Abstract
Anopheles stephensi, a major Asian malaria vector, is invading Africa and has been implicated in recent outbreaks of urban malaria. Control of this species is key to eliminating malaria in Africa. Genetic control strategies, and CRISPR/Cas9-based gene drives are emerging as promising species-specific, environmentally friendly, scalable, affordable methods for pest control. To implement these strategies, a key parameter to optimize for high efficiency is the spatiotemporal control of Cas9 and the gRNA. Here, we assessed the ability of four RNA Pol III promoters to bias the inheritance of a gene drive element inserted into the cd gene of An. stephensi. We determined the homing efficiency and examined eye phenotype as a proxy for non-homologous end joining (NHEJ) events in somatic tissue. We found all four promoters to be active, with mean inheritance rates up to 99.8%. We found a strong effect of the Cas9-bearing grandparent (grandparent genotype), likely due to maternally deposited Cas9.
Similar content being viewed by others
Introduction
Anopheles stephensi mosquitoes have been involved in the transmission of urban malaria in South Asia and the Arabian Peninsula for many decades1. The recent invasion of this species into Africa threatens malaria control efforts by adding a new vector, especially in urban environments where endemic Anopheles mosquitoes were absent2,3,4. In a few years, An. stephensi has spread from East to West Africa and has been implicated in several urban malaria outbreaks5,6,7,8. This is a particular concern both because more people are at risk, and because there are indications that this species is responsible for the spread of drug and diagnosis resistant strains of Plasmodium falciparum5. Control of this species has become a priority, with the WHO releasing an initiative to stop the spread of An. stephensi in Africa in 20233,9,10,11.
Malaria control has reached a plateau in which traditional methods seem less effective, mainly due to the emergence and spread of insecticide resistance. Hence, new mosquito control strategies are needed and are being actively developed. Genetic control methods have emerged as a potentially powerful strategy since they are species-specific, environmentally friendly methods for controlling pests. More specifically, gene drives have emerged as potential genetic biocontrol tools for controlling pest populations threatening global health12,13,14,15. These aim to bias the inheritance of a genetic element such that, under suitable circumstances, it can increase in frequency in a population even if it imposes some fitness cost on individual carriers. For CRISPR/Cas9 based gene drives to be efficient, several parameters need to be optimised, most importantly the spatiotemporal expression of Cas9 and sgRNA. Development of a toolbox for engineering this vector is urgently needed, with most current investigations instead focusing on other important disease vectors such as Aedes aegypti and An. gambiae15,16,17,18,19.
To help fill this toolbox, this study focuses on sgRNA expression, using four different RNA Pol III promoters with Cas9 under the endogenous regulation of zero population growth (zpg) and targeting cardinal (cd), a component of the ommochrome pathway. Ommochrome is the major eye pigment in mosquitoes and cd null individuals display a distinctive, pink-eyed phenotype19. The use of split Cas9 and sgRNA components, each integrated into a specific locus, allows the effect of the Pol III promoter to be investigated accounting for issues due to position effect, a well-known phenomenon with transgenics where chromosome location can alter the expression pattern of promoters within a transgene20,21. The use of the cd gene as a target also allows us to separately measure overall cleavage rates, combining homing (inheritance of the cd integrated, sgRNA expressing element) and NHEJ events (pink-eyed phenotype), making this a useful tool for optimization of both the Cas9 and sgRNA components enabling the determination of which is the limiting factor in the cells at the time at which homing occurs.
The results of this study show that all four RNA Pol III promoters tested were active in both male and female germlines of An. stephensi, inducing mean inheritance bias up to 99.8%. We also reveal a strong effect of the sex of the Cas9-bearing grandparent, likely due to maternal deposition of Cas9. Such maternally-derived Cas9 can then cleave alleles in the developing embryo which are repaired by NHEJ resulting in indels which are resistant to further cleavage. Strategies for gene drives to circumvent this resistance include multiplexing approaches which typically use multiple sgRNAs to target a gene, thus alleles with an indel at one target still have several target sites available for homing16,22,23,24. This study provides multiple functional Pol III promoters which greatly facilitates this type of resistance-management strategy.
