Abstract
Pseudomonas aeruginosa (P. aeruginosa), a member of the ESKAPE family, is the major cause of infections leading to increased morbidity and mortality due to multidrug resistance (MDR). One of the main proteins involved in the Raetz pathway is LpxC, which plays a significant role in anti-microbial resistance (AMR). Our study aimed to identify a novel compound to combat MDR due to the LpxC protein. It involved in silico methods comprising molecular docking, simulations, ADMET profiling, and DFT calculations. First, an ADMET and bioactivity evaluation of the 25 top-hit compounds retrieved from ligand-based virtual screening was performed, followed by molecular docking. The results revealed compound P-2 as the lead compound, which was further subjected to DFT analysis and molecular dynamics (MD) simulations. With these analyses, our in silico study identified P-2, 3-[(dimethylamino)methyl]-N-[(2 S)-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide as a potential lead compound that may behave as a very potent inhibitor of LpxC for the development of targeted therapies against MDR P. aeruginosa.
Similar content being viewed by others
Introduction
The world is currently facing a significant challenge in the development of multidrug-resistant (MDR) bacteria, which have turned into a substantial threat, according to the World Health Organization (WHO)1. Among MDR bacteria, Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (ESKAPE) group of pathogens is of particular interest2. Within this group, P. aeruginosa emerged as one of the most potent MDR pathogens, ranking among the top three3. It is a gram-negative opportunistic bacterium responsible for various infectious diseases4. Currently, antimicrobial resistance (AMR) is estimated to be responsible for about 700,000 deaths annually, a number projected to go as high as 10 million by 20505,6.
Like other gram-negative bacteria, P. aeruginosa is enriched with lipopolysaccharide (LPS), which is the major virulence endotoxin. It comprises three parts: the O-antigen, associated with a core oligosaccharide, which is further anchored to lipid A—the latter being the main human immuno-modulator endotoxin7. P. aeruginosa must maintain the outer covering (the LPS asymmetry) and regulate LPS abundance to survive stresses and face environmental hazards8. Lipid A biosynthesis is crucial for the sustainability of the bacteria; therefore, the enzymes participating in lipid A biosynthesis are also essential for survival and growth9. Hence, the biogenesis of lipid A, which primarily occurs through the Raetz pathway, is highly regulated.
The Raetz pathway comprises six enzymes, all of which are necessary for the synthesis and viability of gram-negative bacteria. They include LpxA, LpxC, LpxD, LpxH, LpxB, and LpxK10. The sequential synthesis of lipid A requires enzymes in each step and is initiated by LpxA, which is responsible for catalyzing the reversible reaction, followed by LpxC, which is considered the main driver and key enzyme in the biogenesis of lipid A11. LpxC (UDP-3-O-((R)-3-hydroxy myristoyl)-N-glucosamine deacetylase) is a zinc-dependent metallo-amidase10. Its expression is strictly regulated in the growth phase and controlled via post-translational degradation by the FtsH protease. However, any imbalance in the cellular level of LpxC results in cellular death12. The LpxC enzyme in P. aeruginosa is highly conserved and lacks homology to mammalian proteins13. The early inhibition of lipid A enzymes such as LpxC has been observed not only to suppress the growth of P. aeruginosa but also to sensitize it to other antibiotics14. Therefore, LpxC is a significant target for substrate mimetic inhibitors in LPS biosynthesis.
Since LpxC irreversibly catalyzes the reaction in the Raetz pathway with minimal side effects, it constitutes a potential target protein for drug design. The design of LpxC inhibitors represented a pioneering clinical development. Many studies have revealed the potency of LpxC inhibitors, demonstrating their efficient bactericidal effects through in vivo experiments9,14,15. Moreover, LpxC inhibitors are not inactivated by resistance mechanisms such as extended-spectrum beta-lactamase or carbapenemase activity16. Extensive research has been conducted to identify novel LpxC inhibitors against P. aeruginosa. Several compounds, such as CHIR-09017, ACHN-97518, and PF-5,081,09019, have been reported as LpxC inhibitors, but many have been terminated due to their potential toxicity and limited efficacy, with only a few entering clinical trials20. However, despite extensive research and efforts by several biopharmaceutical groups to improve their drug-like characteristics, these inhibitors have not yet reached the market21. In such a challenging landscape, there is an immediate need for novel medication.
The rapid development of AMR raises concerns due to the limited efficacy of existing drugs22. To combat AMR, drug discovery and development paved a path involving an efficient in silico approach23. Ligand-based virtual screening (LBVS) and molecular docking are among several in silico methods to identify the inhibitory potential of multiple drug candidates, predict their binding interactions with the amino acid residues of microbial targets, optimize their efficacy, and evaluate their pharmacokinetics and toxicity profiles24. Moreover, density functional theory (DFT) analysis has been widely employed in recent studies25,26 to gain in-depth knowledge of the molecular reactivity of the compound with its target receptors. Several studies have utilized in silico advancements in the form of computational molecular dynamics (MD) simulations and Molecular Mechanics/Poisson-Boltzmann Surface Area approaches to investigate the efficacy of various novel inhibitors27,28,29,30. Implementing such in silico tools and techniques provides a viable route for speeding up progress in solving the challenges posed by AMR.
The present study aimed to employ in silico techniques, including molecular docking, pharmacokinetic evaluation, toxicity prediction, bioactivity assessment, and DFT analysis, along with MD simulation, to identify a potential novel compound that can inhibit the LpxC protein, a potential target in the Raetz pathway, to effectively combat MDR.
Methodology
Target protein retrieval
The crystallized structure of the target LpxC protein was retrieved from the Protein Data Bank (PDB; RCSB PDB: Homepage, accessed on September 4, 2024) in PDB format. The crystal structure of the LpxC protein with the PDB ID 5U3B contains alpha and beta chains of 299 amino acids, with no mutation and a resolution of 2.00Å21.
Ligand-based virtual screening (LBVS)—analog search
SwissSimilarity is an online server (SwissSimilarity) used for LBVS (accessed on September 4, 2024) to identify molecular similarities between query molecules. This web tool is useful in exploring new compounds with bioactive characteristics, analogs with chemically distinct core structures, new compounds for structure-activity relationship (SAR) studies, and drug discovery and development31. In this study, a unique ligand, PubChem CID 59,323,957, was chosen as a reference compound for virtual screening. The string of SMILES was incorporated, and SwissSimilarity analysis was conducted in a single cycle by selecting a ZINC drug-like library, as well as a Chembl drug-like library, for drug discovery combining two-dimensional (2D) and three-dimensional (3D) methods because it is significantly better for drug-like molecules. A cut-off of 90% was set for the compounds against the resultant analogs from the libraries for further analysis.
Assessment of pharmacokinetic characteristics
Computer-aided drug design (CADD) features in pharmacokinetics. This approach assesses the pharmacological properties of compounds, namely absorption, distribution, metabolism, and excretion (ADME). Swiss-ADME (SwissADME), an online web server (accessed on September 4, 2024), predicted the ADME of the compounds for pharmacological evaluation32. It is a user-friendly server that employs molecular fingerprinting metrics, mainly derived from cheminformatics algorithms and fingerprints, to evaluate the pharmacological descriptors of the compounds. The strings of SMILES of all 25 compounds were entered, and ADME analysis was performed.
Toxicity prediction
Toxicity evaluation is an important step in the development of a drug-like candidate. ProTox 3.0 (ProTox-3.0 - Prediction of TOXicity of chemicals (charite.de), accessed on September 4, 2024), a user-friendly online server, was utilized to predict toxicity across various parameters such as potential organ toxicity, toxicological endpoint, and pathways of the selected compounds33. The string of canonical SMILES was used as an input file to run toxicity predictions.
Screening of potential targets
Swiss Target Prediction (SwissTargetPrediction), accessed on September 4, 2024), an online tool, was utilized for ligand-based target prediction to determine the predicted protein target with the most potential for the bioactive molecule34. The compounds were also screened for potential off-targets using a chemogenomics approach35. The string of canonical SMILES of the query molecules was given to predict these targets.
