Abstract
Unravelling the physiological adaptations that enable fish to thrive under diverse environmental stressors is crucial for predicting their resilience to climate change. However, linking molecular mechanisms to ecological contexts remains a significant challenge and such studies are greatly lacking. To close this data gap, we conducted a physiological and molecular mechanisms-integrated comparative study of representative fish species from two ecologically contrasting regions: the Indian Sundarban mangrove estuary and the Sub-Himalayan Terai-Dooars hill streams. We assessed erythropoietic activity, cell cycle dynamics, reactive oxygen species levels, and the expression of genes integral to hypoxia response, electron transport chain regulation, adaptive remodeling, and stress modulation. Our results demonstrate that fish inhabiting the Terai-Dooars region exhibit substantially higher erythropoietic efficiency, elevated reactive oxygen species levels, and significant upregulation of hypoxia-inducible and adaptive remodeling genes as compared to their Indian Sundarban counterparts. These findings suggest that the challenging environmental conditions of the Terai-Dooars—characterized by low temperatures, high water velocities, and physiological hypoxic stress—drive distinctive physiological and genetic adaptations in fish populations. Analysis of variance (ANOVA) on Principal Components (PCs) revealed species-specific cellular variability modulated by regional environmental pressures, emphasizing the interaction of intrinsic and extrinsic factors in shaping adaptive responses. This study bridges critical knowledge gaps by linking physiological traits to ecological contexts, providing insights into community-level adaptation mechanisms. The findings highlight convergent evolutionary strategies and propose molecular and cellular biomarkers for assessing resilience under future climate scenarios. Finally, these ecophysiological insights will enhance the comprehensive understanding of species responses to environmental heterogeneity, with implications for conservation and ecosystem management in the face of global environmental change.
Similar content being viewed by others
Introduction
The adaptive capacity of organisms is governed by a combination of evolutionary history and physiological plasticity, enabling them to survive in diverse and often challenging environmental conditions. Understanding adaptation strategies also requires an integrated exploration of the genetic basis underpinning plasticity and the environmental pressures that shape their evolutionary trajectories1,2. As emphasized by Schulte1,2, this interplay underscores the role of both phenotypic plasticity and local adaptation in shaping organismal responses to changing environment. Aquatic environments are inherently dynamic, exhibiting significant spatial and temporal variations in dissolved oxygen and temperature3,4,5. Populations worldwide exhibit remarkable physiological and genetic adaptability to diverse environments, a key factor in their resilience against the backdrop of climate change3,6,7,8. Oxygen availability and temperature fluctuations represent two of the most critical environmental variables influencing fish physiology9, directly impacting erythropoiesis10, oxidative stress regulation11, metabolic function12, and cellular homeostasis13. In particular, the varied responses to hypoxia and thermal stress across species offer a window into the evolutionary strategies that underpin survival and adaptation8,9,13,14,15,16,17,18,19. Consequently, unravelling these adaptive responses is key to understanding how fish thrive under varying environmental conditions7,8,12,15,20,21,22, which, in turn is essential for predicting their resilience to future climatic shifts7,8.
Thermal stress induces a cascade of physiological and molecular responses in fish, facilitating survival across dynamic environmental conditions. These responses encompass thermoregulatory acclimation, metabolic reprogramming, and behavioural adjustments aimed at optimizing oxygen utilization and energy balance. Under fluctuating thermal regimes, such as those driven by climate variability, fish must dynamically adjust their metabolic pathways to maintain cellular homeostasis and mitigate oxidative stress. Moreover, on similar lines, exposure to hypoxia often drives fish to seek cooler microhabitats, a behavioural thermoregulation strategy that alleviates metabolic strain and enhances survival in energetically demanding conditions. Among them, erythropoiesis, the process of red blood cell formation, is a critical survival mechanism modulated by temperature, oxygen levels, and other environmental factors10,23,24,25. Variations in erythrocyte abundance and cell-cycle progression are tightly regulated, reflecting the adaptive capacity of fish to maintain oxygen homeostasis under stress10,24,25,26,27,28,29. Hypoxic environments trigger metabolic adjustments mediated by hypoxia-inducible factors (HIFs), which regulate downstream genes involved in mitochondrial biogenesis, adipogenesis, and stress responses22,30,31,32,33. These include the NRF-1 (Nuclear Respiratory Factor-1), PGC (Peroxisome Proliferator-Activated Receptor (PPAR) γ Coactivator), PPAR-γ (Peroxisome Proliferator-Activated Receptor (PPAR)-Gamma), and the myocyte enhancer factor-2 (MEF-2), which collectively drive adaptive remodeling processes both at a cellular and molecular level32,33,34,35,36,37,38,39. Additionally, ROS and nitric oxide (NO) act as signaling molecules, orchestrating mitochondrial maintenance and adaptive responses, often mediated by LON protease and cytochrome c oxidase subunit IV (COX-IV) isoforms i1 and i240,41,42,43,44,45.
Research gaps
While fish exhibit well-documented physiological plasticity, the molecular underpinnings driving their adaptive responses across environmental gradients remain insufficiently understood. Although hypoxia- and temperature-induced stress responses in fish have been extensively studied, the majority of investigations have tended to isolate either physiological or molecular mechanisms in separate scenarios, often within controlled experimental frameworks rather than integrated ecological contexts. Hence, it limits a comprehensive understanding of their integrative roles in adaptation. Addressing this gap requires a holistic approach that bridges molecular and physiological frameworks to elucidate mechanisms driving adaptive plasticity in contrasting environments. Previous studies have also advocated the need to elucidate the adaptive significance of physiological traits across taxa and environmental gradients, bridging the gap between physiology, ecology, and adaptation46. This gap is particularly evident in eco-sensitive regions, such as the Indian Sundarbans and the Sub-Himalayan Terai-Dooars, where fish populations experience contrasting environmental conditions. The mangrove estuaries of the Sundarbans, characterized by differential tidal dynamics and fluctuating oxygen levels, imposes different physiological demands as compared to the varying altitudinal geographical terrain, cold, high-velocity streams of the Terai-Dooars, where hypoxia and thermal stress dominate. A comparative study integrating cellular and molecular responses is crucial for deciphering region-specific adaptation strategies. This research endeavour aims to address this gap by integrating cellular physiology with mechanistic molecular analysis, providing a comparative framework for understanding adaptive mechanisms in fish exposed to region-specific environmental pressures.
Objectives
This study hypothesizes that fish species inhabiting environmentally contrasting ecoregions—namely, the Indian Sundarban mangrove estuary and the Sub-Himalayan Terai-Dooars torrential hill streams—exhibit distinct cellular and molecular adaptative traits towards hypoxic and thermal stress. To elucidate the interplay between cellular and molecular adaptation, the current research integrates physiological assessments with gene expression analyses, aiming to uncover region-specific and convergent adaptive responses. Specifically, our study sought to:
-
i.
Characterize erythropoietic dynamics, cell cycle progression, and oxidative stress regulation across fish species from these ecologically divergent environments to determine how oxygen availability and temperature fluctuations shape hematopoietic and metabolic pathways.
-
ii.
Quantify differential gene expression patterns of key molecular regulators—including hypoxia-inducible factors (HIFs), electron transport chain (ETC) modulators, and stress-responsive transcription factors—to delineate the transcriptional landscape governing physiological resilience.
By linking cellular physiology with molecular regulation, this comparative analysis aims to provide novel insights into how fish populations optimize oxygen transport, mitigate oxidative damage, and modulate metabolic functions in response to distinct environmental pressures. These findings will contribute to a broader understanding of vertebrate adaptation mechanisms and offer potential biomarkers for assessing species resilience in the face of climate-driven environmental change.
Materials & methods
Study design
Study area
-
(i)
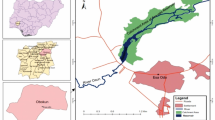
Indian Sundarbans: The Indian Sundarbans, being in the delta region of the Bay of Bengal is the largest mangrove forest and coastal wetland47. The Matla and the Bidyadhari are the two primary tidal rivers forming this estuarine system and owing to its complex topography, this intricate network of upstream rivers is characterized by the spatial and seasonal variations of hydrological parameters. The study area consisted of twenty-five (25) sites randomly selected along the eighty (80) kilometres (km) stretch of the Matla and Bidyadhari rivers along with the Herobhanga rivulet and their adjacently located mudflats (Fig. 1).
Study & sampling sites: Indian (A) and West Bengal (B) Map showing the study areas, Terai-Dooars (C) and Indian Sundarbans (D) region with their respective sampling sites, (E & F).[Software Source: Google Earth Pro: V 7.3.3.7786. Eye alt 78.92 km (Elevation: 143 m) & 93.4 km (Elevation: 23 m). SIO, NOAA, U.S. Navy, NGA, GEBCO. CNES/Airbus 2023, Landsat/Copernicus 2023, Maxar Technologies & TerraMetrics 2023.]
-
(ii)
Sub-Himalayan Terai-Dooars: The Eastern Himalayan region is considered as one of the premiere ‘crisis ecoregions’ and ‘biodiversity hotspots’, being listed in the global 200 ecoregions48. Among the rivers, Murti and Jaldhaka form a riverine network, draining the southern Himalayan slope and eventually debouching on the plains of Terai–Doors49. These torrential hill streams are characterized by tempestuous flow over coarse substratum comprising cobbles, pebbles, sand grains, rocks, etc. and in turn corresponds with the degree of altitudinal slope or gradient with an apparent temperature gradient permitting a significant assemblage of warm and cold-water species49. Fish sampling was performed in nineteen (19) sites along a forty (40) kilometres (km) altitudinal stretch across the gradient of River Murti and Jaldhaka (Fig. 1) that circumambient both the pool and riffle conditions respectively.
Notably, the Sub-Himalayan Terai-Dooars and the Indian Sundarbans represent two hydrographically and physicochemically distinct ecosystems, shaped by contrasting environmental conditions. The Terai-Dooars hill streams (e.g., Murti and Jaldhaka Rivers) are characterized by fast-flowing, freshwater, high-altitude torrents with low salinity, high dissolved oxygen, and coarse substrates comprising gravel and boulders. These streams exhibit significant seasonal variations, influenced by monsoonal precipitation and steep altitudinal gradients, resulting in cold water temperatures and moderate alkalinity50,51,52,53. Conversely, the Sundarbans estuaries (e.g., Matla and the Bidyadhari rivers) form a low-velocity, brackish water network governed by semi-diurnal tides, fluctuating salinities, reduced oxygen levels, and fine silt-clay sediments. The higher water temperature, coupled with estuarine stratification and saline incursions, creates a nutrient-rich and ecologically productive environment54,55,56,57. The Sundarbans also show a higher alkalinity, acidity, and nutrient influx, driven by tidal mixing, compared to the relatively stable ionic composition of the Terai-Dooars. These differences fundamentally influence ichthyofaunal diversity, physiological adaptations, and ecosystem functioning, with fast-swimming, oxygen-demanding species in Terai-Dooars and euryhaline estuarine specialists in the Sundarbans, underscoring their ecological divergence47,51,52,54,57,58,59,60,61,62,63.
