Abstract
Glycogen storage disease (GSD) is a group of rare inherited metabolic disorders characterized by abnormal glycogen storage and breakdown. These disorders are caused by mutations in G6PC1, which is essential for proper glucose storage and metabolism. With the advent of continuous glucose monitoring systems, development of algorithms to analyze and predict glucose levels has gained considerable attention, with the aim of preemptively managing fluctuations before they become problematic. However, there is a lack of research focusing specifically on patients with GSD. Therefore, this study aimed to forecast glucose levels in patients with GSD using state-of-the-art deep-learning (DL) algorithms. This retrospective study utilized blood glucose data from patients with GSD who were either hospitalized or managed at Yonsei University Wonju Severance Christian Hospital, Korea, between August 2020 and February 2024. In this study, three state-of-the-art DL models for time-series forecasting were employed: PatchTST, LTSF N-Linear, and TS Mixer. First, the models were used to predict the patients’ Glucose levels for the next hour. Second, a binary classification task was performed to assess whether hypoglycemia could be predicted alongside direct glucose levels. Consequently, this is the first study to demonstrate the capability of forecasting glucose levels in patients with GSD using continuous glucose-monitoring data and DL models. Our model provides patients with GSD with a more accessible tool for managing glucose levels. This study has a broader effect, potentially serving as a foundation for improving the care of patients with rare diseases using DL-based solutions.
Similar content being viewed by others
Introduction
Glycogen storage disease (GSD) is a rare inherited metabolic disorder characterized by aberrant glycogen storage and mobilization1,2,3,4. The types of GSD range from Type I (Von Gierke Disease) to Type XIV, each manifesting differently and requiring a nuanced management approach. Among these, type I GSD exhibits the most severe symptoms. This condition results from a mutation in G6PC1, which plays a crucial role in glucose storage and degradation. It affects approximately 1 in 20,000 to 40,000 individuals due to its autosomal recessive inheritance. This mutation leads to the inability of the body to consume large amounts of glycogen, resulting in its accumulation in the liver and kidneys. This accumulation causes glucose intolerance, leading to several complications including hypoglycemia, elevated lactic acid levels, and secondary metabolic abnormalities5. Affected individuals may experience growth retardation, osteoporosis, and pulmonary hypertension, with hypokinesis as a potential complication. Moreover, chronic liver and kidney damage can impair their function6. Patients with GSD are prone to severe hypoglycemia because of their inability to produce glucose owing to enzyme deficiency. Although proper dietary intake, including cornstarch, has been used to prevent hypoglycemia, these measures alone have limitations in managing glucose levels. Complications, such as hepatic adenoma, hepatocellular carcinoma, kidney disease, lactic acidosis, and hypertriglyceridemia, which occur in patients with GSD, ultimately arise due to uncontrolled glucose levels. These complications are not inevitable but result from a lack of optimal metabolic control. Therefore, patients with GSD require precise diagnostic and treatment approaches because of the complexity of the disease.
GSD cannot be treated clinically, except for gene therapy, and there are difficulties in the development of effective treatments. However, GSD can be managed by controlling glucose levels. Traditionally, management involves dietary interventions, notably a cornstarch-based diet, to maintain appropriate glucose levels in patients with GSD7. Such dietary management ensures a consistent and stable glucose supply for those who are unable to store or adequately utilize glycogen. Moreover, this continuous dietary approach aids in controlling blood lactate levels, preventing organ damage and supporting normal growth and lifestyle8,9. Despite these efforts, the inherent nature of this genetic condition means that patients may still experience sudden hypoglycemic episodes, in which glucose levels drop sharply, even with diligent dietary management10. To mitigate the risk of hypoglycemia and ensure proper patient care, key clinical metrics, including patient height and weight, along with blood biomarkers such as glucose, cholesterol, and lactic acid levels, are regularly tracked and assessed. Nevertheless, the limited number of medical specialists relative to the number of patients, along with the costs of hospital and outpatient care, make continuous staff-dependent monitoring a challenging and labor-intensive task.
Checking and monitoring glucose levels is the easiest way to managing patients with GSD11,12. This test is often performed using the finger-prick glucose test, a method aimed at preventing hypoglycemia and its associated side effects13. Despite its widespread use, this approach faces challenges in the accurate detection of rapid fluctuations in glucose levels, which can be significantly influenced by the timing of blood sample collection.
The requirement for continuous blood glucose tracking has prompted the development of continuous glucose-monitoring (CGM) systems. Initially designed for individuals with diabetes, the convenience and ability to provide real-time glucose information have broadened its use in effectively managing glucose levels. Moreover, these systems have led to therapeutic advancements in the care of patients with GSD14. In addition, the introduction of CGM systems revolutionized the management process. The CGM system, a wearable device, continuously tracks glucose concentrations at set intervals and displays the data on a dedicated smartphone app or receiver15. This technology offers a detailed view of glucose fluctuations, enabling better management strategies beyond traditional dietary adjustments and symptomatic treatments.
