cynoglossus Semilaevis). Acta Oceanol. Sin 34, 90–97 (2015).
Qu, R. et al. Selection of reference genes for the quantitative Real-Time PCR normalization of gene expression in Isatis Indigotica fortune. BMC Mol. Biol. 20, (2019).
Wang, J., Han, S., Yin, W., Xia, X. & Liu, C. Comparison of reliable reference genes following different hormone treatments by various algorithms for QRT-PCR analysis of metasequoia. Int. J. Mol. Sci. 20, (2019).
Tavares, L. S. et al. Reference genes for gene expression profiling in mouse models of Listeria monocytogenes infection. Biotechniques 76, 104–113 (2024).
Gimeno, J., Eattock, N., Van Deynze, A. & Blumwald, E. Selection and validation of reference genes for gene expression analysis in Switchgrass (Panicum Virgatum) using quantitative Real-Time RT-PCR. PLoS One 9, (2014).
Hossain, M. S., Ahmed, R., Haque, M. S., Alam, M. M. & Islam, M. S. Identification and validation of reference genes for Real-Time quantitative RT-PCR analysis in Jute BMC Mol. Biol. 20, (2019).
Hou, F. et al. Identification and validation of reference genes for quantitative Real-Time PCR studies in long yellow daylily, Hemerocallis citrina Borani PLoS One 12, (2017).
Nong, Q. et al. RNA-Seq-Based selection of reference genes for RT-QPCR analysis of Pitaya. FEBS Open. Bio. 9, 1403–1412 (2019).
Ye, G., Wang, Z., Liu, H. & Li, J. Medicinal and edible Abelmoschus Manihot: Sequencing and characteristic analysis of Full-Length transcriptome. Chin. Agric. Sci. Bull. 39, 29–35 (2023).
Jiang, W. et al. The APETALA2-MYBL2 module represses Proanthocyanidin biosynthesis by affecting formation of the MBW complex in seeds of Arabidopsis Thaliana. Plant. Commun. 5, (2024).
Yu, H. et al. Evaluation of the efficacy of Abelmoschus manihot (L.) on diabetic nephropathy by analyzing biomarkers in the glomeruli and proximal and distal convoluted tubules of the kidneys. Front. Pharmacol. 14, (2023).
Wang, K., Zhang, H., Yuan, L., Li, X. & Cai, Y. Potential implications of Hyperoside on oxidative Stress-Induced human diseases: A comprehensive review. J. Inflamm. Res. 16, 4503–4526 (2023).
Xue, C. et al. Total Flavone of Flowers of Abelmoschus Manihot (L.) medic inhibits the expression of adhesion molecules in primary mesenteric arterial endothelial cells and ameliorates dextran sodium sulphate-induced ulcerative colitis in Mice. Phytomedicine 112 (2023).
Chu, Q. et al. Enzyme-Assisted ultrasonic extraction of total flavonoids and extraction polysaccharides in residue from Abelmoschus manihot (L). Ultrason. Sonochem 104, (2024).
Cai, M., Xiang, Y., Li, Z., Xie, J. & Wen, F. Network Pharmacology and molecular Docking predictions of the active compounds and mechanism of action of Huangkui capsule for the treatment of idiopathic membranous nephropathy. Med. (Baltimore) 102 (2023).
Hou, J. et al. Transcriptome and metabolome analysis reveal the flavonoid biosynthesis mechanism of Abelmoschus manihot L. At different anthesis stages. Metabolites 13 (2023).
Zhou, Y. et al. Combined metabolome and transcriptome analyses reveal the flavonoids changes and biosynthesis mechanisms in different organs of Hibiseu manihot L. Front. Plant. Sci. 13 (2022).
Huang, T. et al. Selection and validation of reference genes for MRNA expression by quantitative Real-Time PCR analysis in Neolamarckia Cadamba Sci. Rep. 8 (2018).
Li, J. et al. Screening of Reference Genes in Real-Time PCR for Radopholus Similis. PeerJ 7 (2019).
Zhu, Z., Yu, J., Tang, X., Xiong, A. & Sun, M. Selection and Validation of Reference Genes in Different Tissues of Okra (Abelmoschus Esculentus L.) Under Different Abiotic Stresses. Genes 14 (2023).
Schmidt, G. W. & Delaney, S. K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana Tabacum) during development and abiotic stress. Mol. Genet. Genom. 283, 233–241 (2010).
Zhang, J. et al. Selection of suitable reference genes for RT-QPCR gene expression analysis in siberian wild rye (Elymus Sibiricus) under different experimental conditions. Genes 10 (2019).
Wang, M., Ren, T., Marowa, P., Du, H. & Xu, Z. Identification and selection of reference genes for gene expression analysis by quantitative real-time PCR in Suaeda Glauca’ S Response to Salinity. Sci. Rep. 11, (2021).
Jiang, Q. et al. Selection of suitable reference genes for QPCR normalization under abiotic stresses in Oenanthe Javanica (BI.) DC. PLoS One 9 (2014).
Jian, B. et al. Validation of internal control for gene expression study in Soybean by quantitative Real-Time PCR. BMC Mol. Biol. 9 (2008).
Abbas, A. et al. Selection and validation of reference genes for RT-QPCR analysis in Aegilops Tauschii (Coss.) under different abiotic stresses. Int. J. Mol. Sci. 22 (2021).
Li, Y. et al. Systematic identification and validation of the reference genes from 60 RNA-Seq libraries in the Scallop Mizuhopecten yessoensis. BMC Genom. 20, 288 (2019).
Chen, J. et al. Identification and validation of the reference genes in the Echiuran worm Urechis unicinctus based on transcriptome data. BMC Genom. 24, 248 (2023).
Zhang, P. et al. Selection and validation of QRT - PCR internal reference genes to study flower color formation in camellia Impressinervis Int. J. Mol. Sci. 25 (2024).
Liu, Y. et al. Identification and validation of reference genes for seashore paspalum response to abiotic stresses. Int. J. Mol. Sci. 18 (2017).
Vandesompele, J. et al. Accurate normalization of Real-Time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3 (2002).
Andersen, C. L., Jensen, J. L. & Orntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-Based tool using Pair-Wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Xie, F., Wang, J., Zhang, B. & RefFinder: A Web-Based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 23, (2023).
Wang, X. et al. Jasmonate mimic modulates cell elongation by regulating antagonistic BHLH transcription factors via brassinosteroid signaling. Plant. Physiol. (2024).
Pfaffl, M. W. A new mathematical model for relative quantification in Real-Time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Jin, Y., Liu, F., Huang, W., Sun, Q. & Huang, X. Identification of reliable reference genes for QRT-PCR in the ephemeral plant Arabidopsis pumila based on Full-Length transcriptome data. Sci. Rep. 9 (2019).
Acknowledgements
This study was supported by the Key Field Research and Development Project of Guangdong Province (2022B0202080002), the Natural Science Foundation of Guangdong Province (2023A1515010341). We thank LetPub (www.letpub.com.cn) for linguistic assistance and pre-submission expert review.
Author information
Authors and Affiliations
Contributions
Q.W. carried out the experiments and wrote the initial draft of the manuscript, M.D. participated in expression analysis, X.Z. provided scientific advice and evaluated the study protocol. J.L. analyzed the results. J.Z. supervised the project and revised the manuscript. All authors reviewed and approved the final draft of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Q., Deng, M., Zhao, X. et al. Screening and validation of optimal real-time PCR reference genes for Abelmoschus Manihot. Sci Rep 15, 11045 (2025). https://doi.org/10.1038/s41598-025-96110-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-96110-7