Sampling
The sampling methodology was tailored to account for the distinct ecological and geomorphological characteristics of the Indian Sundarbans and Sub-Himalayan Terai-Dooars, ensuring effective representation of fish assemblages across contrasting aquatic environments.
In the Indian Sundarbans, a tidal estuarine system with soft mudflat substrates, fluctuating salinity, and variable water depths, a multi-gear approach was employed. Cast nets, effective for shallow, low-velocity waters, targeted smaller, mobile species, while gill nets, with optimized mesh sizes, sampled species of varying morphologies and swimming behaviors. Hook nets facilitated the capture of benthic and demersal species, ensuring a broad taxonomic representation54,57,64,65. In contrast, the Terai-Dooars hill streams, characterized by fast-flowing waters, rocky substrates, and steep altitudinal gradients, required single-pass electrofishing due to the inefficacy of traditional passive and active netting methods under strong currents and variable flow conditions. Electrofishing, widely recognized for its efficacy in lotic systems, generated controlled electrical fields to temporarily immobilize fish, enabling non-destructive collection while preserving the fragile streambed ecosystem. This approach is well-established for reliable fish population assessments in hydrodynamically complex streams51,52,53,59,66,67. The sampling period and intervals were designed to capture ecological variability across both ecoregions. In the Indian Sundarbans, monthly sampling across tidal cycles accounted for salinity fluctuations, tidal inundation, and seasonal shifts, ensuring representation of transient and resident species. In the Sub-Himalayan Terai-Dooars, the sampling regime accommodated monsoonal flow variations. Multi-year sampling minimized interannual biases, while single-pass electrofishing with removal-based estimation provided reliable data in hydrodynamically complex streams, ensuring ecological validity and robust population assessments. Following previous studies51,52,54,59, we conducted samplings at the two ecoregions through the following methods:
-
(i)
Indian Sundarbans: The samplings were conducted five times per month between March 2019 and February 2022. The fishes were sampled on small boats and trawlers with the help of cast net (with a radius of 1.2 m), gill net (20-metre length with 3 centimeters spacing between two adjacent knots) and hook net (50 m with 50 hooks, each at one metre interval) during high tide. On the contrary, during the times of low tide, transects measuring 100-m were prepared and used at each study site in addition to hand nets (dot net) for sampling amphibious fishes.
-
(ii)
Sub-Himalayan Terai-Dooars: The samplings were conducted five times per month for a period ranging between March 2019 and February 2022. Moreover, for every single sampling attempt made within a single range of 90 m both upstream and downstream from the singular sampling focal point, three consecutive efforts were made which was further followed up by a method of removal based on estimation68. Sampling was conducted using both a cast net and a unified single-pass electrofishing (Apparatus: 1020 NP Ultrasonic Inverter Electro Fisher-Fish Machine Shocker) which was attached to a 12 V power system and a current net (mesh size = 2.5 cm x 2.5 cm) by following methods outlined by Bohlin, et al.68.
Following sampling, euthanization with MS-222 (Tricaine Methanesulfonate) in accordance with AVMA guidelines69, the gill, muscle and head kidney tissues were dissected out and subsequently preserved in: (i) RPMI (Roswell Park Memorial Institute)70 to maintain cellular integrity for cellular analysis and flow cytometric studies and (ii) were immediately flash frozen in liquid nitrogen (liq. N₂) to ensure RNA stability for gene expression analysis.
Species selection criteria for cellular and molecular analysis
Given the ecological nature of our study, the selection of species was primarily guided by their relative abundance, dominance, and availability across our various sampling sites. To rigorously maintain consistency among sampled individuals, all cellular and genetic data were derived from the same individual. Consequently, for molecular data collection, five adult individuals/replicates (n = 5) from each of the 18 representative species (i.e., nine from each ecoregion) were taken. The individuals encompassed both males and females and the timing of the specimen collection was kept consistent throughout the year to avoid any potential bias. This comparative arrangement was employed with goal of highlighting the local scale differences, allowing us to emphasize the disparities present in them.
Flow cytometric analysis
A flow cytometric study was employed to investigate the fraction of the erythropoietic cell populations at each cell cycle phase and to quantify the production of reactive oxygen species (ROS) in the gill epithelium. This analysis could provide relevant baseline information about erythropoietic cell development within the head kidney of fishes, as well as the amount of physiological stress (in the form of ROS generation) that is being imparted on the inhabiting fishes across the two ecoregions.
Preparing single cell suspension
An overall generalized procedure (Supplementary File: Sect. 3) was followed as outlined in Reichard and Asosingh71.
Cell cycle analysis
The corresponding cell cycle phases (namely, the Sub-G0, G0/G1, S, G2/M) were identified based on the cell distribution percentages following procedures (Supplementary File: Sect. 3) and interpretations outlined in earlier studies27,29,72.
Reactive oxygen species (ROS) estimation
ROS levels in selected fish species were assessed via DCFH-DA (2′-7′- Dichlorodihydrofluorescein diacetate) probe-based fluorescence, widely used in prior studies73,74. Samples were analyzed using a flow cytometer, with fluorescence proportional to hydrogen peroxide quantity. Unstained cells served as negative controls, while tert-butyl hydroperoxide-treated cells acted as positive controls. Data analysis employed BD FACSuite™ and R software, with 10,000 events analyzed. Histograms depicted cell counts versus DCFDA fluorescence (485 nm excitation, 530 nm detection), enabling ROS quantification.
Erythropoietic study
For erythropoietic investigations, the tissue imprint or impression method was used and improved upon based on earlier related studies72,75,76. Following this, an assessment of the erythropoietic efficiency across selected species was conducted with the help of Nauber’s double hemocytometer as outlined by Homechaudhuri and Jha77. This study primarily focused on the abundance of four erythropoietic developmental stages: SLH (Small Lymphoid haemoblast), BE (Basophilic Erythroblast), YE (Young Reticulocyte), and ME (Mature Erythrocyte)27,29,72,75.
Comparative gene expression analysis
Standardized protocols were followed for total RNA isolation, cDNA synthesis, and subsequent quantitative real-time PCR (qRT-PCR).
RNA isolation and cDNA synthesis
Following the euthanization of fish specimens, gill, head kidney and muscle tissues were dissected and flash-frozen in liquid nitrogen and brought to the laboratory. Subsequently, RNA was extracted from these tissues following which it was reverse transcribed to cDNA following standard protocols (Supplementary File: Sect. 3).
Gene selection
Thirteen genes were selected to explore the molecular and physiological mechanisms underlying fish adaptation based on established roles in critical adaptive pathways, each directly linked to hypoxia and thermal stress responses (see Supplementary File: Sect. 3 & Supplementary Table S4 for more information).
Quantitative PCR (qPCR)
Primers for the respective genes were designed for amplicons ranging from 100 to 200 bp (Supplementary Table S1) (Integrated DNA Technologies, USA) following previous literature44,45,78,79,80,81,82,83,84. Each qRT-PCR reaction had an initial denaturation step of 15 min at 95 °C, followed by an amplification of the target cDNA for 40 cycles, each cycle consisting of denaturation at 95 °C for 15 s, annealing at the appropriate annealing temperature (ranging from 52.5 °C to 55.5 °C depending on the primer combinations as represented in Supplementary Table S1) for 50 s, and elongation at 72 °C for 15 s. In this study, a 21-day acclimatized Labeo rohita specimen, maintained in an experimental aquarium at 25 °C, was utilized as the control (or calibrator) (For additional information see Supplementary File: Sect. 3). Calculations were made in terms of changes in the relative expression (that were expressed as cycle threshold values, i.e., Ct values) which were normalized by taking the geometric mean of two reference/housekeeping genes85, β-actin and Ribosomal protein L-13 (RPL-13) by utilizing the 2−ΔΔCt method86,87,88,89,90. No-template and reverse transcription negative (no-RT) controls were also included to rule out genomic DNA contamination. All samples were run in technical triplicates. Inter-assay calibration was achieved by including the L. rohita calibrator sample on every plate, allowing for correction of plate-to-plate variations and ensuring comparability across all experimental samples.
A comprehensive analysis of differential gene expression was conducted to assess the adaptive physiological and molecular responses of fish populations inhabiting two contrasting ecoregions—the Indian Sundarbans and the Sub-Himalayan Terai-Dooars. This analysis was structured in two key phases:
-
(a)
Fold-change (FC) analysis: This was derived from cycle threshold (Ct) values to estimate the relative upregulation or downregulation of specific genes across the two populations.
-
(b)
Statistical validation using ANOVA: The Ct values were subjected to an analysis of variance (ANOVA) to determine whether the observed differential expression patterns were statistically significant.
While fold-change values provided insights into the relative magnitude of gene expression changes, ANOVA was employed to rigorously evaluate whether the Ct values for each gene exhibited significant differences between populations, thereby establishing the robustness of these transcriptional variations.
Statistical analyses
Statistical analyses, modelling, and graph generation were performed in R statistical platform (v4.2.1)91. Analyses in the R software were done using the “ggstatsplot”92 package. Additionally, for exploring cellular variability across fish species and ecoregions, a multivariate statistical framework was employed, using Python (Version 3.8)93. Key libraries included “pandas”94 for data handling, “scikit-learn”95 for Principal Component Analysis (PCA), “statsmodels”96 for ANOVA, “matplotlib”97 and “seaborn”98 for data visualizations. In order to establish a comparative association of the cellular and genetic traits across different guilds and ecoregions, we employed Welch’s Analysis of Variance (Welch’s ANOVA) (followed by a Games-Howell post-hoc test), Welch’s t-test, and Yuen-Welch’s t-test respectively for understanding the same. The Welch’s ANOVA is slightly different from the traditional Fischer’s ANOVA and in recent times, the former test has been most popularly considered and recommended by many statisticians and researchers99. While the primary goal of identifying significant differences among three or more groups remains, the test has been adapted to reduce reliance on strict assumptions, making it more flexible and broadly applicable—even in complex or less ideal data conditions. One of the crucial advantages of Welch’s ANOVA is that it performs quite well under heteroscedasticity (or unequal variance), while also serving as the foundation strategy for addressing the issue of non-normality that is present in most of the real-world data100. This is followed by a Games-Howell post-hoc test for testing the significant differences between combinations of various groups (i.e., in this case, this post-hoc test was performed between guilds and ecoregions). This test, while being conceptually similar to the widely used Tukey’s test, does not mandate the requirement of either the equal variance or sample sizes respectively as it was strategically formulated for a non-parametric design thereby aiding its flexibility. It was originally conceived with Welch’s degrees of freedom correction in mind, and it employs the studentized range distribution devised by Tukey (being designated “q”). The Games-Howell test, like other nonparametric tests, is run on the ranking variables and is frequently preferred over other procedures such as Tukey’s test due to its flexibility and self-reliance over the mandatory statistical pre-requisites of the parametric tests101,102. To counteract the problem of multiple comparisons during the Games-Howell post-hoc test we had applied the Holm-Bonferroni Method (also called Holm’s Sequential Bonferroni Procedure). We chose this method over the other tests as it balances stringent Type I error control with higher statistical power than Bonferroni, ensuring robust significance detection. Moreover, unlike Benjamini-Hochberg, it controls family-wise error rate (FWER), suiting ecological and physiological studies rigorously103,104,105. Next, when comparing the Welch’s and Yuen-Welch’s t-tests with the classic Student’s t-test, the former measures are most robust, being non-sensitive towards either unequal variance (Welch) or both heteroscedasticity and non-normality (Yuen-Welch) respectively while comparing for significant differences of the means between two groups106,107. The latter Yuen-Welch’s t-test is an extension of the traditional trimmed means t-test whereby not only data sets having low levels of kurtosis (i.e., having a relatively flatter distribution when compared to the normal distribution) can be evaluated for unequal population variances but can also be employed in cases where there has been a violation of the normality rule108. Subsequently, An Analysis of Variance (ANOVA) based Principal Component Analysis (PCA) was performed109,110,111 for understanding the variation accounted for by the different species and the ecoregion-species interaction effect on the cellular parameters.
