Abstract
Severe Acute Respiratory Infection poses a significant threat to human health being a major cause of morbidity and mortality. The rate of co-infection among the underlying pathogens is unknown. During COVID-19 pandemic, reports for respiratory pathogens co-circulations in developing countries were limited. Identification of respiratory pathogens is paramount for effective patient management as early detection decreases the risk of mortality and morbidity. This is the first report to investigate the incidence of respiratory pathogens co-infection among patients with SARI during COVID-19 pandemic in Egypt. Clinically SARI patients were recruited from October 2020 to June 2022. Nasopharyngeal swabs were collected to detect SARS-CoV-2 followed by 33 respiratory pathogens identification using RT-PCR. Of 599 samples tested, 27% (158/599) patients were positive for COVID-19, in which 75.9% (120/158) patients were co-infected with other respiratory pathogens. In total, 31 pathogens were identified with a detection rate of 75% (450/599) among positive and negative COVID-19 patients. Bacterial co-infections rate was 39%, in which Klebsiella pneumoniae (5.3%) is the most common, while viral co-infections rate was 61%, in which Human Coronavirus HKU1 (6.2%) is the most common. Adenovirus, human rhinovirus and RSV were only detected in 70/11.7%, 50/8.3% and 41/6.8% of cases, respectively. Early detection and management of respiratory pathogens co-infection are crucial for effective patient management and preparedness for future pandemics.
Similar content being viewed by others
Introduction
In low- and middle-income countries, the burden of severe acute respiratory infections (SARI) is high, and associated with significant morbidity and mortality1,2. High attention has been paid to the etiological tracing of respiratory tract infection since the advent of COVID-193.
The emerging of the highly pathogenic severe acute respiratory syndrome (SARS-CoV-2) in December 2019 has widely spread to become a major public health threat with a significant number of deaths worldwide4. During COVID-19 pandemic period, co-circulation of respiratory pathogens in developing countries were under-reported5. SARS-CoV-2 screening by real-time polymerase chain reaction (RT-PCR) has been an increased focus among patients with respiratory symptoms in these countries. Testing for other respiratory pathogens that cause SARI has been neglected. SARI clinical symptoms are almost similar to COVID-19 that can obstruct the diagnosis of patients with suspected COVID-196. Detection of other respiratory pathogens is crucial to prevent adverse outcomes, and reduce antibiotic usage for better case management. Disparities in the prevalence of respiratory pathogens co-infection among countries have been reported in several studies, attributed to discrepancies in age groups, geographical location, seasons, and other factors7,8.
Several molecular methods have been described for the diagnosis of respiratory infections9,10,11. Multiplex Real time PCR (rRT-PCR) commercial panels were recently used for detecting a wide range of respiratory pathogens with high sensitivity, efficiency and time saving which made it the method of choice such as FilmArray® Respiratory Panel (Biofire Diagnostics, Salt Lake City, UT), Respiratory Multi Well System MWS r-gene® Range (BioMerieux) and Fast-Track Diagnostics FTD-33. Performance of these new multiplex panels were reported in few studies9,10,12.
Recent studies have shown the presence of respiratory pathogens co-infection during COVID-19 pandemic waves13,14,15. The most common bacterial co-infections reported in COVID-19 patients were Chlamydophila pneumoniae, Mycoplasma pneumoniae, Streptococcus pneumoniae, and Staphylococcus aureus16 and for viral co-infection; influenza A virus (IAV), influenza B virus (IBV), rhinovirus/enterovirus (EV), respiratory syncytial virus (RSV) and metapneumovirus were reported17. Hashemi et al.18 study has detected positive respiratory viruses in most of COVID-19 patients tested in which influenza A virus rate was 22.3%. Roh et al.19 reported 8.8% respiratory pathogens co-infection in which Mycoplasma pneumoniae and adenovirus were detected in COVID-19 patients. In a Chinese study, 5.8% co-infection was detected in COVID-19 patients, and 18.4% co-infection was detected in patients without COVID-1915.
To our knowledge, there was a lack in the studies that described the respiratory pathogen co-infection among SARI patients during COVID-19 pandemic in the region. Only one survey study focused on viral co-infection has been reported among children that was admitted with SARI and negative for COVID-19 in Egypt. This study showed 92.1% viral co-infection rate in which Rhinovirus was the main cause of death among children20.
Here, is the first report that aimed to evaluate the incidence of common respiratory pathogens in SARI patients in Egypt during COVID-19 pandemic, and identify groups at risk of severe disease.