Bioactivity assessment
Molinspiration (Molinspiration Cheminformatics), a web server (accessed on September 4, 2024), was utilized to evaluate the bioactivity scoring of the top-hit compounds across multiple parameters. Each compound was uploaded in MOL format for evaluating interactions36.
Ligand retrieval and preparation
The top-hit virtually screened compounds were selected for ligand preparation for molecular docking. The 2D and 3D structures of the selected compounds were sketched on ChemDraw Professional 16.0 software (Version 16.0.1.4.77) and the 3D structures were saved in SDF format.
The ligands and the CCL were prepared for docking through structural optimization and conformer generation, carried out on the Ligprep tool accessible on Schrödinger’s 2020-3 (Version 12.5.139) computational software. Using the Epik module, desalting and tautomer production were performed at pH 7.0 ± 2, generating a total of 32 poses for each ligand by setting the force field of OPLS3e for minimization37.
Protein preparation
The crystallographic structure of the target protein LpxC (PDB ID: 5U3B) was obtained in PDB format and prepared on Maestro, a suite of Schrödinger 2020-3 (Version 12.5.139) by the Protein Preparation Wizard, a computation tool. The target protein was imported to the interface of the software, and initially, “prime job” was selected to fill the gap within the side chains and missing loops. After that, a het state was generated by utilizing the Epik module (pH = 7.0 ± 2), creating a zero-order state for metals. Further, water molecules that were 3 Å beyond the heteroatoms were removed, and hydrogen atoms were introduced. PROPKA at a pH of 7.0 was employed for optimization. For further improvement, the steric hindrance was eliminated by converging the heavy atom root mean square deviation (RMSD) to 0.30 Å; finally, the target protein was energy-minimized by setting the force field of OPLS3e38.
Glide grid generation
Glide grid generation, a computational approach, is accessible on Schrödinger 2020-3 (Version 12.5.139). It was used to identify favorable interactions among the receptor and ligands for a more precise estimation of the binding score for the ligand configuration39. With the help of the receptor grid generation panel in the Maestro gliding program, the active site residues were specified and selected to create a receptor grid for the protein site in the centroid of the workspace of the CCL40.
Molecular docking
The refined molecular docking of the ligands with the target protein LpxC (PDB ID: 5U3B) was performed on Maestro, Schrödinger 2020-3 (Version 12.5.139). The final files of the energy-minimized ligands of 25 selected compounds and the CCL and output files of the grid-generated receptor were subjected to Ligand Docking for docking purposes41. The molecular docking protocol set an extra precision mode to determine binding modes for flexible ligand sampling. For further improvisation, the per-residue scores were written within 12 Å of grid generation, and RMSD was computed to input ligand geometries. Finally, the interactions were visualized and analyzed on Discovery Studio Visualizer (v21.1.0.20298).
Density functional theory (DFT) calculations
The 3D geometries of the narrowed compounds were designed by GaussView (Version 5.0.8) and further visualized and optimized using the Gaussian 09 W program42. The B3LYP method, a hybrid functional combining Becke’s three-parameter exchange functional with the Lee–Yang–Parr correlation functional, was selected due to its balance of computational efficiency and accuracy in modeling organic molecules. For both gaseous and physiological phases, the Conductor-like Polarizable Continuum Model solvation model was applied to mimic biological environments, typically using water as the solvent, which affects the electron distribution and geometry optimization of the molecules43.
The highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbitals (LUMO) energy levels and their band gaps were computed using the same level of theory. These values were further used to calculate the quantum chemical parameters through Koopman’s theorem using the following equations44,45:
Molecular electrostatic potential surface (MEPS) mapping
The DFT approach analyzed the electron distribution of charges through molecular electrostatic potential surface (MEPS) mapping of the molecules using checkpoint (.chk) files generated by the Gaussian 09 W program. The surface mapping was further visualized on Gauss View 5.0.842. MEPS maps were used to identify electron-rich (nucleophilic) and electron-poor (electrophilic) regions on the molecular surface. Red regions on the map indicate areas of high electron density (negative potential), while blue regions indicate electron-deficient areas (positive potential). This approach helps in predicting potential sites of interaction with biomolecule targets46.
Molecular dynamic (MD) simulations
Desmond, a suite of Schrödinger47,48 was employed to execute the simulations to determine the kinetics and dynamic characteristics of the protein-drug interactions49 using the OPLS 2005 force field algorithm50. Initially, the missing residues of the complexes were filled, followed by the optimization and minimization of the protein using Maestro’s Protein Preparation tool. Before initiating the simulation process, both docked complexes were subjected to the system builder tool using a predefined simple point charge solvent model in an orthorhombic box with dimensions of 10 Å ×10 Å × 10 Å under an OPLS4 force field, 300 K temperature, and 1 atm pressure with 0.15 M NaCl, used for neutralization under physiological circumstances. The simulation was run under Newtonian dynamic equations to predict molecular motions, and the system was further equilibrated using a TIP3P water model and OPLS4 force field in a related comparative analysis51,52. For pressure and temperature control, two coupling schemes were employed, that is, the Martyna–Tuckerman–Klein chain and Nosé–Hoover chain, respectively53. The docked complexes of the CCL and compound P-2 were subjected to simulations for 100 ns by keeping a 100 ps recording interval of the simulation energy, with a force field of OPLS4 and a trajectory recording of 100 ps. The expected changes in the stability and conformation of the complexes were represented as the RMSD and root mean square fluctuation (RMSF) for MD simulations. The radius of gyration (Rgyr) was computed using GROMACS54 by converting the Desmond trajectory to XTC (GROMACS) format via MDTraj39, after which the gmx gyrate command was employed.
Principal component analysis (PCA) and dynamic cross-correlation matrix (DCCM)
For a more detailed examination and understanding of the motion of MD trajectories, especially the internal motion of the protein, principle component analysis (PCA), a dimensionality-reduction statistical method, was applied by creating a covariance matrix using C-alpha (Cα) coordinates from MD simulations. The covariance matrix was constructed and diagonalized using the g_covar command in GROMACS, generating a set of eigenvectors and eigenvalues that described the major collective motions of the protein55. The coordinates were mapped according to the eigenvalues and eigenvectors using GROMACS54. The first two principal components (PC1 and PC2), representing the most significant movements, were extracted to visualize the essential dynamics in a 2D phase space using the g_anaeig command55. Further, a cross-correlation matrix was constructed across all Cα atoms for both complexes during the 100 ns simulation trajectory for the investigation of domain correlations by using the DCCM function in the Bio3D package in R language to compute the DCCM56,57.
Free-energy landscape (FEL)
Free energy landscape (FEL) was utilized to estimate variations in energy during the simulation trajectory, allowing for the identification of the receptor’s metastable conformations and their relative energies in the form of Gibbs free energy. GROMACS54 was employed to create FEL plots.
Free binding energy calculations
Binding free energy calculations were carried out for the docked output receptor-ligand complex on Schrödinger using molecular mechanics with a generalized born surface area (MM/GBSA) methodology. The prime module was chosen to estimate the theoretical binding-free energy of the target receptor LpxC with the lead compound, based on the docked scores58.
Results
Virtual screening
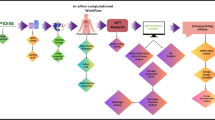
The virtual screening procedure is outlined in the methodology section in (Fig. 1). The analogs generated by the Chembl drug-like library resulted in a similarity score of 82% or less, illustrated in Supplementary File 1. The ZINC drug-like library was also screened and resulted in 400 analogs with a similarity index of 95% or less, highlighted in Supplementary File 2. Based on the similarity score, the top 25 compounds generated through the ZINC drug-like library were chosen against the given ligand (PubChem CID: 59323957) of the target protein (PDB ID: 5U3B), shown in (Fig. 2). All compounds that fell within the cut-off range of ≥ 0.900 were shortlisted for further computational analysis, illustrated in (Supplementary Table 1).
Pharmacokinetic analysis
Various pharmacokinetic properties of the top-screened bioactive compounds were examined with SwissADME, a web tool that gives an overview of their structures and predicts their ADME profiles, as illustrated in (Table 1). ADME was evaluated across the following descriptors: physiochemical parameters, solubility, drug-likeness, medicinal chemistry, and pharmacokinetics.