Results
For each promoter we generated an insertion into cd only differing by the RNA Pol III promoter and terminator sequences used to express the sgRNA. The cd gene encodes a heme peroxidase, an enzyme in the ommochrome pathway19. Like the kmo gene, another gene in this pathway which has been widely used for transgenesis studies, homozygous null mutants for these genes display depigmentation in the mosquito eyes which allows for screening of homing events (fluorescent marker) and non-homologous end joining (NHEJ) events, in the case of cd giving a pink-eyed phenotype. Deposition or somatic expression of Cas9 may result in somatic NHEJ events leading to a mosaic-eyed phenotype. Here, we selected the cd gene, since previous studies showed deleterious effects on the kmo null mutant25.
To study the effect of the promoter, each of the lines (cdsgRNA) was mated to a transgenic line expressing Cas9 under the endogenous control of zpg (zpg3’Cas9). Transheterozygous F1 individuals were then crossed to wild type strain (cd+/+) to measure the inheritance bias and effects of deposited Cas9 or to a cd−/− line which allows assessment of the overall cleavage rate (Fig. 1). This allows the detection of differences in homing and/or cutting efficiencies by screening the F2 offspring for transgene inheritance and eye phenotype.
Pol III promoter affects inheritance rate
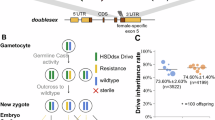
The four promoters assessed were found to be active in both the male and female germlines of An. stephensi (Data file D1). More specifically, those crosses using the U6-C and 7SK promoters had the highest overall rates of inheritance (mean [95% CI]; U6-C, 99.8% [99.6–99.9]; 7SK, 98.7% [98.15–99.1]). Moreover, these two lines also showed an effect depending on the sex of the Cas9-bearing grandparent (U6-C, z = 6.1, p < 0.001; 7SK, z = 6.36, p < 0.001). There was also a significant but reduced effect of the Cas9-bearing parent in U6-C (z = 2.47, p = 0.014). In both instances the highest rates of homing, with means approaching 100%, were observed in the offspring of male Cas9-bearing grandparents (Fig. 2A, Table S2).
Pol III promoters bias the inheritance of a gene drive element in the An. stephensi germline. (A) Open circles represent inheritance rates from a single parent, size indicates the number of offspring per female. Darker, filled points and error bars are estimated means and 95% confidence intervals. (B) Eye phenotypes were determined to be wild type (pink bars) or mosaic/pink eyes (green bars) and are presented as the proportion of the total F2 separated by genotype.
Examination of the eye phenotype showed that mosaicism was dependent on the sex of the Cas9-bearing parent. The results reveal that 100% of the F1 progeny coming from a Cas9-bearing female and carrying the cdsgRNA element displayed mosaic eyes. Furthermore, almost all offspring of the F1 transheterozygous females also showed mosaic eyes, whether they inherited the transgenes or not, while very few mosaics were found in the offspring of the F1 transheterozygous males. This indicates that across all lines, the observed rates of mosaicism could be predicted almost exclusively by the parental genotype (LRT: χ21 = 30915, P < 0.001, Partial R2 = 0.96; Fig. 2B) and supports the conclusion that maternal deposition of Cas9 results in mosaic progeny.
Cleavage rate mediates inheritance bias
In order to determine overall cleavage rates, we also crossed zpg3’Cas9;cdsgRNA transheterozygotes to cd−/−. Progeny of this cross inherit one null cd allele from their cd−/− parent and either the cdsgRNA transgene or the putatively WT (or cut and repaired by NHEJ/homing) allele from the transheterozygote parent. Mosaicism in those progeny not carrying the cdsgRNA indicates that the WT allele was cut somatically (not in all cells). Complete pink eyes would indicate a germline cut repaired by NHEJ into a non-functional allele. Careful observation of the rates of each allows the estimation of the cleavage and NHEJ rates. This is necessarily an approximation, since a very early or high level of embryonic cutting could conceivably give fully pink eyes.