Materials and methods
Patient and sample collection
The study was approved by the Research Ethics Committee at Ain Shams University Faculty of Medicine (IRB no.: FMASU P94a/2020–2021). All methods were performed in accordance with the relevant guidelines and regulations. Patients who met the WHO standard SARI case definition [Patients with acute respiratory infection who have history of fever (or measured fever of ≥ 38 °C), cough and onset of symptoms within the last 10 days and require hospitalization]17 were recruited from the internal medicine, chest department, pediatric hospital, geriatric hospital and intensive care units (ICUs) in Ain Shams University Hospitals, Cairo, Egypt during the period from October 2020 till June 2022. A questionnaire that covered history of fever and/or respiratory symptoms, travelling history, any underlying lung disease, history of chronic or immunocompromised conditions, and outcome was completed for each subject.
A total of 599 nasopharyngeal swab samples were collected from patients after signing informed consents. Samples selected in this study originated from patients with severe acute respiratory symptoms aged less than 15 to more than 60 years old of both genders. Collected samples were stored in 2 ml viral transport media (VTM) and transported to Egypt Center for Research and Regenerative Medicine (ECRRM) and Faculty of Medicine, Ain Shams University laboratories for molecular evaluation.
Nucleic acid purification
Nucleic acid was extracted using Prepito NA Body Fluid Kit (Perkin Elmer, USA) following the manufacturer’s protocol with a final elution of 100 µl. Internal control (supplied with RT-PCR kit) was added to each sample. Extracted nucleic acid concentration and purity was measured by NanoDrop™ One Microvolume UV-Vis Spectrophotometer (ThermoFisher, USA).
Real time polymerase chain reaction (RT-PCR)
Extracted samples were tested for SARS-CoV-2 using ProLab/Cer Test Biotech VIASURE SARS-CoV-2 (N1 + N2) Real Time PCR Detection kit (CerTestBiotec, S.L, Spain), targeting two conserved regions of nucleocapsid protein gene (N1 and N2) following manufacturer’s instructions. Samples were processed with appropriate negative, internal, and positive controls.
Fast Track Diagnostic (FTD®) 33 multiplex kit was used for detection of 33 respiratory pathogens using Real time PCR including influenza A virus (IAV), influenza B virus (IBV), influenza C virus (IAC), influenza A (H1N1) virus swine lineage (IAV (H1N1) swl), human parainfluenza viruses (HPIV) 1, 2, 3 and 4, human coronaviruses (HCOV) NL63, 229E, OC43 and HKU1, human metapneumoviruses A and B (HMPVA and B), human rhinovirus (HRV), respiratory syncytial viruses A and B (HRSVA and HRSVB), human adenovirus (HAdV), enterovirus (EV), human parechovirus (HPeV), human bocavirus (HBoV), Pneumocystis jirovecii (P. Jirovecii), Mycoplasma pneumoniae (M. pneumoniae), Chlamydophila pneumoniae (C. pneumoniae), Streptococcus pneumoniae (S. pneumoniae), Haemophilus influenzae type B (H. Influenza B), Staphylococcus aureus (S. aureus), Moraxella catarrhalis (M. catarrhalis), Bordetella spp., Klebsiella pneumoniae (K. pneumoniae), Legionella pneumophila/ Legionella longbeachae, Salmonella spp. and Haemophilus influenza (H. influenza).
Eight multiplex RT-PCR reactions were prepared; each PCR reaction contained 10uL of nucleic acids template, 12.5µL of the buffer, 1.5µL of primer/probe mix, and 1µL of enzyme. Using an ABI 7500 Real Time PCR System (Applied Biosystems™, USA), reaction mixture was initially incubated at 50 °C for 15 min, 94 °C for 1 min, followed by 40 cycles of: 94 °C for 8 s, and 60 °C for 1 min. Negative and positive controls were added for results verification. The results were considered positive for all sigmoidal curve and cycle thresholds (Ct) < 38 curves following manufacture instruction.
Statistical data management and analysis
Laboratory results of detected pathogens as well as the additional clinical and radiological characteristics were compared using Chi-square test to examine the relationship between two qualitative variables, while Fisher’s Exact test was used to examine the relationship between two qualitative variables when the expected count is less than 5 in more than 20% of cells. Student T Test was used to assess the statistical significance of the difference between two study group means. All statistical analyses and calculations were performed using the SPSS 27.0 statistical software package. Evaluations and comments were made at a significant level of p-value lower than 0.05.