The physiochemical analysis of the compounds revealed that the top-screened compounds had a molecular weight of ≤ 500 Daltons, the total number of H-bond acceptors and donors was not more than 10 and 5 each, respectively, and the log P (octanal-water partition coefficient) of the compounds was ≤ 5, indicating that the compounds fell within the optimal cut-off range of the Lipinski rule of five (LRF). Generally, the compounds that appeared after LRF had better permeability through the gastrointestinal tract and were drug-like molecules59. Moreover, all the compounds had a total polar surface area60 of ≤ 117 Å. All compounds had ≤ 10 rotatable bonds except P-17 and P-4, which contained 11 rotatable bonds (Supplementary Table 2).
The log S value defines the nature of compound solubility in the physiological medium61. The results showed that all compounds had a log S value ≤ 5 except P-9 (Supplementary Table 1). Moreover, the compounds P-1, P-2, P-13, P-14, P-16, and P-23 showed the best log S values, indicating excellent water solubility.
Pharmacokinetic evaluation of compounds is a significant step in drug design and development as it predicts the movement of the drug inside the body62. All compounds demonstrated high gastrointestinal absorption, suggesting good potential for oral bioavailability. The results evidenced that the candidate drug pharmacological profile was remarkably influenced by P-glycoprotein63, showing that of the 25 compounds, most were not the substrate of PGP, indicating a lower incidence of resistance in further in vitro studies64, while a few compounds (P-4, P-9, P-11, P-13, P-14, P-18, P-23, and P-25) were identified as substrates for PGP (PGP-positive).
The drug-likeness of the compounds was assessed based on the rules set forth by the eminent researchers Lipinski, Veber, and Egan, contributing to delineating their molecular properties65,66. All compounds showed zero violation of these rules except P-4 and P-17, which violated the Veber rule (> 9 rotatable bonds). The remaining compounds exhibited the potential to interact with the target of interest effectively67.
Medicinal chemistry revealed that none of the compounds showed any PAINS and Brenk alerts, except P-1 and P-2; however, the CCL showed three Brenk alerts. Moreover, the synthetic accessibility score of the compounds was less than 4; this meant the compound was easily synthesized (regarding the synthetic accessibility profile, a score of 1 indicates easy synthesis, while a score of 10 indicates difficulty68. Finally, bioavailability refers to the absorption rate of drugs entering the circulation in their complete form. The bioavailability scores of all the investigated compounds remained constant at 0.55: as seen in (Fig. 3), the compounds falling in the pink zone of the bioavailability radar had excellent oral bioavailability except for P-4, P-11, P-16, P-17, P-19, and P-25, which possessed zero flexibility based on > 9 rotatable bonds (Supplementary Fig. 1).
Toxicity prediction
ProTox 3.0 was applied to predict compound toxicity. This web server predicted toxicity based on 33 models of in vivo as well as in vitro data69. The results predicting the hepatotoxicity, toxicological endpoints, and toxicological pathways are illustrated in (Supplementary Tables 3–5). They illustrated that all compounds had a non-toxic profile regarding organ toxicity (hepatotoxicity) and toxicity pathways (Tox-21 nuclear receptor signaling pathway and Tox-21 stress response pathway). However, the toxicity endpoints showed unexpected results, as carcinogenicity and cytotoxicity were inactive for all the compounds, while the mutagenicity status was active for P-1, P-2, and P-13, as well as for the reference compound. The immunotoxicity status was active for P-3, P-7, P-12, P-14, P-15, P-17, P-19, P-21, P-22, P-23, and P-24. However, new approach methodologies in immunotoxicity and mutagenicity could be implemented for these compounds to be used as drugs70.
Screening potential targets
Bioactive prediction was performed using the online tool Swiss Target Prediction to estimate the most probable protein targets for the compounds71. Most were predicted as kinase inhibitors and G-protein coupled receptors, as highlighted in (Fig. 4). A few compounds were also predicted to act as enzyme and protease inhibitors, highlighted in (Supplementary Fig. 2). Moreover, the chemogenomics analysis further identified ligand-based targets for the compounds, with a minimal number of predicted off-target interactions (for P-1 and P-2). The results provided additional insight into potential off-target interactions, indicating specificity for the target protein LpxC (Supplementary File 3).
Bioactivity assessment
The bioactivity prediction was further confirmed using a Molinspiration cheminformatics online tool, as the drug is supposed to bind to its potential target. The bioactive probability scoring for organic molecules is as follows: a score > 0 indicates it is active, a score between − 5.0 and 0.0 denotes moderately active, and a score < − 5.0 denotes inactive72. The results in Table 2 (Supplementary Table 6) show that out of all top-hit compounds, P-1 and P-2 exhibited the highest bioactivity against kinase inhibitors that are effective against tumors73, protease inhibitors that are effective against the proteolytic activity of bacteria74, and enzyme inhibitors75.
Molecular docking
The top 25 compounds were further screened by docking analysis to investigate the binding affinities and interactions of each compound with the target protein LpxC (PDB ID: 5U3B). Initially, the CCL (PubChem CID: 59323957) was re-docked within the allosteric site using Schrödinger software to validate docking studies. The process was executed under the guidelines outlined in the methodology. The re-docked superimposition showed an RMSD value of 0.193 Å (Fig. 5), indicating the validity and efficacy of the docking procedure76. The outcomes of the docking analysis of top-hit compounds are illustrated in (Table 3), revealing that the docking score ranged from −7.923 kcal/mol (P-21) as the highest docking score to −2.531 kcal/mol (P-20) as the lowest docking score (Supplementary Table 7).
Interaction analysis
Notably, high binding affinities and interactions of the compounds with the target LpxC were linked to key structural features of the receptor’s active site. SAR studies revealed that interactions of the zinc-binding hydroxamate group with the compounds were crucial for strong binding, ultimately enhancing enzymatic inhibition. The involvement of polar residues near the pocket was responsible for hydrophilic interactions necessary for increased specificity and solubility. Moreover, the hydrophobic moieties in the structure were essential for stabilizing the inhibitor within the active site of the target receptor due to non-polar interactions (Fig. 5B).
The 2D and 3D interactions of top-docked scored compounds were visualized on Discovery Studio Visualizer, as shown in Fig. 6 (Supplementary Fig. 3). P-1, P-2, and P-23 were metal acceptors of ZN501 forming a bond with oxygen, similar to the CCL. The key amino acid residue LYS238 of CCL formed an unfavorable positive-positive bond only with the amine group (-NH2) of P-21. A conventional hydrogen bond was formed in the CCL with the following amino acid residues: THR190, GLU77, MET62, and HIS264 between the carbonyl and amine groups of the ligand. The same type of interaction was also observed in P-1, forming a conventional H-bond with residues THR190 and MET62 between the carbonyl and amine group of P-1. Similarly, the carbonyl and amide groups of P-2 interacted with THR190, GLU77, MET62, and HIS264, along with PHE191 and HIS19, forming an H-bond. The carbonyl group of P-13 interacted with THR190 along with LYS238, while MET62 and PHE191 showed van der Waals interactions. Likewise, the carbonyl and amine groups of P-18 formed a similar H-bond with THR190, along with LYS238 and LYS261, while MET62 and HIS264 showed van der Waals interactions similar to P-13. P-21 only formed an H-bond between the amino acid residue PHE191 and the benzamide group. P-22 formed a conventional H-bond between key amino acid residues THR190 and PHE19 and the hydrogen of the amine group only. The resulting docked ligand P-22 did not act as a metal acceptor; instead, it showed van der Waals interactions with ZN501. Like the CCL, P-23 formed a typical H-bond with the key interacting amino acid residues and the hydrogen of amine groups and oxygen of the carbonyl group.
Binding mode of the complex 5U3B with the best inhibitory compounds. (A,C,E,G,I,K,M) show the visual 3D representations of the binding cavity of 5U3B with the top-scoring ligands P-1, P-2, P-13, P-18, P-21, P-22, and P-23, respectively. (B,D,F,H,J,L,N) depict the 2D plot of the binding interactions between the top inhibitors and LpxC (PDB ID: 5U3B).