We observed inheritance rates similar to those found in the cross to WT from the section above (Data file D1). Again, a very strong effect from the sex of the Cas9-bearing grandparent was detected (U6-C, z = 4.45, p < 0.001; 7SK, z = 6.44, p < 0.001). However, in this set of crosses, we observed a significant but reduced effect of the Cas9-bearing parent in U6-C only (z = 2.14, p = 0.03) (Fig. 3, Table S2). Interestingly, we found that the cleavage rate was equivalent to the inheritance of the drive element for all the promoters except U6A, and for U6C it was nearly 100%. This is likely an indication of expression levels varying with the different promoters. In addition, the high correlation between overall cleavage rate and drive inheritance may also indicate that the expression pattern and/or timing of U6B, U6C and 7SK overlaps with the zpg derived expression of Cas9 during a time window which is most favorable for homology-directed repair (HDR), with very few cleavage events resulting in NHEJ. It may be that for homing gene drives there are optimal pairs/sets of RNA Pol II and RNA Pol III promoters which express at the same time and lead to high drive efficiency, and these may need to be optimised for different species or different drive systems.
Homing and cleavage rates across four independent transgenic lines crossed to pink eyed strain. Open circles represent individual inheritance rates from a single parent, size of the circle indicates the number of offspring. Darker, filled points and error bars are estimated means and 95% confidence intervals. Triangles represent the proportion of the F2 which inherit the cdsgRNA element. Circles represent the proportion of the F2 which have a pink eyed phenotype indicating an indel in cd.
Discussion
Our data are consistent with previous observations of maternally deposited Cas9 resulting in somatic cleavage, and very little paternal deposition26,27,28,29.This indicates that these Pol III promoters were active in early stages of embryonic development, and therefore even if any of them are not deposited maternally, maternal deposition of Cas9 alone can still result in resistance allele formation. To mitigate this effect, methods to further restrict expression of Cas9 to the germline should be investigated. This has been the most problematic issue from the beginning of the development of gene drive strains in An. stephensi. For example, Gantz et al. utilized the vasa promoter to express Cas9 in an autonomous gene drive. They reported near 100% inheritance of the drive through the paternal lineage, however this was significantly reduced through the maternal lineage due to maternal deposition of Cas9 and creation of resistant alleles.
Our results pave the way for future gene drive designs with multiple sgRNAs expressed by multiple RNA Pol III promoters to mitigate resistance. In these systems, multiple sgRNAs are expressed all targeting the same gene, such that alleles which are resistant to one sgRNA may still be cleaved by another sgRNA and the drive is able to spread. Although methods for expressing several sgRNAs from a single transcript such as using tRNAs, ribozymes or Csy4 have been reported in cell culture models, many of these are yet to be validated in mosquitoes and multiple, functional RNA Pol III promoters present a simple alternative. While previous gene drive systems in An. stephensi have mostly utilized a single RNA Pol III promoter, U6A26,28, we have identified three previously uncharacterized highly functional RNA Pol III promoter sequences in An. stephensi which are more active than the U6A promoter commonly used for sgRNA expression. These promoters could all generate up to 100% cutting/biased inheritance in the male and female germlines, although U6C and 7SK outperformed the others with mean inheritance rates near 100%.
Interestingly, we have found U6A to be the least active promoter in conjunction with our zpg expressed Cas9, suggesting that there is an expression level and/or spatiotemporal difference between the different promoters. To our knowledge, the only other study which has shown this was Li et al.18. In their hands, wGDe-U6b when assessed with exu-Cas9, exhibited the highest homing rate compared to the other promoters (U6a, U6b, and U6d) while U6d, which in a previous experiment was shown to be functional, seemed to be inactive when inserted into the target locus. This should not be a surprise, despite being a critical component for mRNA splicing30, there are multiple copies present in the genome and it is likely that some loci may have developed ‘tunability’ to cater to the requirements of different cell types. Further insight into the expression pattern of RNA Pol III promoters, could aid in the optimisation of the homing process. This type of investigation would be challenging, at least for the U6 genes as the RNA they express is identical. Robust characterization of sgRNA expression presents significant technical challenges as these are very short, highly structured RNAs. Little is known as to the ‘correct’ spatiotemporal window for homing to occur and extensive research into this could significantly advance the field. In the meantime, having several options of RNA Pol III promoters to express sgRNAs could aid in this investigation. These multiple, functional promoters for sgRNA expression should be useful for many gene drive applications in this species.