Results
Demographics, clinical characteristics and patient co-morbidities collected are listed in Table 1. The study included 313 (52.3%) Females and 286 males (47.7%). Out of these, 134 (22.4%) patients were > 15 years, 341 (56.9%) patients between 15 and 60 years and 124 (20.7%) patients were more than 60 years. The mean age of the study population was 36.29 years (SE = 0.98), with a median age of 35.10 years and an interquartile range of 36.16 years. The skewness value was 0.046. Most of the patients (58.2%) had symptoms of fever, cough, dyspnea, diarrhea, runny nose, and sore throat. Comorbidities conditions were detected in 46.4% (278/599) of patients, which included hypertension (30.9%), diabetes (26.9%), chronic cardiac disease (26.9%), and Asthma (21.5%).
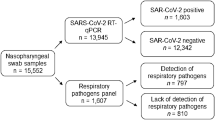
Out of the 599 tested samples, 158 samples showed positive SARS-CoV-2 results. All samples were evaluated for other respiratory pathogens in which 31 pathogens were identified in positive and negative COVID-19 patients, including 12 bacteria, 18 viruses, and 1 fungus with a detection rate of 75% (450/599) (Fig. 1) (Table 2). Bacterial co-infections rate was 39%, in which K. pneumoniae was the most common (5.3%), while viral co-infection rate was 61%, in which HCOV HKU1 (6.2%) was the most common. Solitary infections were detected in the patient samples, 38 (6.3%) with SARS-CoV-2, 41 (6.8%) with other viruses, and 53 (8.9%) with bacteria.
Out of 158 COVID-19 patients, 120 were found to be co-infected with other respiratory pathogens, in which 5.5% were infected with single pathogen, 4.9% were co-infected with two pathogens, and 14.1% were co-infected with three pathogens and more (Table 3). In Negative COVID-19 patients, 330 patients were found to be co-infected with other respiratory pathogens, in which 20.1% were infected with single pathogen, 13.5% were co-infected with two pathogens, 8.4% were co-infected with three pathogens and 25.6% were co-infected with more than three pathogens (Table 4).
Analysis of respiratory pathogens co-infection with age group, comorbidities and symptoms are listed in Table 5. A chi square group difference between number of co-infections among patients with or without COVID-19 and age groups indicated a significant association of moderate effect (χ2 (21, n = 450) = 97.98 p = .001, Cramer’s V = 0.259), while with comorbidities indicated a significant association (χ2 (7, n = 450) = 15.03 p = .036 for hypertension and = 16.79 p = .019 for diabetes; and insignificant association (χ2 (7, n = 488) = 11.81 p = .107 for chronic cardiac diseases.
The most dominant pathogens based on the calculated cumulative score “total infections present in more than 60% of total number of patients” found in all patients are 6.2% HCOV HKU1, 5.3% K. pneumoniae, 4.7% IAV (H1N1) Swl, 4.6% HAdV, 4.4% P. Jirovecii, 4.2% HPIV4, 4.2% EV, 4.1% Bordetella, 4.1% H. Influenza B and 3.3% HCOV NL63 (Fig. 2).
Discussion
Severe acute respiratory infection (SARI), the third leading cause of death globally, represent a challenge for healthcare systems, especially in low- and middle-income countries3,4. A number of respiratory viruses and bacterial etiologies in patients that met SARI criteria have been reported21,22. During COVID-19 pandemic, we experienced a surge of acute respiratory infections caused by several pathogens, including SARS-CoV-2, influenza, RSV, and other seasonal virus and bacteria. The co-circulation of these pathogens increases pressure on healthcare systems, with outbreaks of respiratory infection leading to increased primary care consultations and hospitalizations, particularly for patients with comorbidities and risk factors for severe outcomes18.
After the first confirmed case of COVID-19 in Egypt in February 2020, SARS-CoV-2 were widely spread with increasing in the number of cases as reported in the official website of the Egyptian Ministry of health (https://www.care.gov.eg/EgyptCare/index.aspx)23. Co-infection with bacteria, fungi, and respiratory viruses in SARS-CoV-2 is of particular importance due to the possibility of increased morbidity and mortality21. Therefore, physicians need to be cognizant about excluding other treatable respiratory pathogens22. The rate of co-infection among these pathogens is unknown. Therefore, a positive diagnostic test for another infection does not exclude the need for other micro-organism testing21.