Results
Species identification & selection
Representative species (as explained earlier in sub-Sect. 2.1.3) were identified50,51,52,53,54,59,112,113,114,115 in the Indian Sundarbans, and Sub-Himalayan Terai Dooars regions, respectively and were subsequently selected for the cellular and molecular analysis (Supplementary Table S2).
Evaluating cellular traits across ecoregions
Haematopoietic profiling & analysis
Firstly, across ecoregions, there was a significantly higher population of SLH (p(SLH) = 1.26 × 10− 49) and BE (p(BE) = 1.52 × 10− 39) within the representative fish assemblages of the Indian Sundarbans region while on the contrary, the cell populations of YE (p(YE) = 6 × 10− 8) and ME (p(ME) = 3.08 × 10− 36) dominated the Terai-Dooars region (Fig. 2A). Additionally, in terms of erythropoietic efficiency, the inhabiting fishes of the Terai-Dooars region showed a significantly higher trend (p = 7.37 × 10− 27) as compared to the Indian Sundarbans (Fig. 2B).
Cell cycle & ROS analysis
T-test results showed a significant difference (p < 0.05) with respect to all the cell cycle phases across the two ecoregions. There was a contrasting trend among the cell population distribution patterns with the cell populations across the S (p = 3.27 × 10− 22) and G2/M (p = 1.69 × 10− 13) phase being significantly higher within the fish species of the Terai-Dooars region. However, in contrast, the fishes of the Indian Sundarbans showed a significantly higher cell population within the Sub-G0 (p = 0.001) and G0/G1 (p = 2.98 × 10− 4) phases, respectively. In terms of ROS production, t-test results showed significant variation across the two ecoregions (p = 5.3 × 10− 6) with the representative ichthyofauna of the Sub-Himalayan Terai-Dooars region bearing a higher ROS as compared to the Indian Sundarbans (Fig. 2C,D).
Comparative gene expression analysis between populations
The trends across the different gene classes are as follows:
-
(i)
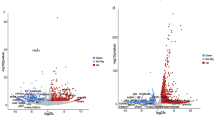
ETC Regulatory Genes: The expression of cytochrome c oxidase subunit IV (COX-IV) isoforms showed distinct ecoregion-specific trends such that COX-IV i1 was upregulated (FC = 2.55) within the Indian Sundarban fishes, and downregulated in the Terai-Dooars fishes (FC = 1.05). In contrast, COX-IV i2 was markedly upregulated (FC = 7.59) in Terai-Dooars fish compared to its 2.49 times FC rise within the Sundarban fishes. The ANOVA results confirmed that the observed differences in Ct values for COX-IV i1 and COX-IV i2 were highly significant (F(2,83.71) = 132.95, p = 1.04 × 10⁻²⁶ for COX-IV i1; F(2,84.89) = 280.32, p = 4.02 × 10⁻³⁸ for COX-IV i2) (Fig. 3 & Supplementary Fig. S1A,B).
-
(ii)
Hypoxia Inducible Genes: Key hypoxia-responsive transcription factors exhibited pronounced ecoregional differences. Fold change analysis indicated that in Terai-Dooars fish, HIF-1α and HIF-2α were upregulated (FC = 5.21 and FC = 5.52), respectively, whereas their expression in Sundarbans fish was comparatively lower (FC = 1.89 and FC = 1.18). Similarly, EPO expression was significantly upregulated (FC = 4.07) in Terai-Dooars, compared to a lower increase (FC = 1.41) within the Sundarban fishes. NRF-1, a key mitochondrial biogenesis regulator, demonstrated markedly elevated expression (FC = 2.89) within the Terai-Dooars fish species, in contrast to a comparatively modest upregulation observed within the Sundarban fish species (FC = 1.42). ANOVA results validated the significance of these expression differences, with HIF-1α (F(2,87.56) = 330.47, p = 1.59 × 10⁻⁴¹), HIF-2α (F(2,87.49) = 402.79, p = 7.32 × 10⁻⁴⁵), EPO (F(2,87.43) = 267.54, p = 5.4 × 10⁻³⁸), and NRF-1 (F(2,87.58) = 150.04, p = 5.12 × 10⁻²⁹) showing highly significant differences between the two populations (Fig. 4 & Supplementary Fig. S1C-F).
-
(iii)
Adaptive Remodelling Genes: Genes encoding stress-induced transcription factors (PGC-1α and PGC-1β) and tissue remodeling regulators (PPAR-γ and MEF-2) followed a similar expression pattern. Fold change analysis showed that PGC-1α and PGC-1β were significantly upregulated in Terai-Dooars fish (FC = 3.52 and FC = 3.61, respectively), while in Sundarbans fish, their expression levels were comparatively lower (FC = 1.38 and FC = 2.14). Similarly, PPAR-γ and MEF-2 were upregulated (FC = 2.53 and FC = 2.92) in Terai-Dooars respectively, whereas in Sundarbans, they exhibited a lower (FC = 1.29 and FC = 1.34) fold change. Statistical validation via ANOVA confirmed that the Ct values of these genes differed significantly across ecoregions (F(2,87.66) = 214.96, p = 1.58 × 10⁻³⁴ for PGC-1α; F(2,87.67) = 212.99, p = 2.2 × 10⁻³⁴ for PGC-1β; F(2,87.64) = 106.28, p = 3.71 × 10⁻²⁴ for PPAR-γ; F(2,87.55) = 153.96, p = 2.15 × 10⁻²⁹ for MEF-2) (Fig. 5 & Supplementary Fig. S1G–J).
-
(iv)
Stress Modulatory Genes: Genes encoding antioxidant and stress-mitigating proteins exhibited significantly higher expression in Terai-Dooars fish, as indicated by fold-change analysis: SOD-2 (FC = 4.73), iNOS (FC = 4.45), and LON (FC = 2.43), compared to Sundarbans fish, which showed 1.25, 1.23, and 1.17 increase, respectively. ANOVA confirmed that these genes exhibited statistically significant differences in Ct values across populations (F(2,87.51) = 324.83, p = 3.19 × 10⁻⁴¹ for SOD-2; F(2,87.47) = 297.36, p = 9.67 × 10⁻⁴⁰ for iNOS; F(2,87.66) = 108.84, p = 1.76 × 10⁻²⁴ for LON) (Fig. 6 & Supplementary Fig. S1 K–M).
Cellular variability
The PCA yielded four primary components (PC 1 to PC 4), collectively explaining 95% of the total cellular variance, with the first component (PC 1) alone capturing 65.4% of the variance (Fig. 7). Subsequently, based on the ANOVA on the PCA scores, PC 1 emerged as the predominant axis of variation, where species identity was the primary determinant (F = 627.2; p < 0.001), further influenced by a significant species-ecoregion interaction (F = 127.7; p < 0.001), suggesting species-specific cellular traits modulated by environmental factors. PC 2 (12%) revealed an additional axis of variability, capturing species-driven differences (F = 4.36; p = 0.003) and significant species-ecoregion interaction (F = 4.29; p < 0.001), indicating that certain cellular traits exhibit plasticity in response to environmental conditions. PC 3 (10.5%) and PC 4 (7.3%) lacked strong species effects but showed significant interaction-driven cellular adaptations (F = 6.0; p < 0.001; F = 4.14; p < 0.001) (Fig. 8).
Principal component analysis with cumulative variance and species clusters: (A) PCA Biplot showing species clustering by ecoregion with variable vectors representing cellular traits, indicating major contributors to cellular differentiation. (B) Cumulative explained variance across principal components, highlighting the proportion of variance captured by each component. For species abbreviations see supplementary table S2.
Distribution of principal components and interaction effects: (A) Boxplot of PC-1 scores across species grouped by ecoregion, illustrating species-specific cellular variability influenced by regional factors. (B) Boxplot of PC-2 scores across species grouped by ecoregion, highlighting secondary species and ecoregion-specific effects. (C) Interaction effect of ecoregion and species on PC-1 scores, showing how environmental context modifies species-specific cellular characteristics. For species abbreviations see supplementary table S2.
Discussion
As per our results, the general trend of cellular traits among the representative fish species varied significantly along the different sites across the two ecoregions.
Mechanistic insights into cellular and molecular adaptations
Haematopoietic patterns and cell cycle regulation
Fish species from the Terai-Dooars region exhibited a significantly higher abundance of mature erythrocytes (YE and ME), reflecting an elevated erythropoietic turnover likely driven by the dual challenge of reduced oxygen partial pressure and increased metabolic demand in cold, fast-flowing hill streams. Conversely, Sundarbans species predominantly displayed immature erythropoietic forms (SLH and BE), suggesting a strategy of maintaining hematopoietic reserves to cope with intermittent hypoxia events. As described in earlier studies72,75,76,116,117, the primary immature stages especially the SLH act as a storage type/precursor cell being housed within the primary erythropoietic tissue of the head kidney whereas the finally differentiated forms (ME) enter circulation to facilitate oxygen transport in response to metabolic demands. Many previous studies27,29,77,118 have suggested the likely cause of this physiological trigger process being attributable to two factors namely, (i) inability to maintain a resting rate of O2 consumption (VO2) that is independent of the ambient [O2] due to a higher physiological oxygen demand118 and (ii) the habitat complexities arising from the differential substratum characteristics coupled with variable water velocity that are being experienced by the fish species of these regions50,52,59,114. At high altitudes, reduced atmospheric pressure lowers the partial pressure of oxygen, limiting its biological availability despite high dissolved concentrations, thereby inducing a chronic physiological hypoxic state within the inhabiting fishes of the Terai-Dooars region17,119,120,121,122. The significantly higher proportion of mature erythrocytes in Terai-Dooars fish suggests an increased hematopoietic turnover, ensuring a steady supply of oxygen carriers to compensate for the low oxygen partial pressure and high metabolic oxygen demand. These findings along with observation made by Jacobsen, et al.17 suggest that while structural adaptations may provide some advantages, physiological modifications such as erythropoietic adjustments are crucial for survival in high-altitude, fast-flowing waters.
