Abstract
Here, a computational model to forecast the population dynamics of a laboratory colony of common marmosets (Callithrix jacchus) is presented. This tool supports decision-making that seeks to maximize welfare and maintain a healthy and genetically diverse colony. The model considers the population in terms of three compartments: breeding adults, their offspring and nonbreeding adults. Natural events are explicitly represented, including births, deaths and the transfer of mature offspring from family housing to adult housing. These events are simulated using rates based on historical data extracted from the colony record-keeping system. Multi-year forecasts of population dynamics are generated, taking full account of interventions such as the implementation of breeding controls, the usage of animals by a portfolio of research projects and relocation to external primate facilities. Model forecasts are validated against real data. Uncertainties in animal usage are propagated through the model. The resulting forecasts provide a realistic range of future stock levels to support colony management and decision-making. The model outputs provide evidence to help assess the impacts of making interventions in the system, for example, breeding control strategies. This evidence-based approach to colony management serves to enhance animal welfare and accountability to regulatory bodies and stakeholders. The model can be adapted to simulate the dynamics of other nonhuman primate colonies.
Similar content being viewed by others
Main
The ability to forecast future population numbers with confidence is important for managing a colony of laboratory animals. In this article, we consider a laboratory colony of common marmosets (Callithrix jacchus), maintained to support a portfolio of biomedical research projects, in a context where animal welfare is paramount and the objectives of the 3Rs are supported1. A host of interacting factors must be considered to ensure that welfare is maximized and a healthy, genetically diverse population is maintained.
For instance, a typical ongoing concern is to implement a breeding strategy that yields the required number of adult animals in the future, while maximizing welfare. The future population must be large enough to serve the scheduled research demand and to preserve sufficient genetic diversity to ensure a healthy population in the long-term, while avoiding the risk of overbreeding. In primate species such as marmosets and tamarins, unrestricted breeding is considered to be enriching for dams2. Furthermore, reducing the number of births through breeding control (for example, contraception) can prevent family groups from developing normal cooperative parenting skills and caregiving behaviors, which can result in increased infant mortality3,4. Breeding strategies must therefore account for these factors.
The situation is further complicated by the finite capacity of animal housing, the complexities of recommended marmoset housing strategies5 and the variable demand from in-house biomedical experiments. Unrestricted breeding could lead to population levels above the maximum acceptable stocking density if unchecked. Conversely, long-term reduction of breeding will lead to a shrinking pool of animals from which to select future breeders. This, in turn, will result in dwindling genetic diversity, increased inbreeding and an increase in mean kinship values, negatively impacting the long-term health of the colony and the suitability of animals for in vivo research.
The scenario sketched above illustrates some of the complexities of colony management. Interactions between natural and controllable factors are nonlinear and time dependent. The impacts of interventions over time are difficult to predict.
Mathematical modeling can provide a valuable aid to evidence-based decision-making. For instance, there is a long history of using population dynamics models in ecology6. An important application of population modeling to conservation biology during the past 40 years has been via the field of population viability analysis (PVA)7. PVA employs models together with real data to assess the likelihood of wildlife population survival or extinction at future times. Several software programs for conducting PVA have been developed, for example, VORTEX8,9, an open-source model that quantifies the probability distribution of fates that a population might experience.
Conditions for captive animal populations, such as in zoos or laboratories, are substantially different to populations in the wild, presenting a promising opportunity for mathematical modeling. In such contexts, colony managers know the population structure in detail. Records of key life events (for example, births and deaths) and important developmental indicators for individual animals10 are maintained. This provides a rich dataset for modeling. However, published examples of models to aid the management of laboratory colonies are scarce. To the best of our knowledge, a paper describing the population modeling of a captive squirrel monkey colony is the only published example11, in which two different spreadsheet-based approaches were tested, one deterministic and the other stochastic. Model inputs were derived from historical data for the squirrel monkey colony. Annual predictions of the colony population were made up to 22 years into the future. However, neither the deterministic nor stochastic model outputs reproduced the real census data well (Fig. 9 in ref. 11). Forecasts from the two models diverged substantially, indicating an inconsistency between them.
In this study, we present a novel stochastic simulation model, based on domain knowledge, which generates multi-year forecasts of the population dynamics of a laboratory colony of marmosets. It represents the population structure and demography-based housing arrangements by separating breeding adults, their offspring and nonbreeding adults into different model compartments. Data to parameterize the mathematical rates of natural events (for example, births and deaths) are extracted directly from the colony record-keeping system. Breeding strategy, an important management intervention, is modeled in detail.