Our results revealed that out of 599 patients presented with SARI from October 2020 till June 2022, 158 samples showed positive SARS-CoV-2 results. Our positivity rate is within the range found in recent studies reported during same time period during COVID-19 pandemic; 23.1% positive for SARS-CoV-2 among Indian patients with SARI/influenza like illness from April to September 202024; 52% positive for SARS-CoV-2 among Irish SARI cases from July 2021 and April 202225; from 10% to 24% in the EU/EEA depending on the week of report during 202312; and 89% among adult SARI patient in United States between March 2020 and April 202326. This disparity in the prevalence of detection of SARS-CoV-2 during this period may be due to regional variability12, the difference in the inclusion criteria of the cases in the different studies, such as the highest trend reported from U.S. may be attributed to age of included patients (median age was 60 years), meanwhile, our study included age group < 15 - >60 years old. Young age group showed significant low SARS-CoV-2 infection, which matches with studies that reported the frequency of positivity on RT-PCR for SARS-CoV-2 was higher in adults than in adolescence and both were higher than in children27. As reported by Goldstein et al. that children aged under 10y have significantly lower susceptibility to SARS-CoV-2 infection, while adults aged over 60y have elevated susceptibility to infection28. Our results showed that the middle-aged and the elderly made up higher percentage of all patients. Similarly, different studies indicated that patients infected with SARS-CoV-2 are mostly middle-aged and elderly15,29.
In the current study, all patients presenting as SARI, either positive or negative PCR COVID-19, had the same common symptoms including fever, cough, sore throat, dyspnea, which matches with other report of same time period30; loss of taste (ageusia) preceding the onset of respiratory symptoms has also been reported to be with SARS-CoV-2 infected patients11, and was significantly linked to SARS-CoV-2 infected group in our study; running nose was significantly linked to SARI patients with SARS-CoV-2 negative test, which in concordance with that reported by Khutade et al.7. Diabetes and hypertension were significantly associated with COVID-19 infection, which matches with that reported by Akhtar et al.31.
After selection of all factors in univariate analysis with a p-value ≤ 0.35, multivariate analysis with different approaches was used to select the most significant factors that influence in-hospital mortality in studies conducted on COVID-19 Egyptian patients during the same period. Being elderly (> 60 years), having a shorter duration of complaint, having a high NLR, and having a higher CT-SS (> 20) were all significant independent predictors of mortality (P < .05 for all), meanwhile, ischemic heart disease, hypertension, hemodialysis and stroke were non- significant32.
Our results showed that COVID-19 patients with certain comorbidities (hypertension, and diabetes) had a significant incidence of coinfection (p-value 0.008 and 0.027 respectively). This matches with Trifonova et al.33, who reported that in co-infected adults with SARS-CoV-2 and other respiratory viruses, the proportion of patients with concomitant diseases was 70%, and the most common were hypertension (48.9%) and diabetes (24.5%). Mukherjee et al.34 found a high prevalence of comorbid conditions among hospitalized patients with laboratory confirmed viral SARI, including 50% with hypertension, 34% with diabetes mellitus, 17% with CKD and 15% with coronary artery disease.
All samples were evaluated for 33 respiratory pathogens in which 31 pathogens were identified (12 bacteria, 18 viruses, and 1 fungus) with a detection rate of positive for any infection 81.5% (488/599), and positive for co-infection 75% (450/599). Zhang et al., reported positive rate of respiratory pathogen strains of 40.18%, and 4.69% of mixed infection were detected during same period of COVID-19 pandemic among patients with respiratory tract infection, whereas the patients were screened for only 13 pathogen3; 72.5% reported by Hanchi et al. from January 2018 to December 2019 among SARIs children for at least one respiratory pathogen and 23.3% of them were co-infected with more than one pathogen35, and 35.5% tested positive for only SARS-CoV-2 and influenza, including 0.8% co-infection34.This disparity in the prevalence of detection of respiratory pathogens may be due to the difference in the inclusion criteria of the cases in the different studies, the population, age groups, the geographical location, the seasons, and the panel used for the molecular diagnosis7,21. The high rate of positivity in our study can be explained by the use of larger panel of respiratory pathogens as well as more sensitive molecular techniques, which detect 33 respiratory pathogens simultaneously.
In our study, 120 positive COVID-19 patients, and 330 negative COVID-19 patients were co-infected with other respiratory pathogens. The results suggest that co-infections rates are significantly higher among SARS-CoV-2 negative SARI group as compared to positive group. This matches with Sapra et al., finding and this may be due to viral interference and competitive advantage of SARS-CoV-2 in modulating the host immunity24 .
For positive co-infection studied samples, bacterial co-infections rate was 39%, in which K. pneumoniae is the most common (5.3%), while viral co-infections rate was 61% in which HCOV HKU (6.2%) was the most common and this is in concordance with other studies that reported HCOV HKU1 as the most dominant virus15,24. These results were contradicting with other research findings in which bacterial pathogens were the dominant co-infections25,36. Similar studies stated that the most common co-pathogens among COVID-19 patients were HCOV HKU115, Klebsiella species (spp.)36, K. pneumoniae29,37, in addition to other pathogens like P. jirovecii29, and EV29,38.