Materials and methods
Plasmids
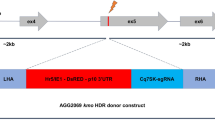
Four insertions into cd expressing the same sgRNA_384 (cut site corresponds to roughly amino acid 384 of cardinal: GCAGCAACTTAATCAAGCCACGG, PAM site italicised) from different RNA pol III promoters were generated. The sgRNA target was identified using CHOPCHOP31 and a high-ranking target site within the desired region, with no variation annotated in Vectorbase32,33 and no off-targets was selected. Three endogenous U6 RNA promoters (ASTE015697-U6A, ASTE015521-U6B, ASTE015587-U6C) as well as the 7SK (ASTE015331) RNA promoter were identified by BLAST using the U6 or 7SK RNA as a query. A CLUSTAL alignment of the promoter sequences used was generated and putative conserved proximal promoter elements (TATA box, PSE) identified (Figure S1). The construct design consists of approximately 1.5 kb of homology 5’ and 3’ of the cut site for the sgRNA to introduce a fluorescent marker (Hr5/IE1-AmCyan-K10) and the sgRNA expression cassette (Pol III promoter–sgRNA–terminator) into the genome by HDR.
The Cas9 is expressed by utilizing the endogenous zpg locus (ASTE011088) as an integral element. Plasmid AGG1590 (zpg3’Cas9) was used to integrate a ubiquitin monomer followed by the Cas9 ORF and marker expression cassette (3xP3-mCherry-SV40) immediately following the zpg coding sequence just upstream of the stop codon by HDR.
Full plasmid sequences are available in NCBI (accession numbers: PQ005639-PQ005643).
Mosquito rearing
All experiments performed for this study were approved by the Biological Agents and Genetic Modification Safety Committee of The Pirbright Institute and are in accordance with all relevant regulations. An. stephensi SDA-500 strain (WT) were maintained at 28 °C, 75–85% relative humidity and 14:10 light/dark cycle34. Larvae were reared as 200 larvae per 500 ml RO water in 6 L trays and fed Seramicron (Sera) as early instars and ExtraSelect (Su-Bridge Pet Supplies Ltd) as later instars. Adults were provided with 10% sucrose ad libitum and blood-fed defibrinated horse blood (TCS Bioscience) using a Hemotek membrane feeder (Hemotek Ltd) with parafilm (Bemis).
Generation of mosquito Transgenic lines
Microinjections on WT embryos were performed 1–2 h post oviposition, as previously described34. Injection mixes contained 300ng/µl Cas9 (PNABio), 50ng/µl in vitro transcribed sgRNA37 and 800ng/µl HDR donor plasmid.
Surviving G0 mosquitoes were pool crossed to WT as 20 G0 males:100 WT females, and 20 G0 females:20 WT males. Females were blood-fed and eggs were collected and hatched. G1 larvae were screened under a Leica MZ165C fluorescence microscope for the presence of the fluorescent marker (Table S1). Positive G1 individuals were mated to WT to continue the line and the insertions confirmed by PCR and Sanger sequencing (Figure S2).
cd−/− mosquitoes used for the cleavage assay crosses were generated by sibling crossing the non-fluorescent F2 progeny from a cdU6A;zpg3’Cas9 homing cross and screening their progeny for eye phenotype. The line was established with two mutations, confirmed by PCR and Sanger sequencing, but after several generations only one mutation remained; this was a 3 bp deletion which resulted in amino acids Q383 and A384 changing to P (Figure S3).
Crosses for drive and cleavage assessment
Males and females, heterozygous for the cdsgRNA element, were crossed reciprocally with heterozygous mosquitoes of the zpg3’Cas9 line (F0). Their progeny (F1) were screened for fluorescence and eye phenotype. For the homing assessment, at least 20 transheterozygous cdsgRNA;zpg3’Cas9 mosquitoes of each sex were crossed to WT mosquitoes of the opposite sex, blood-fed and females separated into individual cups and allowed to lay eggs. The F2 were hatched, reared and screened for fluorescence and eye phenotype. For the cleavage assessment, at least 20 transheterozygous cdsgRNA;zpg3’Cas9 of each sex were crossed to cd−/− of the opposite sex, also allowed to lay individually, and their F2 progeny scored for transgene inheritance indicated by fluorescence and cleavage indicated by eye phenotype.