With the evolution of CGM systems, there has been a surge in the development of algorithms for analyzing and predicting glucose levels to forecast and prevent issues before glucose levels increase or decrease16,17. Owing to its high prevalence, it has developed significantly, and the application of blood glucose prediction models has preceded. Statistically based models such as autoregressive (AR), autoregressive exogenous (ARX), and autoregressive moving average (ARMA) have been proposed18, and machine learning (ML)-based models such as support vector regression (SVR)19 and random forest (RF)20 have been utilized. However, these models often fall short because of their inability to consider a myriad of factors that influence glucose levels, such as lifestyle and physiological changes. To overcome these limitations, the application of deep learning (DL) algorithms is expected to integrate variables affecting blood glucose fluctuations and analyze a wide range of patterns in diabetes.
Despite advances in DL algorithms for diabetes management, research specifically targeting patients with GSD remains lacking. This study aimed to forecast glucose levels in patients with GSD using state-of-the-art (SOTA) DL algorithms in the time series forecasting field.
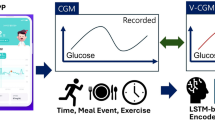
We assessed the predictive accuracy of the model using CGM in patients with GSD as part of their management strategy. Additionally, we trained the models while accounting for patient-specific characteristics to enhance the prevention of sudden hypoglycemia (Fig. 1). The contributions of this study are as follows: patients with GSD can effortlessly track and manage their glucose level through an app or online platform powered by algorithm. This approach promises personalized healthcare tailored to each patient’s glucose data and lifestyle, potentially diminishing the reliance on expensive medical interventions by averting complications.
To the best of our knowledge, this is the first study focusing on forecasting glucose levels in patients with GSD.
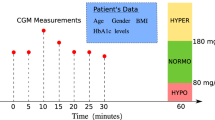
Schematic overview of the study. (a) Preprocessing of the data extracted from the CGM device. (b) Training process for personalized DL models. (c) Input/output and inference process for the blood sugar level prediction task. (d) Input/output and inference process for the hypoglycemia classification task. The data collected from the CGM device undergoes a series of preprocessing steps to ensure accuracy and consistency. Each patient’s data is then used to train a personalized DL model. For the prediction task, the input consists of historical blood glucose data, and the output is the predicted blood sugar level for a specific future time horizon. In the classification task, the output is the probability of hypoglycemia occurring within a specified future time frame. CGM continuous glucose monitoring, DL Deep Learning.
Materials and methods
Data collection
Blood glucose data were collected from patients with GSD who were hospitalized or managed at Yonsei University Wonju Severance Christian Hospital, Wonju, Korea, from August 2020 to February 2024 (IRB-CR324074). A total of 46 patients who wore the CGM system for at least 14 days were included in this study. The maximum duration of the CGM was 524 days (Fig. 2).
The Freestyle Libre (Abbott, Chicago, Illinois, USA) was used. The Freestyle Libre continuously measures glucose concentrations in the interstitial fluid via sensors attached to a patient’s skin. These sensors measure glucose concentration in the interstitial fluid beneath the skin, which closely mirrors the glucose concentration in the blood. The sensor estimates the glucose levels every 15 min based on these data. This device periodically collects glucose concentrations in the interstitial fluid data and provides real-time information to patients who can access the stored information using a reader or smartphone app. The collected data were automatically uploaded to the Libreview platform for analysis and shared with the medical staff. To the best of our knowledge, the dataset used in this study was collected from medical institutions that manage and treat the largest number of GSD patients in Korea.
Preprocessing of CGM data
Preprocessing of the CGM data was conducted in three steps: interpolation, normalization, and slicing. Filtering was not applied during preprocessing to avoid potential distortions in the CGM data pattern, which could complicate the medical interpretation. First, interpolation was performed to address instances in which patients replaced their CGM devices or missed glucose measurements owing to device errors. Referring to the preceding literature21, and sampling rate and reference specifications of the blood glucose meter used in the study, the interpolation period was set to 3 h. The referenced literature utilized a 1-h interval with a 5-min sampling rate. In contrast, our study employed a 15-min sampling rate, warranting a 3-h interval. Consequently, the missing values spanned more than 3 h, these sections were excluded from the slicing process and were not used for model training. For gaps of 3 h or less, quadratic interpolation was applied using the preceding and succeeding 10 data points to fill in missing values. Subsequently, robust scalar normalization was applied to minimize the impact of outliers based on the median and interquartile range of the CGM data. Finally, slicing was performed to create data-label pairs. The look-back window and forecast size for slicing were set to 48 and 4, respectively, based on previous studies and a grid search, allowing prediction of the future 1 h of glucose levels from the past 12 h of data. The collected data included only blood glucose measurement readings and real-time data, without any details on life events such as ingestion or exercise. Therefore, a long length of look-back window was considered to allow the model to capture the morphology and trends in blood glucose fluctuations as effectively as possible. Consequently, during the slicing process, the look-back window was set to 12 h, ensuring that all datasets (training, validation, and testing) included sufficient data for analysis.