This condition in turn triggers a surge in the erythropoietic activity of the inhabiting fishes of the Terai-Dooars region, as is indicated by the predominance of cells in the S and G2/M phases of the cell cycle. This suggests that the primary erythroblasts within these fishes are either in the preparatory or active phases of mitotic division, reflecting a higher level of erythropoiesis27,75. In contrast, fish from the Sundarbans exhibit a higher proportion of cells in the Sub-G0 and G0/G1 phases, reflecting reduced mitotic activity and a correspondingly lower physiological demand under less demanding environmental conditions. Hence, it implies that most of the erythropoietic progenitor cells (for e.g., SLH) stay in an undifferentiated (or immature) state, being rarely induced for mitosis within these resident fish assemblages27,29,72,75.
Patterns of reactive oxygen species (ROS) generation across ecological groups
ROS production was substantially elevated in fish species from the Terai-Dooars region, largely due to elevated metabolic activity coupled with chronic hypoxic exposure in cold, fast-flowing streams49,50,52,53,58,59,114,118. This physiological constraint triggers cellular stress responses aimed at maintaining metabolic homeostasis and ensuring sustained oxygen supply under challenging conditions123. To counteract this hypoxic stress, fish in the Terai-Dooars activate molecular pathways that regulate oxygen utilization and mitochondrial efficiency. Notably, HIF-1α and HIF-2α, which are key regulators of hypoxic response, are significantly upregulated. HIF-1α facilitates the degradation of COX-IV i1 via LON protease, causing a temporary disruption in the electron transport chain (ETC) and leading to transient bursts of ROS generation. This initial ROS production subsequently stimulates a complex regulatory cascade, involving the activation of PGC-1β, MEF-2, and NRF-1, which work cooperatively to upregulate COX-IV i2 expression- an isoform more efficient under hypoxic conditions. The induction of COX-IV i2 re-establishes the ETC, restores mitochondrial function, enhances ATP production, and mitigates excessive ROS accumulation by improving oxidative phosphorylation efficiency. These regulatory cascades are consistent with earlier studies124,125,126,127,128,129demonstrating a strong link between heightened mitochondrial activity, antioxidant defense enzyme expression, and physiological adaptation to hypoxia in aquatic organisms.
Hence, our findings indicate that hypoxia-induced ROS generation in the Terai-Dooars fish populations serves as both a physiological challenge and a regulatory signal for cellular adaptation. The interplay between oxygen availability, metabolic demand, and mitochondrial remodeling highlights how fish optimize energy efficiency and oxidative stress mitigation to survive in environmentally complex habitats. These mechanisms reinforce the broader ecological principle that physiological plasticity is a key determinant of species resilience in fluctuating environments.
Functional divergence in electron transport and hypoxia-responsive genes under ecoregional stressors
The differential expression of cytochrome c oxidase subunit 4 (COX-IV) isoforms—COX-IV i1 and COX-IV i2—across the two ecoregions provides critical insights into the physiological and molecular adaptations of their respective ichthyofaunal communities. Fish inhabiting the Indian Sundarbans exhibited a higher expression of COX-IV i1, which correlates with lower physiological oxygen demands and reduced ROS levels in this relatively stable estuarine environment. In contrast, COX-IV i2 was significantly upregulated in Terai-Dooars fish, aligning with the increased physiological stress imposed by high-altitude, fast-flowing streams. These results are fairly consistent with an earlier study by Semenza, et al.130 that highlighted the role of COX-IV i2 in hypoxia which has been subsequently worked upon by future studies43,131. The differential expression of cytochrome c oxidase subunit 4 isoforms highlights divergent metabolic strategies employed by fish across the two ecoregions. In Terai-Dooars fish, the significant upregulation of COX-IV i2 indicates a physiological shift toward sustaining ATP production under chronic hypoxic conditions, facilitated by enhanced mitochondrial efficiency and remodeling. In contrast, Sundarbans fish exhibited predominant expression of COX-IV i1, aligning with their relatively stable, oxygen-fluctuating estuarine environment and reduced metabolic strain34,43,132.
Furthermore, the concurrent elevation of hypoxia-responsive transcription factors—HIF-1α, HIF-2α, EPO, and NRF-1—in Terai-Dooars species also underscores a comprehensive transcriptional program tailored for hypoxic adaptation. These factors collectively promote erythropoiesis and stimulate mitochondrial biogenesis, reinforcing systemic and cellular resilience under sustained oxygen limitation as has been evidenced by earlier studies22,25,32,33,132,133,134,135,136,137,138.
Functional remodeling of mitochondrial dynamics and stress response pathways
The significant upregulation of adaptive remodeling transcription factors—namely PPARγ, PGC-1α, PGC-1β, and MEF-2—in fish populations from the Terai-Dooars region indicates a finely coordinated molecular adaptation to sustained environmental stressors such as low temperatures, hypoxia, and fluctuating metabolic demands. This expressional elevation coincides with the upregulation of hypoxia-inducible genes, suggesting a concerted physiological response to the complex ecophysiological pressures of cold, fast-flowing hill streams. PPARγ, a central regulator of lipid metabolism, glucose homeostasis, and inflammatory pathways, was markedly expressed, highlighting its role in initiating metabolic adjustments under cold stress by promoting adipogenesis, brown fat recruitment, and glucose utilization—mechanisms consistent with prior findings across diverse vertebrate models45,139,140,141,142. As a master regulator, PPARγ also activates downstream genes such as UCP-1, thereby enhancing thermogenic output and energy balance in response to environmental cues142,143,144.
Parallel upregulation of PGC-1α and PGC-1β further supports this thermogenic remodeling. These co-activators are key orchestrators of mitochondrial biogenesis, facilitating enhanced oxidative capacity and energy production within gill epithelia, a phenomenon widely documented in thermally stressed vertebrates35. Additionally, elevated expression of MEF-2 points to its synergistic role with PGC-1 isoforms and NRF-1 in regulating myocyte differentiation and mitochondrial efficiency under thermal and hypoxic conditions, thereby reinforcing physiological resilience in high-altitude environments45,145,146,147,148.
Complementing these remodeling factors, the stress modulatory genes iNOS, SOD-2, and LON also exhibited significant upregulation in Terai-Dooars fishes relative to Sundarbans counterparts. iNOS and SOD-2 play key roles in managing oxidative damage arising from increased ROS production under metabolic strain, consistent with findings from prior stress-response studies in both vertebrate and invertebrate systems40,149. The heightened expression of LON, a mitochondrial protease with critical chaperonic functions, likely reflects an adaptive response to protein misfolding and mitochondrial dysfunction under prolonged hypoxic and thermal stress, as supported by experimental evidence on its role in the proper functioning and maintenance of the mitochondrial electron transport pathway under hypoxic or thermally stressed conditions132,150,151.
Physiological hypoxic stress and adaptive mechanisms
Despite high dissolved oxygen levels, Terai-Dooars fish experience physiological hypoxia due to lower oxygen partial pressure at high altitudes. The upregulation of HIF-1α and HIF-2α indicates a molecular response to sustain oxygen homeostasis, activating EPO for enhanced erythropoiesis and COX-IV i2, which optimizes electron transport under hypoxic conditions. The higher prevalence of mature erythrocytes supports efficient oxygen transport to meet metabolic demands. Increased ROS production, a byproduct of hypoxia-driven mitochondrial activity, necessitates antioxidant responses, evident from the upregulation of SOD-2 and LON, which counteract oxidative stress while preserving mitochondrial function. In contrast, Indian Sundarbans fish exhibit a hematopoietic reserve strategy, with higher proportions of SLH and BE, ensuring a rapid response to episodic hypoxia. Their reliance on COX-IV i1 and lower ROS levels reflects adaptation to fluctuating oxygen conditions with minimal oxidative stress (Figs. 9 and 10).
Schematic illustration of underlying physiological mechanisms regulating physiological hypoxia & thermal stress in fishes: Figure illustrating the overall mechanism of aquatic adaptation in response to hypoxic and thermal stress through the regulation of several genes that promote adipogenesis (hence as a result boosting mitochondrial biogenesis), myocyte differentiation, ETC regulation by degrading temperature-sensitive COX-IV isoform and inhibiting free radical formation.
Schematic illustration of underlying physiological mechanisms regulating erythropoiesis in fishes in response to physiological hypoxia: Figure illustrating the overall mechanism of aquatic adaptation in response to hypoxic stress through the increased differentiation of the erythropoietic cell lineage starting from the SLH to ME that are eventually released into the blood circulation (in response to a high oxygen demand that forms a characteristic feature of hill stream environment).
Thermal stress and its implications
Terai-Dooars fish face significant thermal stress, reflected in the upregulation of PPARγ, PGC-1α, PGC-1β, and MEF-2, which regulate mitochondrial biogenesis, metabolic remodeling, and adipogenesis. Increased PGC-1α and PGC-1β expression suggests enhanced mitochondrial proliferation, ensuring metabolic efficiency for thermal acclimation. The activation of PPARγ highlights its role in lipid metabolism and adipose tissue remodeling, critical for energy balance under cold stress, mirroring adaptation mechanisms seen in mammals and amphibians. MEF-2 and NRF-1 upregulation further underscores transcriptional regulation supporting stress adaptation. The elevated NRF-1 expression, a key mitochondrial regulator, reinforces the hypothesis that cold-induced stress necessitates robust mitochondrial function for metabolic sustainability. These findings align with previous studies emphasizing mitochondrial efficiency as a core mechanism of thermal acclimation (Figs. 9 and 10).
Based on our results, we summarize the major contributing factors for understanding the possible mechanism of physiological adaptation under hypoxic and thermally stressed conditions. Thermal stress upregulates PGC-1α and 1β, leading to PPAR-γ activation, promoting adipogenesis and mitochondrial biogenesis. Hypoxia activates HIF-1α and 2α, inducing LON protease, degrading COX-IV i1, disrupting ETC, and generating ROS and NO. PGC proteins activate MEF-2 and NRF-1, along with HIF-2α, initiating COX-IV i2, reinstating ETC, mitochondrial replication, and inhibiting free radicals. Finally, in terms of haematopoiesis, PGC-1α, NRF-1, and HIF activate EPO-I genes, initiating erythropoiesis, leading to the differentiation of SLH to mature erythrocytes during hypoxic conditions.