The high rate of fungal co-infection representing P. jirovecii in the studied patients, could be explained by the immunodeficiency brought on by SARS-CoV-2 infection, which may make room for the growth of this opportunistic fungus as reported by Dueñas D et al.29. Co-infection with P. jirovecii and SARS-CoV-2 has been reported in a patient with progressive hypoxemic respiratory failure and CD41 lymphocytopenia39. Therefore, physicians may consider additional diagnostic testing such as serum-beta-D-glucan for P. jirovecii, especially when there are other characteristics supporting co-infections and classical risk factors for P. pneumonia26.
Regarding the profile of the detected respiratory pathogens in the present study, the most common pathogens were SARS-COV2 (158/26.4%), other corona viruses (175/29.2%), para-influenza viruses (118/19.7%), influenza viruses (111/19.7%), adeno-viruses (70/11.7%), gram positive organism (249/41.6%), gram negative organism (89 /14.9%), p. jirovecii (67/11.2%), and mycoplasma pneumoniae (36/6%). Nevertheless, human rhinovirus and RSV were only detected in 50/8.3% and 41/6.8% of cases, respectively. These results are comparable to list of pathogen reported by WHO for differential diagnosis of SARI23. Our study was conducted during COVID pandemic, this makes SARS-CoV-2 at the top of the profile23. A unique wide range of pathogens were screened in our study that make our results more informative about Egyptian patients than, Fahim et al.40, who studied co-infection of only SARS-COV2 and Influenza viruses among acute respiratory infections between 2020 and 2022. Their results showed that 35.5% were positive for viruses, including 0.8% co-infection. Of them, 69.2% were co-infected with Flu A/H, 17.3% Flu-B, and 13.5% Flu A/H13. In recent studies, it was noticeable that Human rhinovirus (HRV) was the most detected respiratory pathogen37,38,41,42. However, this is in contrary to this current profile and may be explained by COVID pandemic throughout the study period.
In SARS-CoV-2 patients specific bacterial co-infecting pathogens were identified, with K. pneumoniae the most common (20.8%). A finding which has previously been reported by Sreenath et al.26 who support the systemic use of antibiotics in patients with severe SARS-CoV-2 pneumoniae based on the higher proportion of patients with co-infections in his cohort result, with rapid de-escalation based on respiratory PCR/culture results22.This finding may reflect high rates of antimicrobial use for admitted COVID-19 patients to treat documented or presumed bacterial co-infections. Thus, it is important to study the occurrence, type, and intended antimicrobial agent use in SARS-COV-2 patients in order to develop additional strategies for the optimal use of antimicrobial agents in this population21.
Co-infections among SARS-COV2 infected group were significantly associate with severity of COVID-19 infection, ICU admission, and higher mortality. Same significant association was raised by Fahim et al.40 and Sharma et al.30. This could be explained by the hypothesis that viruses elevate bacterial co-infection by impairing the host’s immune response, disrupting epithelial barrier integrity, expression of surface receptors and adhesion proteins, direct binding of virus to bacteria, altering nutritional immunity, and affecting the bacterial biofilm. Similarly, the bacteria enhance viral infection by altering the host’s immune response, up-regulation of adhesion proteins, and activation of viral proteins43.
Interestingly, our study demonstrated that SARS-CoV-2 co-infected with influenza A (4/3.33%) and C viruses (2/1.7%), and very low trend co-infected with influenza B virus (1/0.8%), although the latter was found to be a co-infecting virus with all other respiratory viruses tested (12/3.6%). This finding was reported by studies investigating simultaneous circulation of SARS-CoV-2 and influenza for fear of putting an additional burden on health care systems in 2020, 2021 and the first half of 2022. They confirmed a low percentage of patients co-infected with both viruses 2.45% by Boussarsar et al.44 and 0.7% by Dao et al.45. Both studies confirmed more co-infections in critically ill patients compared to mono-infected patients.
Solitary infection was detected in 22% among our studied samples, in which 38 (6.3%) were infected with SARS-CoV-2, 41 (6.8%) with other viruses, and 53 (8.9%) with bacteria. The high proportion of co-infections suggests that testing for SARS-CoV-2 and other common respiratory pathogens should be carried out to ensure accurate diagnoses, prompt patient treatment, and appropriate isolation46.