Data-label pairs where glucose levels were ≥ 300 mg/dL or < 40 mg/dL were retrospectively reviewed and considered outliers due to potential inaccuracies, and thus, were excluded from the dataset. Furthermore, retrospective analysis revealed that external pressure artifacts, particularly during sleep or daily activities, could lead to artificially high or low readings, compromising measurement accuracy. The device used in this study had a lower detection limit of 40 mg/dL, but occasional readings below this threshold were recorded during the first day after CGM placement and toward the end of its lifespan. These readings were deemed erroneous and inconsistent with clinical expectations. Finally, each CGM dataset was sorted in chronological ascending order and then divided at a ratio of 6:2:2 to construct the dataset.
Deep learning models
For the deep-learning model, the latest three state-of-the-art deep-learning models in time-series forecasting were utilized (Fig. 1). First, we employed the PatchTST model proposed for 202222. This model was inspired by the application of transformer models in natural language processing (NLP) and computer vision. The PatchTST architecture processes time-series data in a manner similar to how images are processed by transformers. This effectively captures long-range dependencies through self-attention mechanisms. Second, an LTSF N-linear model was used23. This model emphasizes the development of time series forecasting using multiple linear layers. It features an architecture designed to capture various aspects of time-series data, while maintaining efficient computational complexity and scalability. Multiple linear layers alone have proven to be effective in handling high-dimensional time-series data, which can often be challenging for the other methods. Finally, a TS Mixer is employed24.
The TS Mixer effectively extracts information from time-series data by leveraging a mixing mechanism that integrates both temporal and feature dimensions. This architecture is particularly suitable for multivariate prediction and has demonstrated effectiveness across several benchmarks. The three models (PatchTST, LTSF N-Line, TS Mixer) achieved state-of-the-art performance in 2023, 2022, and 2023, respectively, in time series forecasting. These models were selected as they represented the most recent SOTA approaches available during the stages of research planning and experimentation. The hyperparameters for all the models were set to the default parameters proposed in their respective papers. Specifically, the parameters related to the input and output, such as the lookback window and forecast size, were set to be the same across all three models.
Forecasting of future CGM data
In this study, the patient’s glucose concentrations in the interstitial fluid levels over the future 1 h were predicted using a model (Fig. 1c). Predictions were based on the past blood glucose values of patients with GSD using an independent model for each dataset. The input to the model included both glucose levels and timestamps recorded by the CGM device. Time was segmented into years, months, days, and minutes and encoded using sine and cosine functions to capture the cyclical nature of time over a 24-h period. There were differences in the measurement periods among the patients participating in this study, and there were cases in which the year changed during the measurement. To account for this, all segmented time variables were incorporated as inputs. DL models receive 7-channel multivariate time series data, and because the models used in the study were designed for multivariate prediction, the 7-channel future data were predicted. Optimization was performed by selecting the mean squared error (MSE) loss as the optimization function to minimize the discrepancies between the predicted and actual values of the model. The loss for each channel was calculated and the average loss across all channels was determined. To predict the glucose levels, we defined the optimal model as the one with the lowest mean absolute percentage error (MAPE) between the predicted and actual blood glucose channel values. The model with the lowest MAPE is saved as the optimal model.
Classification of future hypoglycemic event
In this study, a binary classification was conducted to determine the feasibility of predicting hypoglycemia in addition to direct glucose levels (Fig. 1d). Hypoglycemia was defined as a blood glucose level of < 80 mg/dL. Readings below this threshold were labeled as 1, indicating hypoglycemia. The 80 mg/dL threshold was chosen to facilitate the effective use of a cornstarch-based diet, enabling timely interventions to stabilize glucose levels and prevent further declines. This approach ensures a steady glucose release, which is essential for maintaining metabolic stability in patients with GSD. For the model input, 12 h of data was utilized, similar to the forecasting task. In terms of the model structure, the window size was set to 1, and a linear layer was added to process the 7-channel outputs for binary classification. The output node was set to one, with a sigmoid activation function. Optimization was performed using binary cross-entropy loss to minimize the discrepancy between the actual hypoglycemia occurrences and model predictions. In particular, the number of actual hypoglycemic events was significantly lower than the number of normal events, resulting in an imbalance between the two classes. To address this imbalance, class weights are applied to the loss function during the learning process. Finally, the model with the highest area under the receiver operating characteristic (AUROC) curve was selected as the optimal model for predicting hypoglycemia.
Performance metrics
In the forecasting task, the primary outcome of the DL model is the direct prediction of glucose levels. The MAPE was selected as the main indicator because of its interpretability, adaptability to changes in data value sizes, and suitability for comparing the performance of various models. The MAPE provides a probability value between 0 and 100%. The mean absolute error and root mean square error were used as supplementary statistical indicators. For the forecasting task, each model generated predictions for 4-h intervals (15, 30, 45, and 60 min). Therefore, statistical indicators were calculated by comparing the predicted values with the actual values at each time point.
In the classification task, the primary outcome was the model’s predictive power for hypoglycemic events. In binary classification, the AUROC was used as a major indicator and criterion for determining the optimal state of the model. The model’s prediction results were classified into four categories: true positive (TP), false negative (FN), true negative (TN), and false positive (FP), compared to the actual answer. Based on these results, additional statistical indicators, including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and F1 score were used. The optimal cutoff to minimize model misclassification was calculated based on the Youden index (J) within the ROC curve. In the classification task, each model outputs a result at a single time point. Therefore, independent models were developed to predict hypoglycemic events at each future time point. In summary, the models predicting hypoglycemic events after 15, 30, 45, and 60 min were independently trained.
