Abstract
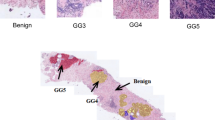
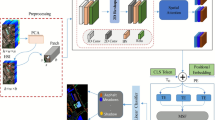
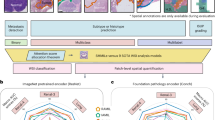
Microscopic hyperspectral imaging (MHSI) of unstained tissue provides quantitative, label-free cues for pathology, but practical diagnosis is hindered by weak morphological contrast and high-dimensional spectra. Patch-wise classification is therefore unstable: discriminative spectral signatures are subtle, spatially sparse, and easily confounded by noise and tissue heterogeneity. To address this, we construct a new unstained breast MHSI dataset and formulate slice-level diagnosis as a multiple instance learning (MIL) problem. We propose a Multi-Scale Hierarchical Attention Network (MS-HAN) tailored to hyperspectral MIL. Each instance (patch) is encoded by an Inception-like multi-branch extractor that operates at a fixed spatial resolution using parallel convolution kernels to capture spectral–spatial patterns at different receptive fields. To reduce high intra-class spectral variability, we introduce a prototype-based clustering regularization that softly assigns instance embeddings to learnable centers and refines the representation. We then apply dual attention directly on the spatial feature map: channel (spectral) attention generates band-wise weights from global spatial descriptors, explicitly modeling inter-band dependencies, followed by spatial attention producing a 2D attention map to localize informative cellular regions. These modules are trained end-to-end with only slice-level labels. Finally, a hierarchical aggregator models inter-patch dependencies via self-attention and performs attention pooling to obtain the slice representation for classification. On a strictly patient-split cohort of 60 patients, MS-HAN achieved 86.7% accuracy and 0.92 AUC, outperforming strong MIL baselines (e.g., TransMIL and DS-MIL). McNemar’s test demonstrated statistically significant improvements over ABMIL (\(p=0.0251\)) and DS-MIL (\(p=0.0198\)), with marginal significance against CLAM and TransMIL (\(p<0.1\)). Ablations verified the necessity of the prototype regularization and hyperspectral-specific attention. Attention visualizations highlighted regions consistent with tumor-related morphology and emphasized informative spectral ranges without pixel-level annotations, pending expert validation. MS-HAN suggests that hyperspectral-specific feature refinement and hierarchical MIL aggregation may improve robust, stain-free breast cancer detection from microscopic MHSI. Further multi-center validation and expert review of attention explanations are needed to establish clinical utility.
Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- MHSI::
-
Microscopic Hyperspectral Imaging
- MIL::
-
Multiple Instance Learning
- CNN::
-
Convolutional Neural Network
- WSI::
-
Whole Slide Image
- ROI::
-
Region of Interest
- AUC::
-
Area Under the Curve
- SD::
-
Standard Deviation
References
Sung, H. et al. Global Cancer Statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74(3), 209–49 (2024).
Gurcan, M. N. et al. Histopathological image analysis: A review. IEEE Rev. Biomed. Eng. 2, 147–71 (2009).
Elmore, J. G. et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313(11), 1122–32 (2015).
Roth, H.R., Lu, L., Seff, A., Cherry, K.M., Wu, J., Lu, S., et al. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. In: Medical Image Computing and Computer-Assisted Intervention-MICCAI 2014. Springer; p. 520–7, (2014).
de Oliveira, A. M. D. C. F., da Mota, P. A. F. C., de Albuquerque, V. H. C. & de Farias, T. M. P. Microscopic hyperspectral imaging: a new tool for pathology. Expert Rev. Med. Devices 19(10), 837–51 (2022).
Shahshahan, M., Tsen, S. D. W., Kj, Bae, Peng, T. Q. & Esfandyarpour, R. Label-free hyperspectral imaging and deep learning for tracking the metabolic response of single cancer cells to chemotherapy. Sci. Rep. 9(1), 18663 (2019).
Li, S. et al. Hyperspectral image classification: A review of recent advances. IEEE Geosci. Remote Sens. Magaz. 8(3), 25–43 (2020).
Lu, G. & Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 19(1), 010901 (2014).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25(8), 1301–9 (2019).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 5(6), 555–70 (2021).
Surface Optics Corporation. SOC-710 Hyperspectral Imager Datasheet. Surface Optics Corporation; 2024. Accessed: 2025–06-18. Available from: https://www.surfaceoptics.com/products/hyperspectral-imaging-systems/soc-710-hyperspectral-imager/.