Our finding of high trend of co-infections, underlines the usefulness of multiplex PCR in the etiological diagnosis of respiratory infections due to their high sensitivity, specificity, and rapidity42, especially when it is difficult to identify the etiologic pathogen on the basis of symptoms only. Indeed, respiratory infections are characterized by low specificity of clinical assessment46. A study conducted by Lee et al.47, showed that the adoption of a molecular respiratory panel with a short turnaround time provides significant benefit and improves decision-making for patient management by reducing antibiotic use and chest X-ray prescription, and finally decreasing the length of inpatients stay. Furthermore, Brendish et al.48 report that the rapid turnaround time was strongly correlated with earlier hospital discharge and early antibiotic discontinuation. They suggest that there is an early “window of opportunity” for the results of the molecular diagnostic tests to change patient management and support the turnaround time as a crucial determinant of medical decision-making. However, the cost-effectiveness of a molecular diagnosis for respiratory pathogens in pediatric patients remains to be determined. Despite the fact that the diagnostic strategy for the respiratory pathogens co-infection in SARI patients based on multiplex PCR is costly, it is likely reduce the overall cost of care as it significantly reduces the length of hospital stay, antibiotic use, better case management and the preparedness for future pandemics48 .
Data availability
All data generated or analyzed during this study are included in this published article.
References
Shi, T. et al. Global disease burden estimates of respiratory syncytial Virus–Associated acute respiratory infection in older adults in 2015: A systematic review and Meta-Analysis. J. Infect. Dis. 222 (Supplement_7), S577–S583 (2020).
Li, Y. et al. Global, regional, and National disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in children younger than 5 years in 2019: a systematic analysis. Lancet Lond. Engl. 399 (10340), 2047–2064 (2022).
Zhang, J., Yang, T., Zou, M., Wang, L. & Sai, L. The epidemiological features of respiratory tract infection using the multiplex panels detection during COVID-19 pandemic in Shandong Province, China. Sci. Rep. 13 (1), 6319 (2023).
Balkhair, A. A. COVID-19 pandemic: A new chapter in the history of infectious diseases. Oman Med. J. 35 (2), e123 (2020).
Parry, M. F., Shah, A. K., Sestovic, M. & Salter, S. Precipitous fall in common respiratory viral infections during COVID-19. Open. Forum Infect. Dis. 7 (11), ofaa511 (2020).
Si, Y. et al. Epidemiological surveillance of common respiratory viruses in patients with suspected COVID-19 in Southwest China. BMC Infect. Dis. 20 (1), 688 (2020).
Khutade, K., Shah, H., Patil, S. & Patel, H. The demographic and clinical characteristics of the Co-infection for the detection of SARSCOV- 2 and influenza A virus by rRT-PCR. J. Adv. Sci. Res. 15 (1), 9–13 (2024).
Dallmeyer, L. K. et al. Epidemiology of respiratory viruses among children during the SARS-CoV-2 pandemic: A systematic review and meta-analysis. Int. J. Infect. Dis. IJID Off Publ Int. Soc. Infect. Dis. 138, 10–18 (2024).
Malhotra, B., Swamy, M. A., Reddy, P. V. J., Kumar, N. & Tiwari, J. K. Evaluation of custom multiplex real - time RT - PCR in comparison to fast - track diagnostics respiratory 21 pathogens kit for detection of multiple respiratory viruses. Virol. J. 13, 91 (2016).
Barratt, K. et al. Comparison of the fast track diagnostics respiratory 21 and seegene allplex multiplex polymerase chain reaction assays for the detection of respiratory viruses. Br. J. Biomed. Sci. 74 (2), 85–89 (2017).
WHO. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected: interim guidance, 13 March 2020. https://iris.who.int/handle/10665/331446 (2020).
Kenmoe, S. et al. Comparison of FTD® respiratory pathogens 33 and a singleplex CDC assay for the detection of respiratory viruses: A study from Cameroon. Diagn. Microbiol. Infect. Dis. 94 (3), 236–242 (2019).
Xing, Q. et al. han, Precautions are Needed for COVID-19 Patients with Coinfection of Common Respiratory Pathogens [Internet]. medRxiv; [cited 2024 Sep 8]. p. 2020.02.29.20027698. Available from: https://www.medrxiv.org/content/. https://doi.org/10.1101/2020.02.29.20027698v2 (2020).
Wee, L. E. et al. Community-acquired viral respiratory infections amongst hospitalized inpatients during a COVID-19 outbreak in Singapore: co-infection and clinical outcomes. J. Clin. Virol. Off Publ Pan Am. Soc. Clin. Virol. 128, 104436 (2020).