Statistical analysis
Analyses were carried out using R version 4.4.035. Estimated means and 95% confidence intervals were calculated by a Generalized Linear Mixed Model, with a binomial (‘logit’ link) error distribution fitted using the glmmTMB package38. Where applicable, initial parameters included transgenic line, sex of the Cas9-bearing G1 parent and G0 grandparent; individual replicates were included as a random effect. The model was summarized with ‘emmeans’39 and model residuals were checked for violations of assumptions using the ‘DHARMa’ package40. Figures were generated with ggplot241. Where applicable a partial McFadden’s R2 was calculated by comparing deviance of full and partial models. Scripts and raw data can be found at Github (https://github.com/Philip-Leftwich/Optimization-sgRNA-Anopheles-stephensi).
Data availability
All data generated for this study is available in the main manuscript and supplementary files. Full plasmid sequences are available from NCBI accession numbers: PQ005639-PQ005643.
References
Ishtiaq, F., Swain, S. & Kumar, S. S. Anopheles stephensi (Asian malaria Mosquito). Trends Parasitol. 37, 571–572 (2021).
Sinka, M. E. et al. A new malaria vector in Africa: Predicting the expansion range of Anopheles stephensi and identifying the urban populations at risk. Proc. Natl. Acad. Sci. 117, 24900–24908 (2020).
Faulde, M. K., Rueda, L. M. & Khaireh, B. A. First record of the Asian malaria vector Anopheles stephensi and its possible role in the resurgence of malaria in Djibouti, Horn of Africa. Acta Trop. 139, 39–43 (2014).
Zhou, G., Zhong, D., Yewhalaw, D. & Yan, G. Anopheles stephensi ecology and control in Africa. Trends Parasitol. 40, 102–105 (2024).
Emiru, T. et al. Evidence for a role of Anopheles stephensi in the spread of drug- and diagnosis-resistant malaria in Africa. Nat. Med. 29, 3203–3211 (2023).
Hawaria, D. et al. First report of Anopheles stephensi from Southern Ethiopia. Malar. J. 22, 373 (2023).
Al-Eryani, S. M. et al. Public health impact of the spread of Anopheles stephensi in the WHO Eastern mediterranean region countries in Horn of Africa and Yemen: need for integrated vector surveillance and control. Malar. J. 22, 187 (2023).
Santi, V. P. et al. Role of Anopheles stephensi mosquitoes in malaria outbreak, Djibouti, 2019 - 27, number 6—June 2021 - Emerging infectious diseases journal - CDC. https://doi.org/10.3201/eid2706.204557
WHO initiative to stop the spread of. Anopheles stephensi in Africa, 2023 update. https://www.who.int/publications/i/item/WHO-UCN-GMP-2022.06
Whittaker, C. et al. Seasonal dynamics of Anopheles stephensi and its implications for mosquito detection and emergent malaria control in the Horn of Africa. Proc. Natl. Acad. Sci. 120, e2216142120 (2023).
Ahmed, A., Abubakr, M., Ali, Y., Siddig, E. E. & Mohamed, N. S. Vector control strategy for Anopheles stephensi in Africa. Lancet Microbe. 3, e403 (2022).
Alphey, L. S., Crisanti, A., Randazzo, F., (Fil) & Akbari, O. S. Standardizing the definition of gene drive. Proc. Natl. Acad. Sci. 117, 30864–30867 (2020).
Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. R Soc. Lond. B Biol. Sci. 270, 921–928 (2003).
Deredec, A., Burt, A. & Godfray, H. C. J. The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179, 2013–2026 (2008).
Hammond, A. et al. Gene-drive suppression of mosquito populations in large cages as a Bridge between lab and field. Nat. Commun. 12, 4589 (2021).
Anderson, M. A. E. et al. A multiplexed, confinable CRISPR/Cas9 gene drive can propagate in caged Aedes aegypti populations. Nat. Commun. 15, 729 (2024).
Kyrou, K. et al. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018).
Li, M. et al. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. 114, E10540–E10549 (2017).
Carballar-Lejarazú, R. et al. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. 117, 22805–22814 (2020).
Markstein, M., Pitsouli, C., Villalta, C., Celniker, S. E. & Perrimon, N. Exploiting position effects and the Gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 (2008).