Woo, S., Park, J., Lee, J.Y., Kweon, I.S. (2018) CBAM: Convolutional Block Attention Module. In: Proceedings of the European conference on computer vision (ECCV); p. 3–19.
Li, B., Li, Y., Eliceiri, K.W. (2021) Dual-stream multiple instance learning network for whole slide image classification with self-supervised contrastive learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; p. 14318–28.
Shao, Z. et al. TransMIL: Transformer based correlated multiple instance learning for whole slide image classification. Adv. Neural. Inf. Process. Syst. 34, 2136–47 (2021).
Ye, Z. et al. HAMIL: Hierarchical attention multi-instance learning for label-free colorectal cancer typing. Laser Photon. Rev. 18(6), 2301072 (2024).
Ye, Z. et al. DFLNet: Disentangled feature learning network for breast cancer ultrasound image segmentation. Digital Signal Process. 145, 104331 (2024).
Li, Y., Zhang, H. & Shen, Q. Spectral-spatial classification of hyperspectral imagery with 3D convolutional neural networks. Remote Sens. 9(1), 67 (2017).
Ahmad, J., Farman, H. & Jan, Z. 3D-convolutional neural networks for medical imaging analysis: A systematic review. Multim. Tools Appl. 82, 27373–415 (2023).
Rajpurkar, P., Chen, E., Banerjee, O. & Chen, Y. Appraising the state of the art in medical AI. Nat. Med. 30(5), 1264–74 (2024).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17(1), 195 (2019).
Bancroft, J. D. & Gamble, M. Bancroft’s Theory and Practice of Histological Techniques 8th edn. (Elsevier, 2019).
Swanson Biotechnology Center, MIT. Pricing | Histology. Massachusetts Institute of Technology; n.d. Available from: https://ki-sbc.mit.edu/histology/pricing.
MUSC College of Medicine. Services & Prices | Histology & Immunohistochemistry. Medical University of South Carolina (MUSC); n.d. https://medicine.musc.edu/departments/pathology-laboratory-medicine/divisions/experimental-pathology/cores/histology-immunohistochemistry/services-prices.
UC Davis Health. Research Histology: Rates. UC Davis Health; n.d. https://health.ucdavis.edu/pathology/research/research_labs/histology/rates.html.
Roche Diagnostics. VENTANA HE 600 automated staining system. Roche Diagnostics; n.d. https://diagnostics.roche.com/us/en/products/instruments/ventana-he-600-ins-4090.html.
Vale-Silva, L. & Rohr, K. A review on deep learning-based methods for multi-modal medical imaging fusion. Expert Syst. Appl. 239, 121542 (2024).
Novis, D. A. & Zarbo, R. J. Interinstitutional comparison of frozen section turnaround time. A College of American Pathologists Q-Probes study of 32868 frozen sections in 700 hospitals. Arch. Patholo. Lab. Med. 121(6), 559–67 (1997).
Chen, Y., Anderson, K. R., Xu, J., Goldsmith, J. D. & Heher, Y. K. Frozen-section checklist implementation improves quality and patient safety. Am. J. Clin. Pathol. 151(6), 607–12 (2019).
World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 310(20), 2191–4 (2013).
Wolff, A. C. et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update. J. Clin. Oncol. 36(20), 2105–22 (2018).
Vo-Dinh, T. editor. Biomedical Photonics Handbook, Second Edition: Biomedical Diagnostics. CRC Press; (2014).
Loshchilov, I., Hutter, F. Decoupled Weight Decay Regularization; (2019). OpenReview paper. International Conference on Learning Representations (ICLR). Available from: https://openreview.net/forum?id=Bkg6RiCqY7.
Ilse, M., Tomczak, J., Welling, M. Attention-based Deep Multiple Instance Learning. In: International Conference on Machine Learning (ICML). PMLR; p. 2127–36, (2018).
Author information
Authors and Affiliations
Contributions
Z.C. and Q.Y. contributed equally to this work. Z.C. and Q.Y. performed the data analysis, developed the model, and wrote the main body of the manuscript. G.Q., X.M., and Z.L. conducted the data acquisition. H.L. and B.S. provided guidance and supervision for the project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki29 and received approval from the Institutional Review Board (IRB) of the Sixth Affiliated Hospital of Sun Yat-sen University (approval number: 2022ZSLYEC-241). Informed consent was obtained from all participants prior to their inclusion.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Z., Yang, Q., Qin, G. et al. MultiScale hierarchical attention network for stain free breast cancer detection in microscopic hyperspectral imaging. Sci Rep (2026). https://doi.org/10.1038/s41598-026-39267-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-39267-z