Integrated physiological adaptation and evolutionary convergence across ecoregions
This study highlights the dynamic and integrative nature of physiological adaptations in freshwater fish species across two contrasting ecoregions—the high-altitude, fast-flowing Terai-Dooars streams and the lowland, tidally influenced Indian Sundarbans. Principal Component Analysis (PCA) revealed strong interspecies clustering along PC1, which accounted for 65.4% of the total variance, emphasizing that species-specific traits predominantly drive physiological differentiation. PC2 through PC4, while accounting for smaller portions of variance, revealed significant species-ecoregion interactions, indicating that environmental context modulates physiological expression beyond intrinsic genetic factors. Species from the Terai-Dooars exhibited elevated erythropoietic efficiency, increased ROS modulation, and a predominance of cells in mitotically active phases, likely reflecting their physiological demand for enhanced oxygen transport under hypobaric and high-velocity stream conditions. Conversely, Sundarbans fish displayed traits optimized for coping with episodic hypoxia, including hematopoietic reserve maintenance and reliance on less energetically demanding pathways. These findings underscore how regional environmental pressures sculpt adaptive strategies, reinforcing the role of cellular plasticity as a critical determinant of survival. Importantly, the convergence of adaptive mechanisms—such as upregulation of HIFs, ROS modulation, and mitochondrial remodeling—across taxonomically diverse species suggests that these are not merely species-specific idiosyncrasies but represent functional adaptations driven by shared ecological stressors thereby supporting the broader ecological theory that species occupying similar environmental niches often evolve parallel physiological traits, regardless of phylogenetic lineage152,153. Moreover, while long-term evolutionary processes undoubtedly underpin species differentiation, acute physiological mechanisms such as HIF activation, oxidative stress regulation, and metabolic remodeling serve as immediate buffers against environmental fluctuations154,155,156,157. The expression of erythropoietic markers, mitochondrial regulators, and stress-response genes such as SOD-2, iNOS, and LON—particularly in Terai-Dooars populations—demonstrates how functional convergence emerges from shared adaptive demands. These findings provide a valuable framework for identifying biomarkers of resilience and forecasting species vulnerability under climate-induced stressors156,158,159,160. Our findings, consistent with Schulte1,2, highlight an interplay of evolutionary history and ecological pressures in shaping adaptive mechanisms. The differential expression of erythropoietic markers, mitochondrial regulators, and stress-modulatory genes underscores functional convergence in response to environmental constraints. These insights emphasize the need to integrate physiological traits into conservation strategies, ensuring the protection of vulnerable populations and ecosystem resilience.
From a conservation standpoint, understanding the ecological and evolutionary underpinnings of such physiological traits is pivotal. Ecosystems like the Eastern Himalayas and Indian Sundarbans represent biodiversity hotspots where regional heterogeneity plays a vital role in sustaining adaptive capacity. Protecting these habitats ensures the preservation of physiological diversity critical for population persistence under future environmental change. Future research should expand this eco-physiological framework by incorporating multi-omics data and cross-ecoregion comparisons to disentangle species-specific responses from community-level convergence. This will enhance our predictive understanding of how environmental pressures shape adaptation across temporal and spatial scales.
Limitations and future directions
While this study provides key insights into fish adaptation to environmental stressors, its correlative approach limits causative inference. The examined conditions offer only a snapshot of adaptation, potentially missing long-term ecological and evolutionary dynamics. Future research should integrate longitudinal environmental data, multi-omics approaches, and controlled experiments to validate observed physiological and genetic responses.
Conclusion
This study establishes a comparative framework for understanding how fish communities adapt to distinct environmental constraints, emphasizing community-level physiological responses over purely phylogenetic influences. By identifying key physiological pathways as potential biomarkers for resilience, we provide a predictive model for assessing species vulnerability under future climate scenarios161. By doing so, we highlight physiological pathways that could serve as predictive markers for assessing vulnerability or adaptability under future climate scenarios. Our findings underscore the interplay between environmental heterogeneity and species-specific physiological traits, highlighting their role in shaping adaptation strategies. This could lead to direct implications for conservation efforts, particularly in biodiversity-rich yet vulnerable regions such as the Eastern Himalayas and coastal mangroves of the Indian Sundarbans. Given the increasing threats posed by climate change, integrating cellular, genetic, and ecological insights is crucial for predicting environmental consequences and identifying vulnerable habitats, such as the Indian Sundarbans and the Terai-Dooars hill streams.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Schulte, P. M. What is environmental stress? Insights from fish living in a variable environment. J. Exp. Biol. 217, 23–34. https://doi.org/10.1242/jeb.089722 (2014).
Schulte, P. M. Responses to environmental stressors in an estuarine fish: interacting stressors and the impacts of local adaptation. J. Therm. Biol. 32, 152–161. https://doi.org/10.1016/j.jtherbio.2007.01.012 (2007). https://doi.org:.
Nikinmaa, M. & Rees, B. B. Oxygen-dependent gene expression in fishes. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 288, R1079–R1090. https://doi.org/10.1152/ajpregu.00626.2004 (2005).
Dallas, H. The effect of water temperature on aquatic organisms: a review of knowledge and methods for assessing biotic responses to temperature. Water Res. Comm. Rep. KV 213 (2009).
Cabrerizo Marco, J. & Emilio, M. Temperature fluctuations in a warmer environment: impacts on microbial plankton. Fac. Rev. 29 https://doi.org/10.12703/r/10-9 (2021).
Zhu, C. D., Wang, Z. H. & Yan, B. Strategies for hypoxia adaptation in fish species: a review. J. Comp. Physiol. B. 183, 1005–1013. https://doi.org/10.1007/s00360-013-0762-3 (2013).
Doney, S. C. et al. Climate change impacts on marine ecosystems. Annual Rev. Mar. Sci. 4, 11–37. https://doi.org/10.1146/annurev-marine-041911-111611 (2011).
Ficke, A. D., Myrick, C. A. & Hansen, L. J. Potential impacts of global climate change on freshwater fisheries. Rev. Fish Biol. Fish. 17, 581–613. https://doi.org/10.1007/s11160-007-9059-5 (2007).
Rodriguez-Dominguez, A., Connell, S. D., Leung, J. Y. S. & Nagelkerken, I. Adaptive responses of fishes to climate change: feedback between physiology and behaviour. Sci. Total Environ. 692, 1242–1249. https://doi.org/10.1016/j.scitotenv.2019.07.226 (2019).
Nikinmaa, M. Environmental regulation of the function of Circulating erythrocytes via changes in age distribution in teleost fish: possible mechanisms and significance. Mar. Genom. 49, 100717. https://doi.org/10.1016/j.margen.2019.100717 (2020).
Dong, X. Y., Qin, J. G. & Zhang, X. M. Fish adaptation to oxygen variations in aquaculture from hypoxia to hyperoxia. J. Fisheries Aquaculture. 2, 23–28 (2011).
Crawshaw, L. I. & Podrabsky, J. E. Encyclopedia of Fish Physiology. 758–764 (eds Farrell, A. P.) (Academic Press, 2011).
Roessig, J. M., Woodley, C. M., Cech, J. J. & Hansen, L. J. Effects of global climate change on marine and estuarine fishes and fisheries. Rev. Fish Biol. Fish. 14, 251–275. https://doi.org/10.1007/s11160-004-6749-0 (2004).
Johnson, M. F. et al. Rising water temperature in rivers: ecological impacts and future resilience. WIREs Water. 11, e1724. https://doi.org/10.1002/wat2.1724 (2024). https://doi.org:.
Barbarossa, V. et al. Threats of global warming to the world’s freshwater fishes. Nat. Commun. 12, 1701. https://doi.org/10.1038/s41467-021-21655-w (2021).
Ibarz, A. et al. Gilthead sea Bream liver proteome altered at low temperatures by oxidative stress. PROTEOMICS 10, 963–975. https://doi.org/10.1002/pmic.200900528 (2010).
Jacobsen, D., Rostgaard, S. & Vásconez, J. J. Are macroinvertebrates in high altitude streams affected by oxygen deficiency? Freshw. Biol. 48, 2025–2032. https://doi.org/10.1046/j.1365-2427.2003.01140.x (2003). https://doi.org:.
Jenny, J. P. et al. Inherited hypoxia: A new challenge for reoligotrophicated lakes under global warming. Glob. Biogeochem. Cycles. 28, 1413–1423. https://doi.org/10.1002/2014GB004932 (2014).
McCarty, J. P., Wolfenbarger, L. L. & Wilson, J. A. Encyclopedia of Life Sciences 1–13 (2017).
Jenny, J. P. et al. Global spread of hypoxia in freshwater ecosystems during the last three centuries is caused by rising local human pressure. Glob. Change Biol. 22, 1481–1489. https://doi.org/10.1111/gcb.13193 (2016).
Jonz, M. G., Buck, L. T., Perry, S. F., Schwerte, T. & Zaccone, G. Sensing and surviving hypoxia in vertebrates. Ann. N. Y. Acad. Sci. 1365, 43–58. https://doi.org/10.1111/nyas.12780 (2016).
Mandic, M., Todgham, A. E. & Richards, J. G. Mechanisms and evolution of hypoxia tolerance in fish. Proc. Royal Soc. B Biol. Sci. 276, 735–744. https://doi.org/10.1098/rspb.2008.1235 (2009).
Lewis, J. M., Hori, T. S., Rise, M. L., Walsh, P. J. & Currie, S. Transcriptome responses to heat stress in the nucleated red blood cells of the rainbow trout (Oncorhynchus mykiss). Physiol. Genom. 42, 361–373. https://doi.org/10.1152/physiolgenomics.00067.2010 (2010).
Lewis, J. M., Klein, G., Walsh, P. J. & Currie, S. Rainbow trout (Oncorhynchus mykiss) shift the age composition of Circulating red blood cells towards a younger cohort when exposed to thermal stress. J. Comp. Physiol. B. 182, 663–671. https://doi.org/10.1007/s00360-012-0650-2 (2012).
Lai, J. C. C., Kakuta, I., Mok, H. O. L., Rummer, J. L. & Randall, D. Effects of moderate and substantial hypoxia on erythropoietin levels in rainbow trout kidney and spleen. J. Exp. Biol. 209, 2734–2738. https://doi.org/10.1242/jeb.02279 (2006).
Götting, M. & Nikinmaa, M. J. Transcriptomic analysis of young and old erythrocytes of fish. Front. Physiol. 8 https://doi.org/10.3389/fphys.2017.01046 (2017).
Chaudhuri, A., Gangopadhyay, K., Podder, A. & Homechaudhuri, S. A quantitative and qualitative evaluation of haematopoiesis in three habitat specialist fishes, Acanthocobitis botia, Devario aequipinnatus and Barilius barna from a hill stream environment. Int. J. Zool. Appl. Biosci. 2, 218–226. https://doi.org/10.5281/zenodo.1312080 (2017).
Murad, A. & Houston, A. H. Maturation of the goldfish (Carassius auratus) erythrocyte. Comp. Biochem. Physiol. Part. A: Physiol. 102, 107–110. https://doi.org/10.1016/0300-9629(92)90019-M (1992). https://doi.org:.