Pi-alkyl interactions were observed in all the compounds. The key amino acid residues involved in pi-alkyl interactions were LEU18, ALA214, ALA206, MET194, VAL216, and ILE197 within the benzamide moiety of the following compounds: P-1, P-2, P-18, P-21, P-22, and P-23. Additionally, these interactions were formed between the cyclohexylamine moiety of P-18, P-21, and P-22 and the cyclopentyl moiety of P-23.
Density functional theory (DFT) analysis
The present study focused on FMOs and quantum descriptors of top-hit compounds using Koopman’s theorem. Such compounds were filtered based on the following criteria: ADMET characterization, bioactivity evaluation, and interactions and binding affinities of the compounds. The comparative DFT analysis between P-1, P-2, and CCL is portrayed in (Table 4).
The FMOs (HOMO and LUMO) are important orbital molecules that describe the optical and electronic properties of the molecules (Fig. 7). The HOMO and LUMO energy describes the ability of a molecule to donate and accept electrons, respectively77. The band-gap energy between these [ΔEGap = ELUMO - EHOMO] represents the energy required for the electronic transition of the compounds. The band-gap energy of the compounds indicated that P-1 and P-2 had lower gas gaps than CCL, reflecting their high reactivity toward the targeted receptors in the physiological state78.
The quantum descriptors were further calculated using the energy gap (ΔEGap). The ionization potential (I) of P-1 (5.94 eV) and P-2 (5.95 eV) was higher than that of CCL (5.07 eV), indicating more chemical inertness and higher stability79. The electron affinity (A) of P-1 and P-2 was 5.32 eV and 5.36 eV, respectively, higher than the affinity of the CCL (1.08 eV), indicating that P-1 and P-2 were more prone to accepting an electron80. The electronegativity (χ) of P-1 was 5.63 eV, that of P-2 was − 5.65 eV, and that of the CCL was 3.08 eV, with P-2 having the highest electronegativity. Moreover, the electrophilicity (ω) of P-2 (54.14 eV) was higher than that of P-1 (51.06 eV), while the CCL had much lower electrophilicity than P-1 and P-2, indicating that P-2 was the most reactive compound. The compounds’ computed chemical hardness and softness described the chemical stability of the compound, wherein a hard compound would be more resistant to the electronic cloud than a soft molecule79. The calculation showed that P-1 and P-2 were softer compounds than the CCL.
Molecular electrostatic potential surface (MEPS) mapping
The MEPS shows the distribution of charge over the entire molecule. The MEPS mapping shown in Fig. 7 represents the charge distribution in compounds P-1 and P-2 in a gradient color-coded progression from red to blue, indicating the energy distribution from negative electrostatic potential to positive electrostatic potential. The overall energy distribution in P-1 and P-2 was scaled as ± 9.392e-3 and ± 9.415e-3, respectively. Green was used for neutral domains. The MEPS revealed a more pronounced nucleophilic region for P-2, indicating the higher reactivity of the compound compared to P1. Additionally, both compounds exhibited positive electrostatic potential, indicating their electrophilicity.
MD simulation
Based on the DFT analysis, the lead compound P-2 was selected to validate the efficacy of protein-ligand interactions by employing MD simulations. MD simulations were conducted for 100 ns to attain an optimum balance between computational efficacy and insight into critical conformational changes within the receptor-ligand complex.
An integral element of our inquiry involved RMSD analysis, which is commonly applied to investigate the system equilibrium during simulations81. The patterns and differences in the mean deviation of the RSMD were studied to determine the synergistic effects of the identified compound within the allosteric site of the receptor. The plot shown in Fig. 8A depicts the RMSD values for Cα atoms for P-2 and CCL. The CCL (represented in blue) exhibited the initiation of equilibration at approximately 1.25 Å, around 25 ns, which was maintained throughout the simulations of 100 ns. Juxtaposing CCL with the P-2 complex (represented in orange), the lead compound P-2 displayed a small fluctuation in RMSD ranging within 2 Å throughout the simulations, suggesting that the complex remained stable during the simulation period and could have inhibitory activity82,83. These fluctuations commenced around 0 ns and persisted until the culmination of simulations at 100 ns. This type of fluctuation indicated significant mobility within the binding region for LpxC in the complex with P-284.
The RMSF plots provide insight into the local changes along the protein chain. The plot presented in (Fig. 8B) presents the RMSF values for the CCL (displayed in blue) and P-2 (displayed in orange) in complex with LpxC. The RMSF value ranged between 0.3 and 2Å for protein residues, showing the stability of protein for 100 ns in simulations. Both complexes showed fluctuations at the N- and C- terminus of the protein tails, which were beta-strands and loops, respectively. However, a pronounced fluctuation was visualized in the alpha-helices of P-2 when compared to the CCL. Out of 299 amino acids in LpxC (PDB ID: 5U3B), 10–30 residues (ILE-11 to PRO-30) of beta-strands and 180–220 residues (SRE-180 to ASN-220) forming alpha-helices, beta-strands, and loops showed fluctuation. Simultaneously, it is noteworthy that the fluctuations visualized lie between 0.4 and 2 Å, owing to the flexibility of the complex85, indicating the consistency and stability of the lead compound within the binding pocket during the 100 ns trajectory of simulations.
Through the RGyr, the overall compactness of the receptor, alteration, and folding status of the protein was determined. The plot shown in Fig. 8C revealed that the receptor was compact and consistent, converging to a single point close to the CCL at the end of the 100 ns trajectory. Yet, for the P-2 complex, a slight fluctuation was observed from 0 to 40 ns, ranging from 18.2 Å to 18.4 Å and 18.4 Å to 18.5 Å for another 10 ns. The possible reason for this fluctuation could be the charged residues in the N-terminus distorting the intramolecular interactions86.
Graphical representation of the simulation. (A) RMSD fluctuation of the CCL (blue) and P-2 (orange), showing the duration on the X-coordinate and RMSD on the Y-coordinate. (B) RMSF of the CCL (blue) and P-2 (orange), showing residues on the X-coordinate and RMSF on the Y-coordinate. (C) RGyr of the CCL (blue) and P-2 (orange), showing time duration on the X-coordinate and RGyr on the Y-coordinate.
PCA and DCCM analysis
The covariance matrix of the coordinate fluctuations was obtained from the principal component projection of the Cα of the protein, explaining its conformational dynamics during the simulation trajectory, shown in the eigenvalue plot (Fig. 9). The eigenvectors were evaluated based on eigenvalues, wherein the higher value controls the overall mobility of the receptor. The plot shows that both complexes showed variation in conformational clustering, with the lead compound P-2 complex showing greater flexibility and mobility during binding, whereas the CCL complex showed less conformational variation. Additionally, Fig. 10C, D illustrates the FEL of the complexes. The plot shows a short basin for the CCL with a narrow well that reflects the lower Gibbs free energy, indicating the rigid conformation of the ligand-binding site, whereas the long basin with a wide well indicates high Gibbs free energy in the P-2 complex, suggesting a flexible binding site conformation of the receptor.
Principal component analysis of the lead compound (P-2) in comparison to the CCL. (A) Combined scatter plots for both complexes (blue = CCL and orange = P-2). (B,D) Individual scatter plots of CCL and P-2, respectively in complex with the target protein 5U3B. (C,E) Plots colored to depict Gibb’s energy.
DCCM was employed to correlate the motion of residues within the protein chain. In RMSD, after 50 ns, the systems reached equilibrium; hence, we extracted those frames for DCCM analysis. The plot displayed in Fig. 10A, B shows varied patterns of DCCM for both complexes. A color-coded scheme was established to visualize the degree of correlation between the mobilities, wherein the blue and pale green colors depict the highly correlated mobility of the residues, while the white color depicts a low correlation between the residues. The CCL indicates a significantly strong correlation, with the residues’ interacting value ranging from 0 to 1.0. Simultaneously, a strong correlation was observed in P-2, with fewer off-diagonal interactions, around 180–240 amino acid residues. The DCCM analysis revealed that the lead compound P-2 exhibited more conformational changes within this region, similar to the fluctuation pattern of RMSD within this complex.