Independent models were trained for each patient to build a personalized predictive model. This approach is expected to effectively capture each patient’s unique blood glucose level variability and enable more accurate predictions. We calculated the statistical metrics for each task by synthesizing the prediction results at each time point. All statistical metrics were reported as point estimates and 95% confidence intervals (CIs). The data were analyzed and visualized using Python 3.9.5 (Python Software Foundation).
Results
Participant characteristics
Table 1 summarizes the demographic and clinical characteristics of the study group. A total of 46 subjects wore CGM devices for blood glucose measurements, with only those who wore the device for at least 14 days included in the study (Fig. 2). The study group comprised 26 male subjects with an average age of 13.27 years, and 20 female subjects with an average age of 14.85 years. The average duration for the male group was 151 days, while that for the female group was 171 days (Table 1). All participating patients were of Asian descent.
Glucose level forecasting result
The results of each model for forecasting glucose levels are summarized in Fig. 3; Table 2. Additionally, the results of the clinical error grid analysis are summarized in Fig S1. To enable a clearer comparison of classification performance across models, the results derived from Equal Error Rates are presented in Table S1. Among the three models, the LTSF N-Linear demonstrated the highest performance. It exhibited low MAPE across all prediction horizons, with a particularly low MAPE of 5.95% (95% confidence interval (CI), 5.64–6.26%) for the 15-min prediction. For the 60-min prediction, which had the longest temporal length, the MAPE was 11.14% (95% CI, 10.39–11.89%), indicating the lowest error at this horizon. In contrast, the TS Mixer exhibited the lowest performance. It exhibited high MAPE values across all prediction horizons, with errors of 16.14% (95% CI, 13.72–18.57%), 16.50% (95% CI, 14.03–18.97%), 16.57% (95% CI, 13.90–19.23%), and 16.77% (95% CI, 13.59–19.95%) for each time point. The Patch TST model demonstrated a relatively moderate performance compared with the other two models.
Model performance for the forecasting task. (a) Linear correlation for the 15-min prediction horizon. (b) Linear correlation for the 30-min prediction horizon. (c) Linear correlation for the 45-min prediction horizon. (d) Linear correlation for the 60-min prediction horizon. The horizontal axis represents the patient’s actual glucose levels, while the vertical axis represents the model’s predicted values. The LTSF N-Linear model demonstrated a relatively high Pearson R across all prediction intervals. Pearson R Pearson correlation coefficient, CI confidence interval.
Hypoglycemia classification result
The results of each model for hypoglycemia prediction are summarized in Fig. 4; Table 3. In the classification task, the TS Mixer model demonstrated relatively high performance among the three models. Notably, the prediction of hypoglycemia 15 min ahead achieved an AUROC of 0.866 (95% CI, 0.829–0.904) with an average sensitivity of 72.27% (95% CI, 63.97–80.56%). Although the predictive power decreased as the temporal horizon was extended, the model still performed reasonably well, with an AUROC of 0.672 (95% CI, 0.635–0.708) for predictions made 60 min in advance. Conversely, the LTSF N-linear model exhibits poor predictive power for classification tasks. Across all prediction horizons, the AUROC values were 0.600 (95% CI, 0.556–0.644), 0.570 (95% CI, 0.539–0.602), 0.562 (95% CI, 0.533–0.591), and 0.566 (95% CI, 0.532–0.587). Again, the Patch TST model exhibited a relatively moderate performance.
Model performance for the classification task. (a) AUROC for hypoglycemia prediction at the 15-min prediction horizon. (b) AUROC for hypoglycemia prediction at the 30-min prediction horizon. (c) AUROC for hypoglycemia prediction at the 45-min prediction horizon. (d) AUROC for hypoglycemia prediction at the 60-min prediction horizon. The thin lines represent the results for individual patients, whereas the thick lines represent the mean. The shaded area on either side of the thick lines indicates the 95% CI. TS Mixer achieved a relatively high AUROC across all prediction horizons. AUROC area under receiver operating characteristics curve, CI confidence interval, ERR equal error rate.
Discussion
Traditionally, patients with GSD have had to be hospitalized or rely on fingerprick tests at home to monitor and manage their glucose levels. However, these methods have limitations because they do not provide a comprehensive view of glucose levels throughout the day. Although the CGM was originally designed for patients with diabetes with blood glucose issues, it is also considered applicable to patients with GSD. Derks’s research on 15 patients with GSD demonstrated that an in-depth analysis using CGM data can effectively evaluate glucose management25. Additionally, a study in 2022 involving 10 adult patients with GSD showed that CGM may be beneficial26. Another study demonstrated the ability of an artificial neural network to predict glucose levels in patients with type 1 diabetes using an artificial neural network27,28.
Therefore, we hypothesized that metabolic predictions could be made based on the results of previous studies and data from patients with GSD accumulated at the Yonsei University Wonju Severance Christian Hospital. Our objective was to validate and analyze this hypothesis using the results generated by the DL models. This study yielded meaningful outcomes, confirming that by analyzing the CGM data from patients with GSD, it is possible to manage and predict their glucose levels. This finding supports the feasibility of precision medicine tailored to each individual, highlighting the potential for customized care that addresses the unique metabolic needs of each patient.