Wang, M. et al. Clinical diagnosis of 8274 samples with 2019-novel coronavirus in Wuhan [Internet]. Infectious Diseases (except HIV/AIDS); Feb [cited 2023 Oct 29]. https://doi.org/10.1101/2020.02.12.20022327 (2020).
Massey, B. W., Jayathilake, K. & Meltzer, H. Y. Respiratory microbial Co-infection with SARS-CoV-2. Front. Microbiol. 11, 2079 (2020).
Chen, X. et al. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 104 (18), 7777–7785 (2020).
Hashemi, S. A., Safamanesh, S., Ghasemzadeh-moghaddam, H., Ghafouri, M. & Azimian, A. High prevalence of SARS-CoV-2 and influenza A virus (H1N1) coinfection in dead patients in Northeastern Iran. J. Med. Virol. 93 (2), 1008–1012 (2021).
Roh, K. H. et al. Coinfections with respiratory pathogens among COVID-19 patients in Korea. Can. J. Infect. Dis. Med. Microbiol. J. Can. Mal Infect. Microbiol. Médicale. 2021, 6651045 (2021).
Kandeel, A. et al. Multicenter study to describe viral etiologies, clinical profiles, and outcomes of hospitalized children with severe acute respiratory infections, Egypt 2022. Sci. Rep. 13 (1), 21860 (2023).
Assane, D. et al. Viral and bacterial etiologies of acute respiratory infections among children under 5 years in Senegal. Microbiol. Insights. 11, 1178636118758651 (2018).
Nguyen, H. K. L. et al. Surveillance of severe acute respiratory infection (SARI) for hospitalized patients in Northern Vietnam, 2011–2014. Jpn J. Infect. Dis. 70 (5), 522–527 (2017).
ECDC. Acute respiratory infections in the EU/EEA: epidemiological update and current public health recommendations [Internet]. 2023 [cited 2024 Sep 8]. https://www.ecdc.europa.eu/en/news-events/acute-respiratory-infections-eueea-epidemiological-update-and-current-public-health.
Saied, A. A., Metwally, A. A., Madkhali, N. A. B., Haque, S. & Dhama, K. Egypt’s COVID-19 recent happenings and perspectives: A Mini-Review. Front. Public. Health. 9, 696082 (2021).
Alhumaid, S. et al. Coinfections with bacteria, fungi, and respiratory viruses in patients with SARS-CoV-2: A systematic review and Meta-Analysis. Pathogens 10 (7), 809 (2021).
Sreenath, K. et al. Coinfections with other respiratory pathogens among patients with COVID-19. Microbiol. Spectr. 9(1). https://doi.org/10.1128/spectrum.00163-21.
WHO. Clinical care of severe acute respiratory infections – Tool kit [Internet]. 2022 [cited 2024 Sep 8]. https://www.who.int/publications/i/item/clinical-care-of-severe-acute-respiratory-infections-tool-kit.
Sapra, M. et al. Respiratory viral infections other than SARS CoV-2 among the North Indian patients presenting with acute respiratory illness during the first COVID-19 wave. VirusDisease 33 (1), 57–64 (2022).
Dueñas, D., Daza, J. & Liscano, Y. Coinfections and superinfections associated with COVID-19 in Colombia: A narrative review. Med. (Mex) 59 (7), 1336 (2023).
Sharma, A. et al. Clinical features and mortality in COVID-19 SARI versus Non COVID-19 SARI cases from Western Rajasthan, India. J. Fam Med. Prim. Care. 10 (9), 3240–3246 (2021).
Akhtar, Z. et al. SARS-CoV-2 and influenza virus coinfection among patients with severe acute respiratory infection during the first wave of COVID-19 pandemic in Bangladesh: a hospital-based descriptive study. BMJ Open. 11 (11), e053768 (2021).
Elgohary, M. A. et al. Development and validation of a predictive scoring system for in-hospital mortality in COVID-19 Egyptian patients: a retrospective study. Sci. Rep. 12 (1), 22352 (2022).
Trifonova, I. et al. Clinical significance and role of coinfections with respiratory pathogens among individuals with confirmed severe acute respiratory syndrome coronavirus-2 infection. Front Public Health [Internet]. 2022 [cited 2023 Oct 26];10. https://www.frontiersin.org/articles/. https://doi.org/10.3389/fpubh.2022.959319.
Mukherjee, V. et al. COVID-19 across pandemic variant periods: the severe acute respiratory Infection-Preparedness (SARI-PREP) study. Crit. Care Explor. 6 (7), e1122 (2024).
Hanchi, A. L. et al. Epidemiology of respiratory pathogens in children with severe acute respiratory infection and impact of the multiplex PCR film array respiratory panel: A 2-Year study. Int. J. Microbiol. 2021, 2276261 (2021).