Binding free energy calculation
The MM/GBSA energy calculations were performed on the Schrödinger suite to estimate the overall binding free energy of the lead compound P-2 complexed with the target receptor LpxC compared to the CCL complex over a 100 ns simulation trajectory. The plot in Fig. 11 revealed that the CCL complex consistently showed a minimum energy level ranging from approximately −45 to −50 kcal/mol, indicating a much stronger and more stable binding affinity of the complex. On the other hand, the lead compound P-2 complexed with 5U3B showed a slightly higher energy value, spanning −35 to −40 kcal/mol. The varying energy level of the P-2 complex indicated stable and moderate binding affinity with the target receptor, as observed from the docking scores.
Discussion
AMR is an emerging threat to global health. MDR pathogens prioritized by the WHO include the ESKAPE group of gram-negative bacteria, of which P. aeruginosa exhibits significant resistance against various classes of antibiotics. The discovery of new antibiotics effective against gram-negative bacteria is challenging due to the formidable permeability barrier associated with the two-membrane structure (i.e., the inner cytoplasmic membrane and the outer LPS membrane) of P. aeruginosa. One enzyme involved in the Raetz pathway for the production and assembly of the LPS in P. aeruginosa is uridine diphosphate-3-O-(hydroxymyristoyl)-N-acetylglucosamine deacetylase (LpxC), which is highly conserved87; thus, it can be targeted while minimizing the risk of off-targets and the development of potential resistance mechanisms.
The present study targeted LpxC for the discovery of novel compounds to combat MDR due to its highly conserved nature among gram-negative bacteria88. LBVS was conducted against the CCL of the target protein (PDB ID: 5U3B) using a ZINC drug-like library to identify the top-hit bioactive analogs for further in silico analysis. Further top-hit compounds were subjected to ADMET analysis. A major advantage of employing ADMET filters was the revelation of a potential candidate for drug discovery89. The top-hit compounds based on LRF all demonstrated acceptable ADMET profiles, focusing on physicochemical features, pharmacokinetics, solubility, drug-likeness, and medicinal chemistry toxicities. However, a few compounds were reported as borderline cases that could be optimized through structural modifications. Such modifications may improve their stability and absorption. Moreover, the bioactivity assessment of the compounds revealed P-1 and P-2 as the most potent compounds, with bioactivity similar to the CCL, suggesting their potential as LpxC inhibitors. Numerous other studies have been performed on MDR bacteria to identify such novel compounds. Sudhir Kumar et al.90 employed a similar approach on gram-negative bacteria, suggesting indole-based LpxC inhibitors as novel anti-AMR compounds. The available experimental research on LpxC suggested that analogs retrieved from the ZINC library exhibited a remarkable ADMET profile, proving themselves as potential candidates against MDR pathogens91.
Subsequently, molecular docking studies were carried out with advanced computational tools based on molecular binding affinities to predict and evaluate the spatial poses and interactions between ligands and proteins, providing comprehensive insight into the complex interactions among the amino acid residues and the ligands92. The compounds with top docking scores, ranging from −7.923 kcal/mol (P-21) to −6.019 kcal/mol (P-13) showed interactions with key amino acid residues also involved in the CCL. However, compounds P-1 and P-2 exhibited more significant interactions than the CCL with the target protein (PDB ID: 5U3B), aligning with a study conducted on marine-bioactive compounds against P. aeruginosa for the dual inhibition of target proteins involved in the Raetz pathway93. Damele et al.94 and Bhaskar et al.95 also targeted the proteins involved in the Raetz pathway in search of compounds that could be used as inhibitors to combat MDR P. aeruginosa. Notably, P-2 demonstrated a suitable binding affinity and stability due to its interactions. Key interactions included the direct coordination of P-2’s hydroxamate group with the zinc ion (ZN501) in the active site, an essential interaction for enzymatic inhibition, as observed in the CCL. Additionally, P-2 formed hydrogen bonds with residues THR190, GLU77, MET62, HIS264, and PHE191, enhancing the ligand’s stability and specificity within the binding pocket. The benzamide moiety of P-2 also engaged in pi-alkyl interactions with LEU18, ALA214, ALA206, and MET194, contributing to the overall stability of the protein-ligand complex. Overall, compound P-2 possessed specificity for the target receptor due to structural and functional adaptation to the active domain of the receptor. Although the hydroxamate group has the potential to react with other metalloproteins, the complementary structural features were absent in these receptors, further minimizing the risk of off-target interactions.
Furthermore, the SAR analysis of the lead compound P-2 revealed that incorporating carboxylate or hydroxamic chelating groups could strengthen zinc metal coordination, as shown in (Supplementary Fig. 4). Lipophilicity and hydrophobicity could be improvised by using bulky aliphatic groups and other hydrophobic substitutes, respectively, to form more stable interactions with the non-polar pocket residues (such as leucine and alanine). Favorable polar hydrogen interactions could be optimized by incorporating hydroxyl or amine groups to strongly interact with histidine and glutamine residues, further repositioning the existing groups to enhance stability. Additionally, the residues involved in favorable water interactions should be retained for solvent accessibility. Optimizing the balance of hydrophilicity and hydrophobicity will ultimately improve permeability and solubility to enhance the binding affinity of the lead compound P-2. Nevertheless, the selectivity of P-2 must be confirmed through experimental assays.
To gain further insight into the reactivity and chemical stability of the lead compound, DFT calculations—a computational quantum methodology for investigating the applications of a compound in theoretical chemistry96—were made due to their cost-effectiveness and greater accuracy. This analysis represents a remarkable compromise between the computational time and the standard quality of output results97. Now, it is a highly used approach for investigating the electronic structures of compounds in computational chemistry. One such study conducted by Kumar and Bhardwaj98 investigated the stability of chrysin-cyclodextrin through DFT calculations and simulations, providing structural insight into the important atoms within chrysin-cyclodextrin. Our results, illustrated in (Table 4), showed a smaller energy gap, and quantum descriptors indicated the molecular reactivity of the lead compound toward the target receptor. MEPS mapping also revealed the electrophilicity and nucleophilicity of the lead compound P-2. The lead compound showed electrophilicity, particularly around its amine group. Notably, this functional group was exclusively involved in the formation of polar interactions, specifically conventional hydrogen bonds. Therefore, this functional group could be crucial in enhancing the ligand’s binding affinity for the key residues of the target receptor, LpxC (PDB ID: 5U3B). The electron distribution also played a vital role in the pharmacological profile of the compounds. The DFT results across various parameters indicated a balanced polarity of the compound, which is essential for efficient absorption and distribution, suggesting the efficacy of the lead compound as a potential drug candidate to combat AMR. Likewise, in silico research on the compounds retrieved from virtual screening against proteins involved in the biosynthesis of lipid A showed significant electrophilic and nucleophilic characteristics93. Moreover, research for assessing the chemical reactivity and stability of compounds against the anti-carbapenem resistance mechanisms of P. aeruginosa showed promise in combating MDR99.
Finally, MD simulations were performed comparing the lead compound P-2 and the CCL to observe receptor-ligand interactions in a dynamic setting100. While docking provided a static view of the interactions, MD simulations captured the dynamic behavior and stability of the complex over time. These simulations supported the docking results and provided additional insights. The RMSD plots demonstrated that the compound did not leave the binding pocket of the target protein throughout the simulations, with minimal fluctuation during the trajectory, which suggested the flexibility of the complex. Furthermore, the RMSF highlighted the minimal residual movement of critical binding pocket residues such as THR190, GLU77, and HIS264, further confirming the stability of these interactions. Maximum fluctuation was observed only at the N- and C-terminal regions, which were distant from the binding site and had no impact on ligand binding. On the other hand, the RGyr showed that the complex remained steady and compact throughout the 100 ns simulations. Thus, the lead compound exhibited remarkable flexibility and compactness compared to CCL within the allosteric site of the target protein (PDB ID: 5U3B). Advanced research identifying LpxC inhibitors for MDR pathogens revealed that LBVS compounds exhibited potential inhibitory activity in a dynamic, real-time setting101. Likewise, Ahmad et al.102 targeted LpxC inhibitors for MDR bacteria to observe their behavior in trajectories. Moreover, in silico studies for the discovery of novel compounds against MDR P. aeruginosa showed stability and consistency of the compound during simulation trajectories103. Subsequently, the MM/GBSA estimated the binding free energy of the lead compound P-2. This approach is most popular because of its accuracy relative to any docking score system. It has been widely employed in various in silico studies29,104.