From a big data-based perspective, the data is typically divided by the number of individuals, followed by learning and testing. However, to predict the aspects related to human metabolism, we considered individualized data learning to be more appropriate. Therefore, we adopted an approach in which an independent model was trained for each study subject and quantification indicators were subsequently calculated. Given that the number of models to be tested increases the time and computational costs exponentially, we focused our efforts on deriving research results using the latest models that have achieved SOTA performance in the field of time-series forecasting.
We conducted both forecasting and classification tasks to assess the performance of the model in predicting and managing immediate blood glucose levels (Fig. 1). In the forecasting task, the LTSF N-Linear model demonstrated relatively strong forecasting ability. For blood glucose level prediction 15 min ahead, the Pearson’s R correlation coefficient was 0.887 (95% CI, 0.886–0.888), indicating a high predictive accuracy (Fig. 3). Even at the 30- and 45-min prediction horizons, the Pearson’s R values remained relatively high than those of the compared models, with values of 0.721 (95% CI, 0.719–0.724) and 0.617 (95% CI, 0.614–0.621), respectively. However, at the 60-min prediction horizon, the predictive power significantly decreased, with a Pearson’s R of 0.561 (95% CI, 0.557–0.565). The LTSF N-Linear focuses solely on the temporal relationships between data points in a linear manner. It performs well when complex modeling of the data structure is not required, owing to the simplicity of its architecture. The participants in this study continuously managed their diet to maintain stable blood sugar levels. Since changes in metabolic blood sugar primarily occur through food intake29,30, it is likely that the LTSF N-Linear benefited structurally from this process. The blood sugar data in this study followed a relatively linear trend over short prediction periods with few complex nonlinear relationships. As a result, LTSF N-Linear delivered excellent short-term prediction results, as demonstrated in this study; however, its performance for long-term predictions indicates areas for improvement. However, only the variables directly related to blood sugar levels and time were used in this study. As the input data were already time-series data, temporal information may have been inherently included, making it difficult for the model to capture additional dependencies, particularly those related to metabolism. Furthermore, the relatively complex architecture of the model may have led to overfitting, as it may have been more sensitive to noise than to learning meaningful patterns. Patch TST, which employs a patch-based approach, was designed to capture both short- and long-term trends. Although its performance was slightly lower than that of the LTSF M Linear, the difference between the two models was not significant. The patch-based TST produced reasonably predictable results, demonstrating its capability to handle tasks effectively.
In the classification task, the TS Mixer demonstrated a strong classification performance. Notably, an AUROC of 0.866 (95% CI, 0.829–0.904) for the 15-min prediction horizon (Fig. 4). Although the predictive power decreased for the 45- and 60-min prediction horizons, it still achieved a specificity of 74.09% (95% CI, 66.97–81.22%) and 75.42% (95% CI, 69.75–81.1%) for these horizons, respectively. Additionally, the model maintained a NPV of 91.21% (95% CI, 86.52–95.90%) and 92.72% (95% CI, ` 90.72–94.71%), even at longer time intervals (Table 3). Conversely, the LTSF N-Linear model showed little classification ability, with AUROC values close to 0.5. Specifically, it recorded an AUROC of 0.556 (95% CI, 0.600–0.644) for the near-term prediction horizon. For the TS Mixer, the model constructs a predictive decision boundary for binary classification based on nonlinear patterns. Conversely, LTSF N-Linear, which operates on a simple linear basis, is well suited for trend prediction but struggles in classification tasks where nonlinear patterns are crucial. In this study, even after adding an additional linear layer, the performance deteriorated, with AUROC values close to 0.5. This outcome reflects the challenges that linear classifiers face in achieving good performance in binary classification31,32. The patch TST sits between the relatively simple LTSF N-Linear and the more complex TS Mixer, and its quantitative performance indicators fall in the middle range. While Patch TST is capable of capturing dependencies and nonlinear patterns in waveforms, it is likely to be less effective at considering global patterns compared with the TS Mixer, as it relies on patch-based processing. Additionally, when analyzing the overall metrics, certain models demonstrated effective performance in hypoglycemia classification. However, a slight low PPV was observed in some cases. This reduction could result in inaccurate predictions, leading to false alarms or unnecessary treatments or interventions, highlighting the need for further refinement and improvement.