Chaudhuri, A., Gangopadhyay, K. & Homechaudhuri, S. Understanding haematopoiesis as survival strategy in two characteristic habitat specialist fishes inhabiting a hill stream environment. J. Environ. Biol. 38, 477–482. https://doi.org/10.22438/jeb/38/3/MS-245 (2017).
Heise, K. et al. Effects of seasonal and latitudinal cold on oxidative stress parameters and activation of hypoxia inducible factor (HIF-1) in zoarcid fish. J. Comp. Physiol. B. 177, 765–777. https://doi.org/10.1007/s00360-007-0173-4 (2007).
Rytkönen, K. T., Ryynänen, H. J., Nikinmaa, M. & Primmer, C. R. Variable patterns in the molecular evolution of the hypoxia-inducible factor-1 alpha (HIF-1α) gene in teleost fishes and mammals. Gene 420, 1–10. https://doi.org/10.1016/j.gene.2008.04.018 (2008).
Mandic, M., Best, C. & Perry, S. F. Loss of hypoxia-inducible factor 1α affects hypoxia tolerance in larval and adult zebrafish (Danio rerio). Proc. Royal Soc. B Biol. Sci. 287, 20200798. https://doi.org/10.1098/rspb.2020.0798 (2020).
Guan, L., Chi, W., Xiao, W., Chen, L. & He, S. Analysis of hypoxia-inducible factor alpha polyploidization reveals adaptation to Tibetan plateau in the evolution of schizothoracine fish. BMC Evol. Biol. 14, 192. https://doi.org/10.1186/s12862-014-0192-1 (2014).
Porplycia, D. S. Expression Patterns of Cytochrome C Oxidase Subunit 4 Paralogs in Relation To Development and Environmental Challenges Master of Science thesis, (Queen’s University, 2015).
Scarpulla, R. C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Et Biophys. Acta (BBA) - Mol. Cell. Res. 1813, 1269–1278. https://doi.org/10.1016/j.bbamcr.2010.09.019 (2011). https://doi.org:.
Ganassi, M. et al. Distinct functions of alternatively spliced isoforms encoded by zebrafish mef2ca and mef2cb. Biochimica et Biophysica Acta (BBA). Gene Regul. Mech. 1839, 559–570. https://doi.org/10.1016/j.bbagrm.2014.05.003 (2014).
He, M. et al. Common carp mef2 genes: evolution and expression. Genes (Basel) 10 (2019).
Lv, F., Zhu, C., Yan, X., Wang, X. & Liu, D. Generation of a mef2aa:EGFP Transgenic zebrafish line that expresses EGFP in muscle cells. Fish Physiol. Biochem. 43, 287–294. https://doi.org/10.1007/s10695-016-0286-3 (2017).
Sengupta, A., Mukherjee, S., Bhattacharya, S., Saha, S. K. & Chattopadhyay, A. Expression pattern of myogenic regulatory transcription factor mRNAs in the embryo and adult Labeo Rohita (Hamilton, 1822). Int. J. Zool. 2014 (259685). https://doi.org/10.1155/2014/259685 (2014).
Jeon, M. J. et al. Mitochondrial dysfunction and activation of iNOS are responsible for the palmitate-induced decrease in adiponectin synthesis in 3T3L1 adipocytes. Exp. Mol. Med. 44, 562–570. https://doi.org/10.3858/emm.2012.44.9.064 (2012).
Knott, A. B. & Bossy-Wetzel, E. Impact of nitric oxide on metabolism in health and age-related disease. Diabetes Obes. Metab. 12 (Suppl 2), 126–133. https://doi.org/10.1111/j.1463-1326.2010.01267.x (2010).
Pinti, M. et al. Emerging role of Lon protease as a master regulator of mitochondrial functions. Biochim. Et Biophys. Acta (BBA) - Bioenergetics. 1857, 1300–1306. https://doi.org/10.1016/j.bbabio.2016.03.025 (2016). https://doi.org:.
Hüttemann, M., Lee, I., Liu, J. & Grossman, L. I. Transcription of mammalian cytochrome C oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 274, 5737–5748. https://doi.org/10.1111/j.1742-4658.2007.06093.x (2007).
Little, A. G., Kocha, K. M., Lougheed, S. C. & Moyes, C. D. Evolution of the nuclear-encoded cytochrome oxidase subunits in vertebrates. Physiol. Genom. 42, 76–84. https://doi.org/10.1152/physiolgenomics.00015.2010 (2010).
Duggan, A. T. Control of Cytochrome c oxidase Biosynthesis in the Thermal Remodeling of White Muscle of Two Cyprinid Minnows Master of Science thesis, (Queen’s University, 2010).
Weiher, E. et al. Advances, challenges and a developing synthesis of ecological community assembly theory. Philosophical Trans. Royal Soc. B: Biol. Sci. 366, 2403–2413. https://doi.org/10.1098/rstb.2011.0056 (2011).
Mukherjee, S., Chaudhuri, A., Kundu, N., Mitra, S. & Homechaudhuri, S. Comprehensive analysis of fish assemblages in relation to seasonal environmental variables in an estuarine river of Indian sundarbans. Estuaries Coasts. 36, 192–202. https://doi.org/10.1007/s12237-012-9558-z (2013).
Brooks, T. M. et al. Global biodiversity conservation priorities. Science 313, 58–61. https://doi.org/10.1126/science.1127609 (2006).
Rudra, K. Rivers of the Ganga-Brahmaputra-Meghna Delta: A Fluvial Account of Bengal XVII Geography of the Physical Environment Ch. 3, 27–47 (Springer, Cham, Springer International Publishing, 2018).
Panja, S. et al. Comparative assessment of piscine beta diversity profile and key determinant environmental factors in two freshwater rivers of variable Spatial scale in Dooars, West Bengal, India. Trop. Ecol. 62, 589–599. https://doi.org/10.1007/s42965-021-00171-4 (2021).
Panja, S., Podder, A., Chakrabarty, M. & Homechaudhuri, S. Species conservation target for freshwater fishes inhabiting Bengal sub-tropical montane rivers of Eastern Himalayas: an indexed value approach for priority determination. Aquat. Ecol. 56, 1323–1346. https://doi.org/10.1007/s10452-022-09973-7 (2022).
Panja, S., Podder, A. & Homechaudhuri, S. Evaluation of aquatic ecological systems through dynamics of ichthyofaunal diversity in a Himalayan torrential river. Murti Limnol. 82, 125779. https://doi.org/10.1016/j.limno.2020.125779 (2020).
Panja, S., Podder, A. & Homechaudhuri, S. Modeling the climate change impact on the habitat suitability and potential distribution of an economically important hill stream fish, neolissochilus Hexagonolepis, in the Ganges–Brahmaputra basin of Eastern Himalayas. Aquat. Sci. 83, 66. https://doi.org/10.1007/s00027-021-00820-9 (2021).
Podder, A. et al. Patterns of morphological traits shaping the feeding guilds in the intertidal mudflat fishes of the Indian sundarbans. J. Fish Biol. 99, 1010–1031. https://doi.org/10.1111/jfb.14800 (2021).
Chaudhuri, A., Mukherjee, S. & Homechaudhuri, S. Diet composition and digestive enzymes activity in carnivorous fishes inhabiting mudflats of Indian Sundarban estuaries. Turkish J. Fish. Aquat. Sci. 12 https://doi.org/10.4194/1303-2712-v12_2_11 (2012).
Chaudhuri, A., Mukherjee, S. & Homechaudhuri, S. Food partitioning among carnivores within feeding guild structure of fishes inhabiting a mudflat ecosystem of Indian sundarbans. Aquat. Ecol. 48, 35–51. https://doi.org/10.1007/s10452-013-9464-x (2013).
Chaudhuri, A., Mukherjee, S. & Homechaudhuri, S. Seasonal dynamics of fish assemblages in an intertidal mudflat of Indian sundarbans. Scientia Mar. 77, 301–311. https://doi.org/10.3989/scimar.03766.15A (2013).
Panja, S., Podder, A. & Homechaudhuri, S. Understanding the impact of future Climatic scenarios upon key environmental factors that determine piscine assemblage of a torrential upland river of Eastern Himalayas, India. Curr. Sci. 120, 1471–1481. https://doi.org/10.18520/cs/v120/i9/1471-1481 (2021).
Podder, A. et al. Role of morphology in shaping the feeding niche of fishes inhabiting Eastern Himalayan torrential rivers of West Bengal, India. Ecol. Freshw. Fish. 30, 444–465. https://doi.org/10.1111/eff.12596 (2021).
Chakrabarty, M. & Homechaudhuri, S. Analysis of trophic gradient through environ-mental filter influencing fish assemblage structure of the river Teesta in Eastern Himalayas. (2014).
Chakrabarty, M., Panja, S. & Homechaudhuri, S. Predictive modeling of a Fish-Based index of biotic integrity in assessing environmental degradation in the lower Teesta river basin of Eastern Himalayan foothills, India. Proc. Zool. Soc. 76, 22–35. https://doi.org/10.1007/s12595-023-00464-7 (2023).
Chaudhuri, A., Podder, A., Biswas, M., Roy, A. & Homechaudhuri, S. Characterization of chloride cells coupled with immunolocalization of Na + K + ATPase in fishes of different migratory guilds of Indian sundarbans. Reg. Stud. Mar. Sci. https://doi.org/10.1016/j.rsma.2021.101980 (2021).
Kundu, N., Chaudhuri, A., Mukherjee, S., Sen, S. & Homechaudhuri, S. Seasonal fish diversity under tidal influence in the intertidal mudflats of Indian sundarbans. Indian J. Fisheries. 59, 43–52 (2012).
Chaudhuri, A., Mukherjee, S. & Homechaudhuri, S. Food partitioning among carnivores within feeding guild structure of fishes inhabiting a mudflat ecosystem of Indian sundarbans. Aquat. Ecol. 48, 35–51. https://doi.org/10.1007/s10452-013-9464-x (2014).
Hoque Mozumder, M. M., Shamsuzzaman, M. M., Rashed-Un-Nabi, M. & Karim, E. Social-ecological dynamics of the small scale fisheries in Sundarban Mangrove forest, Bangladesh. Aquaculture Fisheries. 3, 38–49. https://doi.org/10.1016/j.aaf.2017.12.002 (2018).
Goutte, A., Molbert, N., Guérin, S., Richoux, R. & Rocher, V. Monitoring freshwater fish communities in large rivers using environmental DNA metabarcoding and a long-term electrofishing survey. J. Fish Biol. 97, 444–452. https://doi.org/10.1111/jfb.14383 (2020).
Pottier, G. et al. Electrofishing in streams of low water conductivity but high biodiversity value: challenges, limits and perspectives. Fish. Manag. Ecol. 27, 52–63. https://doi.org/10.1111/fme.12384 (2020).
Bohlin, T., Hamrin, S., Heggberget, T. G., Rasmussen, G. & Saltveit, S. J. Electrofishing — Theory and practice with special emphasis on salmonids. Hydrobiologia 173, 9–43. https://doi.org/10.1007/bf00008596 (1989).