The energy trajectory for the lead compound P-2 showed a suitable energy value, with fluctuations around − 40 kcal/mol, which is indicative of its moderately stable binding interactions with the target receptor LpxC. However, minimal structural optimization could be performed for the lead compound to attain a more stable binding affinity and further enhance its inhibitory efficacy. Together, these findings validate P-2’s strong potential as an LpxC inhibitor and highlight the value of integrating static docking analyses with dynamic simulations for a comprehensive assessment of ligand efficacy. Considering multiple evaluation parameters, the lead compound P-2 emerged as a promising novel compound exhibiting potential inhibitory activity against MDR P. aeruginosa.
Conclusion
The present study was exclusively based on in silico analysis, emphasizing the significant inhibitory potential of the novel compounds retrieved through LBVS, against MDR P. aeruginosa. Various computational tools indicated the potential pharmacological and toxicological profiles of the lead compound, with minimal off-target effects. The lead compound P-2 demonstrated considerable binding affinity (−6.263 kcal/mol) for the highly conserved non-mammalian LpxC target protein of the bacterium, suggesting its inhibitory potential against the target. Moreover, the simulations confirmed the retention of critical interactions, including zinc coordination and hydrogen bonding, and offered unique insights into the flexibility of the binding pocket and protein-ligand dynamics. These findings underscore the potential of the lead compound P-2 as an LpxC inhibitor to combat MDR P. aeruginosa. However, in vitro and in vivo studies are recommended to further authenticate the efficacy of this compound. The current research could also pave pathways for the design and development of next-generation inhibitors to combat MDR among ESKAPE members.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files].
References
Horcajada, J. P. et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32, (2019).
Amera, G. M. et al. Computer aided ligand based screening for identification of promising molecules against enzymes involved in peptidoglycan biosynthetic pathway from Acinetobacter baumannii. Microb. Pathog. 147, 104205 (2020).
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018).
Beswick, E., Amich, J. & Gago, S. Factoring in the complexity of the cystic fibrosis lung to understand Aspergillus fumigatus and Pseudomonas aeruginosa interactions. Pathog (Basel Switzerland) 9, (2020).
Shevade, A. & Naik, S. Mitigation of antimicrobial resistance (AMR) in G20. Indian Public. Policy Rev. 4, 1–29 (2023).
Islam, S. et al. Antibacterial potential of propolis: molecular docking, simulation and toxicity analysis. AMB Express 14, 81 (2024).
Simpson, B. W. & Trent, M. S. Pushing the envelope: LPS modifications and their consequences. Nat. Rev. Microbiol. 17, 403–416 (2019).
Bertani, B. & Ruiz, N. Function and biogenesis of lipopolysaccharides. EcoSal Plus 8, (2018).
Zhou, P. & Zhao, J. Structure, Inhibition, and regulation of essential lipid A enzymes. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1862, 1424–1438 (2017).
Dowhan, W. The Raetz pathway for lipid A biosynthesis: Christian Rudolf Hubert Raetz, MD phd, 1946–2011. J. Lipid Res. 52, 1857–1860 (2011).
Emiola, A., George, J. & Andrews, S. S. A complete pathway model for lipid A biosynthesis in Escherichia coli. PLoS One 10, e0121216 (2014).
Schäkermann, M., Langklotz, S. & Narberhaus, F. FtsH-mediated coordination of lipopolysaccharide biosynthesis in Escherichia coli correlates with the growth rate and the alarmone (p)ppGpp. J. Bacteriol. 195, 1912–1919 (2013).
Jackman, J. E., Raetz, C. R. & Fierke, C. A. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase of Escherichia coli is a zinc metalloenzyme. Biochemistry 38, 1902–1911 (1999).
Zhou, P. & Hong, J. Structure- and ligand-dynamics-based design of novel antibiotics targeting lipid A enzymes LpxC and LpxH in gram-negative bacteria. Acc. Chem. Res. 54, 1623–1634 (2021).
Niu, Z. et al. Small molecule LpxC inhibitors against gram-negative bacteria: advances and future perspectives. Eur. J. Med. Chem. 253, 115326 (2023).
Zhao, J. et al. Preclinical safety and efficacy characterization of an LpxC inhibitor against gram-negative pathogens. Sci. Transl Med. 15, eadf5668 (2023).
McClerren, A. L. et al. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to Ciprofloxacin. Biochemistry 44, 16574–16583 (2005).
Krause, K. M. et al. Potent LpxC inhibitors with in vitro activity against multidrug-resistant Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 63, (2019).
Tomaras, A. P. et al. LpxC inhibitors as new antibacterial agents and tools for studying regulation of lipid A biosynthesis in gram-negative pathogens. MBio 5, e01551–e01514 (2014).
Cohen, F. et al. Optimization of LpxC inhibitors for antibacterial activity and cardiovascular safety. ChemMedChem 14, 1560–1572 (2019).
Piizzi, G. et al. Design, synthesis, and properties of a potent inhibitor of pseudomonas aeruginosa deacetylase LpxC. J. Med. Chem. 60, 5002–5014 (2017).
Romano, K. P. & Hung, D. T. Targeting LPS biosynthesis and transport in gram-negative bacteria in the era of multi-drug resistance. Biochim. Biophys. Acta Mol. Cell. Res. 1870, 119407 (2023).
Muteeb, G., Rehman, M. T., Pani, B., Khan, R. H. & Editorial Novel drug-designing approaches to combat antimicrobial resistance. Front. Mol. Biosci. 10, 1342702 https://doi.org/10.3389/fmolb.2023.1342702 (2023).
Selvaraj, C., Chandra, I. & Singh, S. K. Artificial intelligence and machine learning approaches for drug design: challenges and opportunities for the pharmaceutical industries. Mol. Divers. 26, 1893–1913 (2022).
Kumar, P., Bhardwaj, V. K., Shende, P. & Purohit, R. Computational and experimental analysis of Luteolin-β-cyclodextrin supramolecular complexes: insights into conformational dynamics and phase solubility. Eur. J. Pharm. Biopharm. Off J. Arbeitsgemeinschaft fur Pharm. Verfahrenstechnik E V 205, 114569 (2024).
Kumar, P., Kumar Bhardwaj, V. & Purohit, R. Molecular and quantum mechanical insights of conformational dynamics of maltosyl-β-cyclodextrin/formononetin supramolecular complexes. J. Mol. Liq. 397, 124196 (2024).
Bhardwaj, V. K. & Purohit, R. A comparative study on inclusion complex formation between Formononetin and β-cyclodextrin derivatives through multiscale classical and umbrella sampling simulations. Carbohydr. Polym. 310, 120729 (2023).
Bhardwaj, V., Singh, R., Singh, P., Purohit, R. & Kumar, S. Elimination of bitter-off taste of stevioside through structure modification and computational interventions. J. Theor. Biol. 486, 110094 (2020).
Singh, R., Bhardwaj, V. K. & Purohit, R. Inhibition of nonstructural protein 15 of SARS-CoV-2 by golden Spice: A computational insight. Cell. Biochem. Funct. 40, 926–934 (2022).
Rauf, A. et al. Aldose reductase inhibitory evaluation and in Silico studies of bioactive secondary metabolites isolated from Fernandoa. Adenophylla (Wall. Ex G. Don). J. Mol. Struct. 141308 (2025).
Bragina, M. E., Daina, A., Perez, M. A. S., Michielin, O. & Zoete, V. The SwissSimilarity 2021 web tool: novel chemical libraries and additional methods for an enhanced ligand-based virtual screening experience. Int. J. Mol. Sci. 23, (2022).
Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717 (2017).
Banerjee, P., Kemmler, E., Dunkel, M. & Preissner, R. ProTox 3.0: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 52, W513–W520 (2024).
Daina, A., Michielin, O. & Zoete, V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364 (2019).
Antunes, S. S., Won-Held Rabelo, V. & Romeiro, N. C. Natural products from Brazilian biodiversity identified as potential inhibitors of PknA and PknB of M. tuberculosis using molecular modeling tools. Comput. Biol. Med. 136, 104694 (2021).
Ahsan, M. J., Govindasamy, J., Khalilullah, H., Mohan, G. & Stables, J. P. POMA analyses as new efficient bioinformatics’ platform to predict and optimise bioactivity of synthesized 3a,4-dihydro-3H-indeno[1,2-c]pyrazole-2-carboxamide/carbothioamide analogues. Bioorg. Med. Chem. Lett. 22, 7029–7035 (2012).
Mishra, A., Mulpuru, V. & Mishra, N. An interaction network driven approach for identifying cervical, endometrial, vulvar carcinomic biomarkers and their multi-targeted inhibitory agents from few widely available medicinal plants. Appl. Biochem. Biotechnol. 195, 6893–6912 (2023).
Modanwal, S., Mishra, A. & Mishra, N. An integrative analysis of GEO data to identify possible therapeutic biomarkers of prostate cancer and targeting potential protein through Zea mays phytochemicals by virtual screening approaches. J. Biomol. Struct. Dyn. 43, 709–729 (2025).
Rajput, D., Jain, D. & Kashaw, S. K. & Patil, U. K. Molecular docking studies on phytoconstituent isolated from Nyctanthes arbortristis Linn. Int. J. Pharm. Investig. 14, (2024).
Seeliger, D. & de Groot, B. L. Conformational transitions upon ligand binding: holo-structure prediction from apo conformations. PLoS Comput. Biol. 6, e1000634 (2010).
Ikram, S., Ahmad, F., Ahmad, J. & Durdagi, S. Screening of small molecule libraries using combined text mining, ligand- and target-driven based approaches for identification of novel granzyme H inhibitors. J. Mol. Graph. Model. 105, 107876 (2021).
Raczyńska, E. D. et al. Nitriles with high gas-phase basicity—Part II transmission of the push–pull effect through Methylenecyclopropene and Cyclopropenimine scaffolds intercalated between different electron donor (s) and the cyano N-protonation site. Molecules 27, 4370 (2022).
Rauf, A. et al. Anticancer potential of flavonoids isolated from Pistacia chinensis against glioblastoma (U87) cell line: extensive in vitro and in Silico research. Eurasian J. Med. Oncol. 5768 (2025).
Tsuneda, T., Song, J. W., Suzuki, S. & Hirao, K. On Koopmans’ theorem in density functional theory. J. Chem. Phys. 133, 174101 (2010).
Alruwaili, M. et al. In silico identification and characterization of potent laccase Inhibitors against cryptococcus neoformans: A multi-scale computational study.
Rauf, A., Khan, M. U. & Akram, Z. Lipoxygenase inhibitory potential of secondary metabolites isolated from Pistacia integerrima: a comprehensive in vitro analysis integrating molecular docking, ADMET and DFT studies. Tradit Med. Res. 10, 52 (2025).
Bhrdwaj, A. et al. Structure-based virtual screening, molecular docking, molecular dynamics simulation of EGFR for the clinical treatment of glioblastoma. Appl. Biochem. Biotechnol. 195, 5094–5119 (2023).
Mukherjee, S. et al. Structure-based virtual screening, molecular docking, and molecular dynamics simulation of VEGF inhibitors for the clinical treatment of ovarian cancer. J. Mol. Model. 28, 100 (2022).
De Vivo, M., Masetti, M., Bottegoni, G. & Cavalli, A. Role of molecular dynamics and related methods in drug discovery. J. Med. Chem. 59, 4035–4061 (2016).
Bandaru, S. et al. Molecular dynamic simulations reveal suboptimal binding of salbutamol in T164I variant of Β2 adrenergic receptor. PLoS One 12, e0186666 (2017).
Yadav, M. et al. Structure-based virtual screening, molecular docking, molecular dynamics simulation and Pharmacokinetic modelling of cyclooxygenase-2 (COX-2) inhibitor for the clinical treatment of colorectal cancer. Mol. Simul. 48, 1081–1101 (2022).
Nayarisseri, A. et al. Potential inhibitors of VEGFR1, VEGFR2, and VEGFR3 developed through deep learning for the treatment of cervical cancer. Sci. Rep. 14, 13251 (2024).
Khan, M. F. et al. Exploring optimal drug targets through subtractive proteomics analysis and pangenomic insights for tailored drug design in tuberculosis. Sci. Rep. 14, 10904 (2024).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Nayarisseri, A. et al. Shape-based machine learning models for the potential novel COVID-19 protease inhibitors assisted by molecular dynamics simulation. Curr. Top. Med. Chem. 20, 2146–2167 (2020).
Skjærven, L., Yao, X. Q., Scarabelli, G. & Grant, B. J. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinform. 15, 399 (2014).
Prajapati, L., Khandelwal, R., Yogalakshmi, K. N., Munshi, A. & Nayarisseri, A. Computer-aided structure prediction of bluetongue virus coat protein VP2 assisted by optimized potential for liquid simulations (OPLS). Curr. Top. Med. Chem. 20, 1720–1732 (2020).
Hosen, M. A. et al. A computational investigation of Galactopyranoside esters as antimicrobial agents through antiviral, molecular docking, molecular dynamics, pharmacokinetics, and bioactivity prediction. J. Biomol. Struct. Dyn. 42, 1015–1030 (2024).
Doak, B. C., Over, B., Giordanetto, F. & Kihlberg, J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem. Biol. 21, 1115–1142 (2014).
Liu, M. et al. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J. Pathol. 221, 221–228 (2010).
Sardar, H., Shareef, U. & Khan, H. Molecular Docking & in Silico ADME analysis of 5-O-methyl-11-O-acetylalkannin. Phytopharm Commun. 4, 17–28 (2024).
Isyaku, Y., Uzairu, A. & Uba, S. Computational studies of a series of 2-substituted phenyl-2-oxo-, 2-hydroxyl- and 2-acylloxyethylsulfonamides as potent anti-fungal agents. Heliyon 6, e03724 (2020).
Zhang, H. et al. Chemical molecular-based approach to overcome multidrug resistance in cancer by targeting P-glycoprotein (P-gp). Med. Res. Rev. 41, 525–555 (2021).
Mirzaei, S. et al. Advances in Understanding the role of P-gp in doxorubicin resistance: molecular pathways, therapeutic strategies, and prospects. Drug Discov. Today 27, 436–455 (2022).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Veber, D. F. et al. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 45, 2615–2623 (2002).
Alshehri, M. M. et al. Computer-aided drug discovery of c-Abl kinase inhibitors from plant compounds against chronic myeloid leukemia. J. Biomol. Struct. Dyn. 1–21. https://doi.org/10.1080/07391102.2024.2329297 (2024).
Parrot, M. et al. Integrating synthetic accessibility with AI-based generative drug design. J. Cheminform. 15, 83 (2023).
Banerjee, P., Eckert, A. O., Schrey, A. K. & Preissner, R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 46, W257–W263 (2018).
Maddalon, A., Iulini, M., Melzi, G., Corsini, E. & Galbiati, V. New approach methodologies in immunotoxicology: challenges and opportunities. Endocr. Metab. Immune Disord. Drug Targ. https://doi.org/10.2174/1871530323666230413081128 (2023).
Riaz, R., Parveen, S., Shafiq, N., Ali, A. & Rashid, M. Virtual screening, ADME prediction, drug-likeness, and molecular docking analysis of Fagonia indica chemical constituents against antidiabetic targets. Mol. Divers. https://doi.org/10.1007/s11030-024-10897-7 (2024).