This study has some limitations. First, there is a lack of input data for predicting changes in glucose levels; human metabolism is highly complex, and incorporating additional information, such as meal timing, physical activity, and other events, could improve the model’s predictive accuracy. For example, in the OhioT1DM dataset33, a well-known dataset for blood glucose prediction, biometric information, such as insulin dosage and heart rate, and behavioral factors, such as meal intake, sleep, and physical activity, were systematically monitored. In contrast, this study relied solely on the blood glucose data obtained from the CGM device and time annotations. Although the patients in this study had already maintained optimal metabolic control, the data were retrospective, and it was difficult to obtain additional inputs owing to the limitations of the CGM device. As a result, information such as biometric data or behavioral factors could not be incorporated into this study. Because34,35, the absence of this information is considered one of the primary reasons for the poor predictive power of the model over longer prediction horizons in this study. In summary, several studies on glucose levels prediction have achieved acceptable or expected results using blood glucose data alone. However, given the characteristics of GSD patients, who frequently require dietary control, it is expected that incorporating additional information could help compensate for the model’s limited predictive capability. Second, this study was conducted using retrospective data. Future studies should incorporate prospective data to verify the applicability of the model to real-world GSD cases. If future studies can predict glucose levels using CGM data from patients who are not yet in optimal metabolic control, this could lead to a groundbreaking management method. Third, a relatively high sampling rate was used in this study, and the CGM device was set to measure blood glucose every 15 min in real time. In general, a shorter sampling rate21,27,36. Furthermore, this study aimed to explore the potential of DL models to help patients with GSD efficiently manage their glucose levels in daily life and proactively prevent possible hypoglycemic symptoms. Therefore, the sampling rate used in this study is considered a factor that directly influences the model’s predictive power. In future research, it will be important to adjust the sampling rate, considering both model performance and real-life applicability, and to evaluate whether the adjusted rate enhances efficiency37,38. Fourth, the general applicability and interpretability of error grid analysis are limited. Various analysis methods, such as the clinical error grid methodology, have been proposed to validate39,40. In contrast, this study was conducted in a group of patients with GSD, a condition with an extremely low prevalence. The type 2 diabetes is typically associated with elevated blood sugar levels, while hypoglycemia is a major concern in type 1 diabetes. In contrast, GSD is characterized by low blood sugar levels caused by genetic mutations affecting glucose storage and release due to genetic mutations. Consequently, the blood sugar range in patients with GSD differs significantly from that in patients41, making it difficult to apply the analytical frameworks designed for diabetes to GSD. When performing clinical error grid methodology on the model used in this study, it was observed that most characteristics of GSD patients fell within Region A (Fig. S1). However, the sizes of Regions B and D, which are critical for identifying in-application treatment failures or instances of hypoglycemia, differed from the average blood glucose range of GSD patients. While some quantification results demonstrated excellent accuracy, the overall persuasiveness of the methodology is limited by the constraints. These challenges limit the applicability of previously proposed glycemic analysis methodologies for GSD and consequently impose restrictions on the analysis of model outcomes in this study. Fifth, further research on interpretability is needed to understand how patient-specific factors influence the model’s performance. To investigate whether pattern-specific factors, such as gender, affect glucose level fluctuations and subsequently impact model outcomes, we analyzed test results by gender (Tables 4 and 5). However, no significant differences were observed between the two groups across any task. One potential explanation for these findings is that statistical analysis of Table 1 revealed no significant differences between the two groups. This trend is likely reflected in the model results. Additionally, due to the limitations of the retrospective dataset, the collected data lacks explanatory power, as it only includes glucose levels and does not provide additional relevant information. Consequently, these constraints make it challenging to analyze patient-specific factors and limit the examination of other variables beyond gender. In the future, follow-up studies should not only refine the research design but also enhance model interpretability and incorporate additional analyses using Explainable AI methods. Finally, this study requires external validation. Although many DL models produce promising results based on specific datasets, it is crucial to verify whether they generalize well to data from other42. This study was no exception. Although an independent model was developed for each individual to achieve personalized optimization, all data used for learning were obtained from a single hospital. In particular, owing to the rarity of GSD, there is a bias in the age range of the participants, and the study population was limited to Asian populations. The data used in this study were primarily collected from medical institutions in Korea that diagnose and treat the largest number of GSD patients. In contrast, other medical institutions rarely manage patients with rare diseases such as GSD, making it relatively difficult to collect external datasets. This limitation may reduce reliability due to the small number of contributors and raises. Additionally, there is a potential bias related to the CGM device used, particularly with respect to the sampling rate. Therefore, external validation is necessary to address these biases and verify the model’s performance across a more diverse set of subjects.
Despite these limitations, this study is significant, as it is the first to demonstrate the ability to forecast glucose levels using CGM and DL in patients with GSD. GSD is a genetic disorder for which no definitive treatment is currently available. Even if gene therapy becomes available in the future, its cost may be prohibitive for many patients. However, the use of CGM, which is relatively affordable and easily accessible, combined with DL-based personalized blood glucose management, can offer an accessible method for managing this condition. If this method is further refined, hypoglycemia can be predicted in advance and alerts can be provided, enabling better management of cornstarch intake, diet, and exercise based on blood glucose predictions. Although patients must continue to consume cornstarch and follow an appropriate diet, this approach is highly beneficial from a cost perspective. In this study, predictions were conducted using a single variable, glucose levels. While some DL models demonstrated promising results depending on the task, it is crucial to acknowledge that fluctuations in glucose levels can be highly sensitive to patient-specific factors such as fat metabolism, physical activity, and cornstarch intake. Follow-up studies are planned to address these variables. Additionally, the consideration of interpretability is being planned by leveraging explainable machine learning techniques. This approach is expected to provide clinicians with intuitive and actionable insights through the development of visualization dashboards.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to privacy and ethical restrictions but are available from the corresponding author on reasonable request with permission from the Ethics Committee (irb@yonsei.ac.kr; 82-033-741-1715).