Leary, S. et al. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition121 (American Veterinary Medical Association (AVMA), 2020).
Renshaw, S. A. et al. The addition of RPMI significantly improves the cellularity of cerebrospinal fluid cytology specimens over time. Cancer Cytopathol. 121, 271–274. https://doi.org/10.1002/cncy.21243 (2013).
Reichard, A. & Asosingh, K. Best practices for Preparing a single cell suspension from solid tissues for flow cytometry. Cytometry Part. A. 95, 219–226. https://doi.org/10.1002/cyto.a.23690 (2019).
Gangopadhyay, K., Bhattacharyya, M. & Homechaudhuri, S. Pesticide-induced alterations of Hemopoietic functions in Anabas testudineus inhabiting wetlands in agricultural landscape. Toxicol. Environ. Chem. 95, 806–813. https://doi.org/10.1080/02772248.2013.808846 (2013).
Eruslanov, E. & Kusmartsev, S. Advanced Protocols in Oxidative Stress II. 57–72 (eds Armstrong, D.) (Humana, 2010).
Mukherjee, S., Ray, M. & Ray, S. Shift in aggregation, ROS generation, antioxidative defense, lysozyme and acetylcholinesterase activities in the cells of an Indian freshwater sponge exposed to washing soda (sodium carbonate). Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 187, 19–31. https://doi.org/10.1016/j.cbpc.2016.05.001 (2016).
Gangopadhyay, K. & Homechaudhuri, S. Descriptive characteristics of Haemopoietic cell lineages in a facultative air breathing fish Clarias Batrachus (L). Turkish J. Zool. 35, 737–746. https://doi.org/10.3906/zoo-0905-37 (2011).
Kondera, E. Haematopoiesis in the head kidney of common carp (Cyprinus Carpio L.): a morphological study. Fish Physiol. Biochem. 37, 355–362. https://doi.org/10.1007/s10695-010-9432-5 (2011).
Homechaudhuri, S. & Jha, A. A technique to evaluate the erythropoietic efficiency in fish. Asian Fisheries Sci. 14, 453–455 (2001).
Boskovic, S. et al. Characterization of zebrafish (Danio rerio) muscle Ankyrin repeat proteins reveals their conserved response to endurance exercise. PLOS ONE. 13, e0204312. https://doi.org/10.1371/journal.pone.0204312 (2018).
Kim, J. O., Oh, S. Y., Oh, M. J. & Nishizawa, T. Universal primers targeting beta actin genes of several fish species. Fish. Pathol. 48, 56–58. https://doi.org/10.3147/jsfp.48.56 (2013).
Lang, X., Wang, L. & Zhang, Z. Stability evaluation of reference genes for real-time PCR in zebrafish (Danio rerio) exposed to cadmium chloride and subsequently infected by bacteria Aeromonas hydrophila. Aquat. Toxicol. 170, 240–250. https://doi.org/10.1016/j.aquatox.2015.11.029 (2016).
Lin, Y. F., Sam, J. & Evans, T. Sirt1 promotes tissue regeneration in zebrafish through regulating the mitochondrial unfolded protein response. iScience 24 https://doi.org/10.1016/j.isci.2021.103118 (2021).
Rahman, M. S. & Thomas, P. Effects of hypoxia exposure on hepatic cytochrome P450 1A (CYP1A) expression in Atlantic croaker: molecular mechanisms of CYP1A Down-Regulation. PLOS ONE. 7, e40825. https://doi.org/10.1371/journal.pone.0040825 (2012).
Thomas, P., Rahman, M. S., Khan, I. A. & Kummer, J. A. Widespread endocrine disruption and reproductive impairment in an estuarine fish population exposed to seasonal hypoxia. Proc. Royal Soc. B: Biol. Sci. 274, 2693–2702. https://doi.org/10.1098/rspb.2007.0921 (2007).
He, M. et al. Common carp mef2 genes: evolution and expression. Genes (Basel). 10, 588. https://doi.org/10.3390/genes10080588 (2019).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, (2002).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2 – ∆∆CT method. Methods 25, 402–408. https://doi.org/10.1006/meth.2001.1262 (2001).
Hellemans, J. & Vandesompele, J. Quantitative Real-Time PCR: Methods and Protocols (eds Roberto Biassoni & Alessandro Raso) 19–26 (Springer New York, 2014).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. https://doi.org/10.1007/s13353-013-0173-x (2013).
Rojas-Hernandez, N., Véliz, D. & Vega-Retter, C. Selection of suitable reference genes for gene expression analysis in gills and liver of fish under field pollution conditions. Sci. Rep. 9, 3459. https://doi.org/10.1038/s41598-019-40196-3 (2019).
Sahoo, P. K., Parida, S., Parida, S., Parida, P. & Paul, A. Stability evaluation and validation of appropriate reference genes for real-time PCR expression analysis of immune genes in the Rohu (Labeo rohita) skin following argulosis. Sci. Rep. 13, 2660. https://doi.org/10.1038/s41598-023-29325-1 (2023).
R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. (2020).
Patil, I. Visualizations with statistical details: the ‘ggstatsplot’ approach. J. Open Source Softw. 6, 1–5. https://doi.org/10.21105/joss.03167 (2021).
Van Rossum, G. & Drake, F. L. Python 3 Reference Manual (CreateSpace, 2009).
McKinney, W. SciPy 51–56.
Pedregosa, F. et al. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Seabold, S. & Perktold, J. Statsmodels: econometric and statistical modeling with python. SciPy 7 (2010).
Hunter, J. D. & Matplotlib A 2D graphics environment. Comput. Sci. Eng. 9, 90–95. https://doi.org/10.1109/MCSE.2007.55 (2007).
Waskom, M. L. Seaborn: statistical data visualization. J. Open Source Softw. 6 https://doi.org/10.21105/joss.03021 (2021).
Celik, N. & Welch’s, A. N. O. V. A. Heteroskedastic skew-t error terms. Commun. Stat. Theory Methods 1–12. https://doi.org/10.1080/03610926.2020.1788084 (2020).
Wilcox, R. R. Applying Contemporary Statistical Techniques (ed Rand R. Wilcox) Ch. 9, 285–328 (Academic Press, 2003).
Games, P. A. & Howell, J. F. Pairwise multiple comparison procedures with unequal N’s and/or variances: A Monte Carlo study. J. Educational Stat. 1, 113–125. https://doi.org/10.3102/10769986001002113 (1976).
Sauder, D. C. & DeMars, C. E. An updated recommendation for multiple comparisons. Adv. Methods Practices Psychol. Sci. 2, 26–44. https://doi.org/10.1177/2515245918808784 (2019).
Haynes, W. Encyclopedia of Systems Biology (eds Werner Dubitzky, Olaf Wolkenhauer, Kwang-Hyun Cho, & Hiroki Yokota) 902–902 (Springer New York, (2013).
Hommel, G. A stagewise rejective multiple test procedure based on a modified bonferroni test. Biometrika 75, 383–386. https://doi.org/10.1093/biomet/75.2.383 (1988).
Giacalone, M., Agata, Z., Cozzucoli, P. C. & Alibrandi, A. Bonferroni-Holm and permutation tests to compare health data: methodological and applicative issues. BMC Med. Res. Methodol. 18, 81. https://doi.org/10.1186/s12874-018-0540-8 (2018).
Keselman, H. J., Othman, A. R., Wilcox, R. R. & Fradette, K. The new and improved Two-Sample t test. Psychol. Sci. 15, 47–51 (2004).
Laura Lu, Z. & Yuan, K. H. Encyclopedia of Research Design. 1, 1620–1623 (eds Salkind, N. J.) (SAGE, 2010).
Wilcox, R. R. Introduction to Robust Estimation and Hypothesis Testing (ed Rand R. Wilcox) Ch. 5, 145–234 (Academic Press, 2017).
Abapihi, B. et al. ANOVA on principal component as an alternative to MANOVA. Journal of Physics: Conference Series 1899, 012103 (2021). https://doi.org/10.1088/1742-6596/1899/1/012103
Harrington, P. B. et al. Analysis of variance–principal component analysis: A soft tool for proteomic discovery. Anal. Chim. Acta. 544, 118–127. https://doi.org/10.1016/j.aca.2005.02.042 (2005). https://doi.org:.
Jolliffe, I. T. & Cadima, J. Principal component analysis: a review and recent developments. Philosophical Trans. Royal Soc. A: Math. Phys. Eng. Sci. 374, 20150202. https://doi.org/10.1098/rsta.2015.0202 (2016).
Talwar, P. K. & Jhingram, A. G. Inland Fishes of India and Adjacent Countries 1 & 2 (A.A. Balkema, 1991).
Day, F. The Fishes of India; Being a Natural History of the Fishes Known to Inhabit the Seas and Fresh Waters of India, Burma and Ceylon: Text and atlas. 1 852 1878 [1876–1878], (Bernard Quaritch, 1878).
Panja, S., Podder, A., Chakrabarty, M. & Homechaudhuri, S. Spatial pattern of freshwater habitats and their prioritization using additive partitions of beta diversity of inhabitant piscine assemblages in the Terai–Dooars ecoregion of Eastern Himalayas. Limnology 23, 57–72. https://doi.org/10.1007/s10201-021-00666-y (2021).
Shaw, G. E. & Shebbeare, E. O. The Fishes of Northern Bengal. 3 1-137 (1937).
Kondera, E. Haematopoiesis and Haematopoietic organs in fish. Sci. Annals Pol. Soc. Anim. Prod. 15, 9–16 (2019).
Kondera, E. Cell composition of the head kidney of European Chub (Squalius cephalus L). Fisheries Aquat. Life. 22, 271–280. https://doi.org/10.2478/aopf-2014-0029 (2015).
Nilsson, G. E. & Östlund-Nilsson, S. Hypoxia in paradise: widespread hypoxia tolerance in coral reef fishes. Proc. Royal Soc. London Ser. B Biol. Sci. 271, S30-S33. https://doi.org/10.1098/rsbl.2003.0087 (2004).
Spicer & Gaston. Amphipod gigantism dictated by oxygen availability? Ecol. Lett. 2, 397–403. https://doi.org/10.1046/j.1461-0248.1999.00105.x (1999).
Smit, H., Amelink-Koutstaal, J. M., Vijverberg, J. & Von Vaupel-Klein, J. C. Oxygen consumption and efficiency of swimming goldfish. Comp. Biochem. Physiol. Part. A: Physiol. 39, 1–28. https://doi.org/10.1016/0300-9629(71)90343-4 (1971).
Svetlichny, L. & Obertegger, U. Influence of temperature on swimming performance and respiration rate of the cold-water cyclopoid copepod Cyclops vicinus. J. Therm. Biol. 109, 103320. https://doi.org/10.1016/j.jtherbio.2022.103320 (2022).