Abd El-Halim, M., Hafez, R., Albahet, H. & Sherif, I. Respiratory co-infections in COVID-19-positive patients. Eur. J. Med. Res. 28 (1), 317 (2023).
Sulayyim, H. J. A., Ismail, R., Hamid, A. A. & Ghafar, N. A. Antibiotic resistance during COVID-19: A systematic review. Int. J. Environ. Res. Public. Health. 19 (19), 11931 (2022).
Uhteg, K., Amadi, A., Forman, M. & Mostafa, H. H. Circulation of Non-SARS-CoV-2 respiratory pathogens and coinfection with SARS-CoV-2 amid the COVID-19 pandemic. Open. Forum Infect. Dis. 9 (3), ofab618 (2022).
Menon, A. A. et al. A case of COVID-19 and Pneumocystis jirovecii coinfection. Am. J. Respir Crit. Care Med. 202 (1), 136–138 (2020).
Fahim, M. et al. Epidemiology, disease severity and outcome of severe acute respiratory syndrome coronavirus 2 and influenza viruses coinfection seen at Egypt integrated acute respiratory infections surveillance, 2020–2022. Can. J. Infect. Dis. Med. Microbiol. J. Can. Mal Infect. Microbiol. Medicale. 2022, 7497500 (2022).
Pan, F., Wang, B., Zhang, H., Shi, Y. & Xu, Q. The clinical application of filmarray respiratory panel in children especially with severe respiratory tract infections. BMC Infect. Dis. 21 (1), 230 (2021).
Juliana, A. E. et al. Viral causes of severe acute respiratory infection in hospitalized children and association with outcomes: A two-year prospective surveillance study in Suriname. PLoS ONE. 16 (2), e0247000 (2021).
Lei, C. et al. Viral etiology and epidemiology of pediatric patients hospitalized for acute respiratory tract infections in Macao: a retrospective study from 2014 to 2017. BMC Infect. Dis. 21 (1), 306 (2021).
Boussarsar, M. et al. Epidemiology and burden of severe acute respiratory infections (SARI) in the aftermath of COVID-19 pandemic: A prospective Sentinel surveillance study in a Tunisian medical ICU, 2022/2023. PLOS ONE. 18 (12), e0294960 (2023).
Dao, T. L., Hoang, V. T., Colson, P., Million, M. & Gautret, P. Co-infection of SARS-CoV-2 and influenza viruses: A systematic review and meta-analysis. J. Clin. Virol. Plus. 1 (3), 100036 (2021).
Morales-Jadán, D. et al. Coinfection of SARS-CoV-2 with other respiratory pathogens in outpatients from Ecuador. Front Public Health [Internet]. 2023 Oct 27 [cited 2024 Sep 8];11. https://www.frontiersin.org/journals/public-health/articles/. https://doi.org/10.3389/fpubh.2023.1264632/full.
Lee, B. R., Hassan, F., Jackson, M. A. & Selvarangan, R. Impact of multiplex molecular assay turn-around-time on antibiotic utilization and clinical management of hospitalized children with acute respiratory tract infections. J. Clin. Virol. Off Publ Pan Am. Soc. Clin. Virol. 110, 11–16 (2019).
Brendish, N. J., Malachira, A. K., Beard, K. R., Ewings, S. & Clark, T. W. Impact of turnaround time on outcome with point-of-care testing for respiratory viruses: a post hoc analysis from a randomised controlled trial. Eur Respir J [Internet]. Aug 1 [cited 2024 Sep 8];52(2). https://erj.ersjournals.com/content/52/2/1800555 (2018).
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
This research was funded by Egypt Center for Research and Regenerative Medicine (ECRRM) (Grant number: 1/2021).
Author information
Authors and Affiliations
Contributions
KA, SAG: Create original concept of research. SAG, WAH: Methodology design and implementation. AAE, SHAA, HH, FME, OMA, TMH, and YKF: Processed samples and performed lab analysis. AYA, ISH: Interpretation of data (statistical analysis). NTF: compiled the data, and wrote the manuscript. NTF, AYA, ISH: prepared the figures and tables. MAE, SAG, HE, OIM, AOM, and MEM: manuscript reviewing & editing. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fahmy, N.T., Elgohary, M.AS., Hassan, W.A. et al. Incidence of common respiratory pathogens among patients with severe acute respiratory infection during COVID-19 pandemic in Egypt. Sci Rep 15, 15711 (2025). https://doi.org/10.1038/s41598-025-98907-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98907-y