References
Fukuda, T. et al. Blood glucose trends in glycogen storage disease type Ia: A cross-sectional study. J. Inherit. Metab. Dis. 46, 618–633 (2023).
Hendriksz, C. J. & Gissen, P. Glycogen storage disease. Paediatr. Child. Health 25, 139–144 (2015).
Özen, H. Glycogen storage diseases: New perspectives. World J. Gastroenterol. 13, 2541 (2007).
Shin, Y. S. Glycogen storage disease: Clinical, biochemical, and molecular heterogeneity. In Seminars in Pediatric Neurology, Vol. 13 115–120 (Elsevier, 2006).
Weinstein, D. A., Steuerwald, U., De Souza, C. F. & Derks, T. G. Inborn errors of metabolism with hypoglycemia: Glycogen storage diseases and inherited disorders of gluconeogenesis. Pediatr. Clin. 65, 247–265 (2018).
Rossi, A. et al. A prospective study on continuous glucose monitoring in glycogen storage disease type IA: Toward glycemic targets. J. Clin. Endocrinol. Metab. 107, e3612–e3623 (2022).
Saban, O. S. et al. Glycogen storage disease type IA refractory to cornstarch: Can next generation sequencing offer a solution? Eur. J. Med. Genet. 65, 104518 (2022).
Dahlberg, K. R. et al. Cornstarch requirements of the adult glycogen storage disease Ia population: A retrospective review. J. Inherit. Metab. Dis. 43, 269–278 (2020).
Ross, K. M. et al. Dietary management of the glycogen storage diseases: Evolution of treatment and ongoing controversies. Adv. Nutr. 11, 439–446 (2020).
Dambska, M., Labrador, E., Kuo, C. & Weinstein, D. Prevention of complications in glycogen storage disease type Ia with optimization of metabolic control. Pediatr. Diabetes. 18, 327–331 (2017).
Vashist, S. K. Continuous glucose monitoring systems: A review. Diagnostics 3, 385–412 (2013).
Peeks, F. et al. Research priorities for liver glycogen storage disease: An international priority setting partnership with the James Lind alliance. J. Inherit. Metab. Dis. 43, 279–289 (2020).
Herbert, M. et al. Role of continuous glucose monitoring in the management of glycogen storage disorders. J. Inherit. Metab. Dis. 41, 917–927 (2018).
Kasapkara, C. S., Cinasal Demir, G., Hasanoglu, A. & Tumer, L. Continuous glucose monitoring in children with glycogen storage disease type I. Eur. J. Clin. Nutr. 68, 101–105 (2014).
Peeks, F. et al. Clinical and biochemical heterogeneity between patients with glycogen storage disease type IA: The added value of CUSUM for metabolic control. J. Inherit. Metab. Dis. 40, 695–702 (2017).
Montaser, E., Díez, J. L. & Bondia, J. Glucose prediction under variable-length time-stamped daily events: A seasonal stochastic local modeling framework. Sens. (Basel). 21 (9), 3188. https://doi.org/10.3390/s21093188 (2021).
Liu, K. et al. Machine learning models for blood glucose level prediction in patients with diabetes mellitus: Systematic review and network meta-analysis. JMIR Med. Inf. 11, e47833. https://doi.org/10.2196/47833 (2023).
Eren-Oruklu, M., Cinar, A., Quinn, L. & Smith, D. Estimation of future glucose concentrations with subject-specific recursive linear models. Diabetes Technol. Ther. 11, 243–253 (2009).
Georga, E. I. et al. Multivariate prediction of subcutaneous glucose concentration in type 1 diabetes patients based on support vector regression. IEEE J. Biomed. Health Inf. 17, 71–81 (2012).
Li, J. & Fernando, C. Smartphone-based personalized blood glucose prediction. ICT Express. 2, 150–154 (2016).
Li, K. et al. A deep learning framework for accurate glucose forecasting. IEEE J. Biomed. Health Inf. 24, 414–423 (2019).
Nie, Y., Nguyen, N. H., Sinthong, P. & Kalagnanam, J. A time series is worth 64 words: Long-term forecasting with transformers. arXiv preprint arXiv:2211.14730 (2022).
Zeng, A., Chen, M., Zhang, L. & Xu, Q. Are transformers effective for time series forecasting? Proc. AAAI Conf. Artif. Intell. 37, 11121–11128 (2023).
Chen, S. A., Li, C. L., Yoder, N., Arik, S. O. & Pfister, T. Tsmixer: An all-mlp architecture for time series forecasting. arXiv preprint arXiv:2303.06053 (2023).
Peeks, F. et al. A retrospective in-depth analysis of continuous glucose monitoring datasets for patients with hepatic glycogen storage disease: Recommended outcome parameters for glucose management. J. Inherit. Metab. Dis. 44, 1136–1150 (2021).
Isaacs, S. & Isaacs, A. Endocrinology for the hepatologist. Curr. Hepatol. Rep. 23, 99–109 (2024).