Roberts, J. A. et al. Targeted transgene integration overcomes variability of position effects in zebrafish. Dev. Camb. Engl. 141, 715–724 (2014).
Edgington, M. P., Harvey-Samuel, T. & Alphey, L. Population-level multiplexing: A promising strategy to manage the evolution of resistance against gene drives targeting a neutral locus. Evol. Appl. 13, 1939–1948 (2020).
Yang, E. et al. A homing suppression gene drive with multiplexed gRNAs maintains high drive conversion efficiency and avoids functional resistance alleles. G3 GenesGenomesGenetics 12, jkac081 (2022).
Hou, S. et al. A homing rescue gene drive with multiplexed gRNAs reaches high frequency in cage populations but generates functional resistance. J. Genet. Genomics. 51, 836–843 (2024).
Bottino-Rojas, V. et al. Beyond the eye: kynurenine pathway impairment causes midgut homeostasis dysfunction and survival and reproductive costs in blood-feeding mosquitoes. Insect Biochem. Mol. Biol. 142, 103720 (2022).
Adolfi, A. et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat. Commun. 11, 5553 (2020).
Verkuijl, S. A. N., Anderson, M. A. E., Alphey, L. & Bonsall, M. B. Daisy-chain gene drives: the role of low cut-rate, resistance mutations, and maternal deposition. PLOS Genet. 18, e1010370 (2022).
Gantz, V. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl. Acad. Sci. 112, E6736–E6743 (2015).
Pham, T. B. et al. Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLOS Genet. 15, e1008440 (2019).
Das, G., Henning, D., Wright, D. & Reddy, R. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J. 7, 503–512 (1988).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174 (2019).
Giraldo-Calderón, G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 (2015).
Giraldo-Calderón, G. I. et al. VectorBase.org updates: bioinformatic resources for invertebrate vectors of human pathogens and related organisms. Curr. Opin. Insect Sci. 50, 100860 (2022).
Southworth, J. et al. Expanding the transgene expression toolbox of the malaria vector Anopheles stephensi. Insect Mol. Biol. 34, 104–110 (2025).
Core Team, R. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Available from: https://www.r-project.org/
Catteruccia, F., Benton, J. P. & Crisanti, A. An Anopheles Transgenic sexing strain for vector control. Nat. Biotechnol. 23, 1414–1417 (2005).
Bassett, A. & Liu, J. L. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69, 128–136 (2014).
Brooks, M. GlmmTMB balances speed and flexibility among packages for Zero-inflated generalized linear mixed modeling. R J. 9, 378 (2017).
Lenth, R. V. & emmeans Estimated Marginal Means, aka Least-Squares Means. (2023).
Hartig, F. DHARMa: residual diagnostics for hierarchical (multi-level/mixed) regression models. (2022).
Wickham, H. Ggplot2 (Springer International Publishing, 2016). https://doi.org/10.1007/978-3-319-24277-4
Acknowledgements
This publication is based on research funded by the Gates Foundation (INV-008549). The conclusions and opinions expressed in this work are those of the author(s) alone and shall not be attributed to the Foundation. Under the grant conditions of the Foundation, a Creative Commons Attribution 4.0 License has already been assigned to the Author Accepted Manuscript version that might arise from this submission. This work was also supported by strategic funding from the UK Biotechnology and Biological Sciences Research Council (BBSRC) to The Pirbright Institute (BBS/E/I/00007033, BBS/E/I/00007038, and BBS/E/I/00007039).
Author information
Authors and Affiliations
Contributions
MAEA, JXDA, and LA conceived and designed the research. EG, JXDA, KN, LS, and MLG performed the research. PTL analyzed the data. LA contributed with funding acquisition and project supervision. MA and EG wrote the first draft of the manuscript and all authors contributed to reviewing and editing the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
LA is an adviser to Synvect Inc and Biocentis Ltd, with equity and/or financial interest in those companies. All remaining authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gonzalez, E., Anderson, M.A.E., Ang, J.X.D. et al. Optimization of SgRNA expression with RNA pol III regulatory elements in Anopheles stephensi. Sci Rep 15, 13408 (2025). https://doi.org/10.1038/s41598-025-98557-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98557-0