Verma, A. Lead finding from Phyllanthus Debelis with hepatoprotective potentials. Asian Pac. J. Trop. Biomed. 2, S1735–S1737 (2012).
Jan, H. et al. Phytochemical analysis and versatile in vitro evaluation of antimicrobial, cytotoxic and enzyme Inhibition potential of different extracts of traditionally used Aquilegia pubiflora wall. Ex royle. BMC Complement. Med. Ther. 21, 165 (2021).
Li, Y. et al. Identification of trypsin-degrading commensals in the large intestine. Nature 609, 582–589 (2022).
Bibens, L., Becker, J. P., Dassonville-Klimpt, A. & Sonnet, P. A review of fatty acid biosynthesis enzyme inhibitors as promising antimicrobial drugs. Pharmaceuticals (Basel) 16, (2023).
da Fonseca, A. M. et al. Screening of potential inhibitors targeting the main protease structure of SARS-CoV-2 via molecular docking, and approach with molecular dynamics, RMSD, RMSF, H-bond, SASA and MMGBSA. Mol. Biotechnol. 66, 1919–1933 (2024).
Ahamed, F. M. M. et al. Molecular dynamics simulation, QSAR, DFT, molecular docking, ADMET, and synthesis of Ethyl 3-((5-bromopyridin-2-yl) Imino) butanoate analogues as potential inhibitors of SARS-CoV-2. Polycycl. Aromat. Compd. 44, 294–312 (2024).
Balogun, T. A. et al. Computational evaluation of bioactive compounds from Colocasia affinis Schott as a novel EGFR inhibitor for cancer treatment. Cancer Inf. 20, 11769351211049244 (2021).
Ejaz, S. A., Aziz, M., Zafar, Z., Akhtar, N. & Ogaly, H. A. Revisiting the inhibitory potential of protein kinase inhibitors against NEK7 protein via comprehensive computational investigations. Sci. Rep. 13, 4304 (2023).
Rahman, J. et al. Biological evaluation, DFT calculations and molecular Docking studies on the antidepressant and cytotoxicity activities of cycas pectinata buch.-ham. Compounds. Pharmaceuticals (Basel) 13, (2020).
Danazumi, A. U. & Umar, H. I. You must be flexible enough to be trained, Mr. Dynamics simulator. Mol. Divers. 28, 2731–2733 (2024).
Altayb, H. N. Fludarabine, a potential DNA-dependent RNA polymerase inhibitor, as a prospective drug against Monkeypox virus: A computational approach. Pharmaceuticals (Basel) 15, (2022).
Khan, M. U. et al. Identification of novel natural compounds against CFTR P. Gly628Arg P.thogenic variant. AMB Express 14, 99 (2024).
do Carmo, A. L. et al. Competition between phenothiazines and BH3 peptide for the binding site of the antiapoptotic BCL-2 protein. Front. Chem. 8, 235 (2020).
Mini, M., Jayakumar, D. & Kumar, P. In-silico and in-vitro assessment of the antibiofilm potential of Azo dye, carmoisine against Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 42, 6700–6710 (2024).
Dao, T. N. P. et al. Phytotherapeutic potential of compounds identified from fractionated extracts of Morus alba L., as an inhibitor of interleukin-6 in the treatment of rheumatoid arthritis: computational approach. J. Biomol. Struct. Dyn. 1–14. https://doi.org/10.1080/07391102.2024.2330713 (2024).
Silver, L. L. A gestalt approach to gram-negative entry. Bioorg. Med. Chem. 24, 6379–6389 (2016).
Amudala, S., Aidhen, I. S. & Sumit & LpxC Inhibition: potential and opportunities with carbohydrate scaffolds. Carbohydr. Res. 537, 109057 (2024).
Kralj, S., Jukič, M. & Bren, U. Molecular filters in medicinal chemistry. Encyclopedia 3, 501–511 (2023).
Kumar Pal, S. & Kumar, S. Indole-based LpxC (UDP-3-O-(R-3-hydroxyacyl)-N-acetylglucosaminedeacetylase) inhibitors for Salmonella Typhi: rational drug discovery through in Silico screening. 3 Biotech. 13, 281 (2023).
Ahmed, M. Z., Alqahtani, A. S., Shukla, Kumar, P., Pal, S. K. & S. & Pharmacophore-based approach for the identification of potent inhibitors against LpxC enzyme from Salmonella Typhi. Chem. Phys. Impact. 9, 100729 (2024).
Torres, P. H. M., Sodero, A. C. R. & Jofily, P. & Silva-Jr, F. P. Key topics in molecular docking for drug design. Int. J. Mol. Sci. 20, (2019).
Alamri, M. A., Prinsa, Kawsar, S. M. A. & Saha, S. Exploring marine-derived bioactive compounds for dual Inhibition of Pseudomonas aeruginosa LpxA and LpxD: integrated bioinformatics and cheminformatics approaches. Mol. Divers. https://doi.org/10.1007/s11030-024-10888-8 (2024).
Damale, M. G., Pathan, S. K., Patil, R. B. & Sangshetti, J. N. Pharmacoinformatics approaches to identify potential hits against tetraacyldisaccharide 4’-kinase (LpxK) of Pseudomonas aeruginosa. RSC Adv. 10, 32856–32874 (2020).
Bhaskar, B. V. et al. Structure-based virtual screening of Pseudomonas aeruginosa LpxA inhibitors using pharmacophore-based approach. Biomolecules 10, (2020).
Peverati, R. & Truhlar, D. G. Quest for a universal density functional: the accuracy of density functionals across a broad spectrum of databases in chemistry and physics. Philos. Trans. Ser. Math. Phys. Eng. Sci. 372, 20120476 (2014).
Bursch, M., Mewes, J. M., Hansen, A. & Grimme, S. Best-practice DFT protocols for basic molecular computational chemistry. Angew Chem. Int. Ed. Engl. 61, e202205735 (2022).
Kumar, P., Bhardwaj, V. K. & Purohit, R. Dispersion-corrected DFT calculations and umbrella sampling simulations to investigate stability of Chrysin-cyclodextrin inclusion complexes. Carbohydr. Polym. 319, 121162 (2023).
Ogbodo, S. E. et al. Computational investigation of amide derivative as potential anti-carbapenem-resistant Pseudomonas aeruginosa. J. Indian Chem. Soc. 101, 101184 (2024).
Wu, X., Xu, L. Y., Li, E. M. & Dong, G. Application of molecular dynamics simulation in biomedicine. Chem. Biol. Drug Des. 99, 789–800 (2022).
Zoghlami, M., Oueslati, M., Basharat, Z., Sadfi-Zouaoui, N. & Messaoudi, A. Inhibitor assessment against the LpxC enzyme of antibiotic-resistant Acinetobacter baumannii using virtual screening, dynamics simulation, and in vitro assays. Mol. Inf. 42, e2200061 (2023).
Ahmad, S. et al. Subtractive genomics, molecular Docking and molecular dynamics simulation revealed LpxC as a potential drug target against multi-drug resistant Klebsiella pneumoniae. Interdiscip Sci. 11, 508–526 (2019).
Mohammed, E. Z. et al. Design, synthesis, and molecular dynamic simulations of some novel benzo[d]thiazoles with anti-virulence activity against Pseudomonas aeruginosa. Eur. J. Med. Chem. 279, 116880 (2024).
Singh, R., Bhardwaj, V. K., Sharma, J., Das, P. & Purohit, R. Identification of selective cyclin-dependent kinase 2 inhibitor from the library of pyrrolone-fused benzosuberene compounds: an in Silico exploration. J. Biomol. Struct. Dyn. 40, 7693–7701 (2022).
Funding
This work was funded by the Deanship of Graduate Studies and Scientific Research at Jouf University under Grant No. (DGSSR-2023-01-02388).
Author information
Authors and Affiliations
Contributions
All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alanazi, A., Younas, S., Khan, M.U. et al. A combined in silico and MD simulation approach to discover novel LpxC inhibitors targeting multiple drug resistant Pseudomonas aeruginosa. Sci Rep 15, 16900 (2025). https://doi.org/10.1038/s41598-025-99215-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-99215-1