Zhu, T. et al. Enhancing self-management in type 1 diabetes with wearables and deep learning. NPJ Digit. Med. 5, 78 (2022).
van Doorn, W. P. et al. Machine learning-based glucose prediction with use of continuous glucose and physical activity monitoring data: The Maastricht study. PLoS One. 16, e0253125 (2021).
White, F. J. & Jones, S. A. The use of continuous glucose monitoring in the practical management of glycogen storage disorders. J. Inherit. Metab. Dis. 34, 631–642 (2011).
Kaiser, N. et al. Glycemic control and complications in glycogen storage disease type I: Results from the Swiss registry. Mol. Genet. Metab. 126, 355–361 (2019).
Zhao, X., Niu, L. & Shi, Y. Kernel based simple regularized multiple criteria linear programs for binary classification. In 2013 IEEE/WIC/ACM International Joint Conferences on Web Intelligence (WI) and Intelligent Agent Technologies (IAT), vol. 3 58–61 (IEEE, 2013).
Kuhn, M., Johnson, K., Kuhn, M. & Johnson, K. Nonlinear classification models. In Applied Predictive Modeling, 329–367 (2013).
Marling, C. & Bunescu, R. The OhioT1DM dataset for blood glucose level prediction: Update 2020. In CEUR Workshop Proceedings, Vol. 2675 71 (NIH Public Access, 2020).
Livesey, G., Taylor, R., Hulshof, T. & Howlett, J. Glycemic response and health—A systematic review and meta-analysis: Relations between dietary glycemic properties and health outcomes. Am. J. Clin. Nutr. 87, 258S–268S (2008).
Nansel, T. R., Lipsky, L. M. & Liu, A. Greater diet quality is associated with more optimal glycemic control in a longitudinal study of youth with type 1 diabetes. Am. J. Clin. Nutr. 104, 81–87 (2016).
Hovorka, R. et al. Blood glucose control by a model predictive control algorithm with variable sampling rate versus a routine glucose management protocol in cardiac surgery patients: A randomized controlled trial. J. Clin. Endocrinol. Metab. 92, 2960–2964 (2007).
Zimmet, P., Alberti, K. & Shaw, J. Global and societal implications of the diabetes epidemic. Nature 414, 782–787 (2001).
Deshpande, A. D., Harris-Hayes, M. & Schootman, M. Epidemiology of diabetes and diabetes-related complications. Phys. Ther. 88, 1254–1264 (2008).
Clarke, W. L., Cox, D., Gonder-Frederick, L. A., Carter, W. & Pohl, S. L. Evaluating clinical accuracy of systems for self-monitoring of blood glucose. Diabetes Care 10, 622–628 (1987).
Sengupta, S. et al. Clarke error grid analysis for performance evaluation of glucometers in a tertiary care referral hospital. Indian J. Clin. Biochem. 37, 199–205 (2022).
Rajas, F., Labrune, P. & Mithieux, G. Glycogen storage disease type 1 and diabetes: Learning by comparing and contrasting the two disorders. Diabetes Metab. 39, 377–387 (2013).
Amir, G., Maayan, O., Zelazny, T., Katz, G. & Schapira, M. Verifying generalization in deep learning. In International Conference on Computer Aided Verification 438–455 (Springer, 2023).
Acknowledgements
This research was supported and funded by the SNUH Lee Kun-hee Child Cancer & Rare Disease Project, Republic of Korea (FP-2022-00007-012). Additionally, it was supported by the “Regional Innovation Strategy (RIS)” through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (MOE) (2022RIS-005).
Funding
This research was supported and funded by the SNUH Lee Kun-hee Child Cancer and Rare Disease Project, Republic of Korea (FP-2022-00007-012). Additionally, this research was supported by the “Regional Innovation Strategy (RIS)” through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (MOE) (2022RIS-005).
Author information
Authors and Affiliations
Contributions
All authors have contributed significantly to this article and agree with the intellectual content of the manuscript and its submission to npj Digital Medicine. Author Contributions Conceptualization: Sejung Yang, Yunkoo Kang Data curation: Ji Seung Ryu, Jang Hoon Ru Formal analysis: Ji Seung Ryu Funding acquisition: Sejung Yang, Yunkoo Kang Investigation: Ji Seung Ryu Methodology: all authors. Project administration: Sejung Yang, Yunkoo Kang Resources: Yunkoo Kang Software: Ji Seung Ryu Supervision: Sejung Yang, Yunkoo Kang Validation: all authors. Visualization: all authors. Writing—original draft: Ji Seung Ryu, Yunkoo Kang Writing—review and editing: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Study approval and accordance statement
All methods in this study were conducted in accordance with relevant guidelines and regulations, with approval from the Ethics Committee. Informed consent was obtained from all subjects involved in the study and/or their legal guardians.
Ethics statement
This study was approved and overseen by the Yonsei University Institutional Review Board (IRB-CR324074).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ryu, J.S., Ru, J.H., Kang, Y. et al. A deep learning approach for blood glucose monitoring and hypoglycemia prediction in glycogen storage disease. Sci Rep 15, 13032 (2025). https://doi.org/10.1038/s41598-025-97391-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97391-8