Wang, B. et al. Influences of oxygen and temperature interaction on the antibacterial activity, antioxidant activity, serum biochemical indices, blood indices and growth performance of crucian carp. PeerJ 11, e14530. https://doi.org/10.7717/peerj.14530 (2023).
Wang, C. et al. Hypoxia-inducible factor 1α from a high-altitude fish enhances cytoprotection and elevates nitric oxide production in hypoxic environment. Fish Physiol. Biochem. 46, 39–49. https://doi.org/10.1007/s10695-019-00694-7 (2020).
Filho, D. W. & Boveris, A. Antioxidant defences in marine fish—II. Elasmobranchs. Comp. Biochem. Physiol. Part. C: Pharmacol. Toxicol. Endocrinol. 106, 415–418. https://doi.org/10.1016/0742-8413(93)90155-E (1993). https://doi.org:.
Filho, D. W., Giulivi, C. & Boveris, A. Antioxidant defences in marine fish—I. Teleosts. Comp. Biochem. Physiol. Part. C: Pharmacol. Toxicol. Endocrinol. 106, 409–413. https://doi.org/10.1016/0742-8413(93)90154-D (1993).
Filho, D. W., Torres, M., Marcon, J., Fraga, C. & Boveris, A. Comparative antioxidant defenses in vertebrates-emphasis on fish and mammals. Trends Comp. Biochem. Physiol. 7, 33–45 (2000).
Filho, D. W. Reactive oxygen species, antioxidants and fish mitochondria. Front. Bioscience-Landmark. 12, 1229–1237. https://doi.org/10.2741/2141 (2007).
Filho, D. W. Effect of different oxygen tensions on weight gain, feed conversion, and antioxidant status in Piapara, leporinus elongatus (Valenciennes, 1847). Aquaculture 244, 349–357 (2005).
Nikinmaa, M. Oxygen-dependent cellular functions—why fishes and their aquatic environment are a prime choice of study. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 133, 1–16. https://doi.org/10.1016/S1095-6433(02)00132-0 (2002).
Semenza, G. L. et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1 *. J. Biol. Chem. 271, 32529–32537. https://doi.org/10.1074/jbc.271.51.32529 (1996).
Hüttemann, M., Kadenbach, B. & Grossman, L. I. Mammalian subunit IV isoforms of cytochrome C oxidase. Gene 267, 111–123. https://doi.org/10.1016/S0378-1119(01)00385-7 (2001). https://doi.org:.
Fukuda, R. et al. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122. https://doi.org/10.1016/j.cell.2007.01.047 (2007). https://doi.org:.
Gao, G. & Moyes, C. D. Evaluating the role of NRF-1 in the regulation of the goldfish COX4-1 gene in response to temperature. J. Exp. Biol. 219, 3019–3027. https://doi.org/10.1242/jeb.141184 (2016).
Dhar, S. S., Ongwijitwat, S. & Wong-Riley, M. T. T. Nuclear respiratory factor 1 regulates all ten Nuclear-encoded subunits of cytochrome C oxidase in neurons**. J. Biol. Chem. 283, 3120–3129. https://doi.org/10.1074/jbc.M707587200 (2008).
Bremer, K. & Moyes, C. D. Origins of variation in muscle cytochrome C oxidase activity within and between fish species. J. Exp. Biol. 214, 1888–1895. https://doi.org/10.1242/jeb.053330 (2011).
Chou, C. F., Tohari, S., Brenner, S. & Venkatesh, B. Erythropoietin gene from a teleost fish,fugu rubripes. Blood 104, 1498–1503. https://doi.org/10.1182/blood-2003-10-3404 (2004).
Kulkarni, R. P., Tohari, S., Ho, A., Brenner, S. & Venkatesh, B. Characterization of a hypoxia-response element in the Epo locus of the pufferfish, Takifugu rubripes. Mar. Genom. 3, 63–70. https://doi.org/10.1016/j.margen.2010.05.001 (2010).
Paffett-Lugassy, N. et al. Functional conservation of erythropoietin signaling in zebrafish. Blood 110, 2718–2726. https://doi.org/10.1182/blood-2006-04-016535 (2007).
Nicholls, D., Cunningham, S. & Wiesinger, H. Mechanisms of thermogenesis in brown adipose tissue. Biochem. Soc. Trans. 14, 223–225. https://doi.org/10.1042/bst0140223 (1986).
Rosen, E. D. & Spiegelman, B. M. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 276, 37731–37734. https://doi.org/10.1074/jbc.R100034200 (2001).
Lemberger, T., Desvergne, B. & Wahli, W. Peroxisome proliferator-activated receptors: A nuclear receptor signaling pathway in lipid physiology. Annu. Rev. Cell Dev. Biol. 12, 335–363. https://doi.org/10.1146/annurev.cellbio.12.1.335 (1996).
Tontonoz, P. & Spiegelman, B. M. Fat and beyond: the diverse biology of PPARγ. Annu. Rev. Biochem. 77, 289–312. https://doi.org/10.1146/annurev.biochem.77.061307.091829 (2008).
Tontonoz, P., Hu, E., Graves, R. A., Budavari, A. I. & Spiegelman, B. M. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8, 1224–1234 (1994).
Wafer, R., Tandon, P. & Minchin, J. E. N. The role of peroxisome proliferator-activated receptor gamma (PPARG) in adipogenesis: applying knowledge from the fish aquaculture industry to biomedical research. Front. Endocrinol. 8 https://doi.org/10.3389/fendo.2017.00102 (2017).
LeMoine, C. M. R., Lougheed, S. C. & Moyes, C. D. Modular evolution of PGC-1α in vertebrates. J. Mol. Evol. 70, 492–505. https://doi.org/10.1007/s00239-010-9347-x (2010).
Czubryt, M. P., McAnally, J., Fishman, G. I. & Olson, E. N. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc. Natl. Acad. Sci. U S A. 100, 1711–1716. https://doi.org/10.1073/pnas.0337639100 (2003).
Ramachandran, B., Yu, G. & Gulick, T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J. Biol. Chem. 283, 11935–11946. https://doi.org/10.1074/jbc.M707389200 (2008).
Wu, Z. et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98 115–124. https://doi.org:https://doi.org/10.1016/S0092-8674(00)80611-X (1999).
Padmini, E. & Vijaya Geetha, B. Usha Rani, M. Pollution induced nitrative stress and heat shock protein 70 overexpression in fish liver mitochondria. Sci. Total Environ. 407, 1307–1317. https://doi.org/10.1016/j.scitotenv.2008.09.038 (2009).
Bota, D. A., Van Remmen, H. & Davies, K. J. A. Modulation of Lon protease activity and aconitase turnover during aging and oxidative stress. FEBS Lett. 532, 103–106. https://doi.org/10.1016/S0014-5793(02)03638-4 (2002).
Voos, W. & Pollecker, K. The mitochondrial Lon protease: novel functions off the beaten track?? Biomolecules 10, 253 (2020).
Pianka, E. R. On r- and K-Selection. Am. Nat. 104, 592–597 (1970).
Losos, J. B. Convergence, adaptation, and constraint. Evolution 65, 1827–1840. https://doi.org/10.1111/j.1558-5646.2011.01289.x (2011).
Somero, G. N. The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 213, 912–920. https://doi.org/10.1242/jeb.037473 (2010).
Pörtner, H. O. & Farrell, A. P. Physiology and climate change. Science 322, 690–692. https://doi.org/10.1126/science.1163156 (2008).
Guderley, H. Functional significance of metabolic responses to thermal acclimation in fish muscle. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 259, R245–R252. https://doi.org/10.1152/ajpregu.1990.259.2.R245 (1990).
Guderley, H. Metabolic responses to low temperature in fish muscle. Biol. Rev. 79, 409–427. https://doi.org/10.1017/S1464793103006328 (2004).
De Almeida-Val, V. M. F., Gomes, C. & Lopes, N. P. A. R. Fish Physiol. 21 443–500 (Academic Press, 2005).
Abdel-Tawwab, M., Monier, M. N., Hoseinifar, S. H. & Faggio, C. Fish response to hypoxia stress: growth, physiological, and immunological biomarkers. Fish Physiol. Biochem. 45, 997–1013. https://doi.org/10.1007/s10695-019-00614-9 (2019).
Bickler, P. E. & Buck, L. T. Hypoxia tolerance in reptiles, amphibians, and fishes: life with variable oxygen availability. Annu. Rev. Physiol. 69, 145–170. https://doi.org/10.1146/annurev.physiol.69.031905.162529 (2007).
Nagelkerken, I. et al. The effects of climate change on the ecology of fishes. PLOS Clim. 2, e0000258. https://doi.org/10.1371/journal.pclm.0000258 (2023).
Acknowledgements
The authors sincerely acknowledge the financial assistance from Human Resource Development Group (HRDG), Council of Scientific and Industrial Research (CSIR), Ministry of Science & Technology, Government of India (Grant Number: 09/028(1101)/2019-EMR-I; Grant Recipient: Anupam Podder), DST-INSPIRE AORC, Department of Science & Technology (D.S.T.), Government of India (Grant Number: DST/INSPIRE Fellowship/2016/IF160059; Grant Recipient: Soumyadip Panja), Department of Science & Technology and Biotechnology, Government of West Bengal, (1765(Sanc.)/ST/P/S&T/1G-30/2017; Grant Recipient: Krishna Gangopadhyay) and Women Scientist Scheme A (WOS-A), Department of Science and Technology (D.S.T.), Government of India (Sanction No. SR/WOS-A/LS-693/2016(G); Grant Recipient: Atreyee Chaudhuri). The authors are thankful to Mrs. Missidona Biswas and Mrs. Anwesha Roy for helping in sampling efforts and manuscript review. The authors would also like to genuinely thank the coordinators, Dr. Ritesh Tiwari & Dr. Sneha Mitra, Centre for Research in Nanoscience and Nanotechnology (CRNN), University of Calcutta, for providing the analytical and infrastructural facilities pertaining to flow cytometry and the Dean, Faculty Council for Post-Graduate Studies in Science for providing the necessary instrumentation facilities in the UGC- (UPE) Phase II- Centre for Modern Biology, S.N. Pradhan Centre for Neurosciences, University of Calcutta. Finally, the authors also acknowledge the infrastructural support from the Department of Zoology, University of Calcutta and the local people and fishermen of the Indian Sundarbans & Terai-Dooars region of West Bengal, India regarding their assistance in various activities related to sampling.
Author information
Authors and Affiliations
Contributions
AP and SH conceived and designed the study. AP, SP, KG and AC performed sampling, data collection and experiments. AP and SP analyzed the data. AP and SH wrote the manuscript; All authors reviewed the manuscript and provided editorial advice.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Podder, A., Panja, S., Gangopadhyay, K. et al. Comparative study of fish adaptation at cellular and genetic levels across eco-sensitive zones with contrasting oxygen and temperature regimes. Sci Rep 15, 35252 (2025). https://doi.org/10.1038/s41598-025-97749-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97749-y