Crucially, the planned issuing of animals to a portfolio of biomedical research projects is incorporated into the model, to allow the interactions among interventions, research requirements and housing constraints to be analyzed. Uncertainty in project likelihood and timing is factored into the predictions, to provide decision-makers with a realistic future picture of stock levels, including best and worst cases, to aid colony management. The model also includes alternative outlets, such as relocation to external primate facilities, providing another means of balancing project demand and population growth. These capabilities make the model a tool to support the reduction of animals bred for research purposes and the refinement of husbandry procedures, aligning this work with the objectives of the 3Rs12.
The model is written in R13, which is freely available, simple to install and ubiquitous across research organizations. This renders the model transparent to users, customizable to the structures and needs of different colonies and amenable to analysis and postprocessing. A set of R scripts containing the model have been made available for reuse and modification on GitHub14.
Results
In general, the model is parameterized using real data from a past observation period. Predictions of population levels month-by-month for a future forecast period are made, taking into account the breeding controls, issues and relocations planned during the forecast period. In general, a target number of breeders is maintained in the colony, so the number of breeders in the model is assumed constant during forecasts. By simulating the stochastic model many times, distributions of credible outcomes for the numbers of offspring and nonbreeding adults (hereafter simply ‘adults’, unless a distinction is required) over time are determined, which are then summarized by statistical measures (for example, median, quantiles and range).
Model reproduces historical data
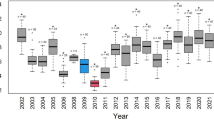
We verified the ability of the model to reproduce real data extracted from the colony record-keeping system. For each year \(Y=\) 2012–2022 inclusive, the model was simulated 200,000 times using the initial conditions, transition rates, issues, relocations and breeding control data for year \(Y\). Model outputs for offspring and adults were generated, quantiles were calculated across all model runs and the minimum and maximum values of the model outputs (that is, the range) were calculated at each monthly time point. Results for each year \(Y\) were calculated as the percentage change from the initial numbers of offspring and adults at the start of the year.
Model performance was quantified by calculating the root mean square error (r.m.s.e.) for each year. Two r.m.s.e. were calculated in each case. First, the r.m.s.e. between each individual realization and the observed data was calculated. The realization closest to the data was identified by finding the realization with the smallest r.m.s.e. (r.m.s.e.Min). Second, the r.m.s.e. between the median model prediction and the observed data was calculated (r.m.s.e.Med) for each year. These metrics facilitate comparison between the model medians and the closest realizations to the data, as well as a comparison between model performance for different years. The results of the verification runs are shown in Fig. 1.
For each verification year, median model predictions are depicted by solid colored lines, 95% credible intervals by shaded regions and ranges by dotted lines. All quantiles were calculated from 200,000 model realizations. The initial observed values for each verification period (circles filled white) were used as initial conditions for model simulations. The r.m.s.e.Med and r.m.s.e.Min values quantify the r.m.s.e. of the subsequent observations (circles filled black) with the median model prediction or the realization closest to the observed data, respectively.
In Fig. 1, the median, 95% credible interval and range depict statistical summaries of the distribution of model outputs at each time point. For instance, the shaded 95% credible interval represents the region encompassing 95% of simulation outputs. The unshaded region between the 95% credible interval and the range encompasses the most outlying 5% of model outputs. The model outputs all of the raw data for each individual realization and for ease, the user can specify the quantiles that are automatically calculated. In this way, the output is customizable. For instance, a user can request other statistical summaries, such as quartiles, deciles or any other required percentile, allowing the results to be interpreted in probabilistic terms.
The results in Fig. 1 verify that the model reproduces the observed data well for offspring and adults each year. Overall, 83% of the observed data points are within the 95% credible interval and every data point is captured within the range of model predictions. In addition, the model generates individual realizations that closely reproduce the observed data for both offspring and adults each year, indicating that the population dynamics are predictable and verifying that the model generates credible simulations, as intended.
Model generates credible 2-year forecasts
The primary use case for the model is to generate 2-year forecasts of the number of adult animals over time, taking into account planned breeding controls, issues and relocations. This capability was tested using historical data by calculating transition rates based on an observation period, year \(Y\), and running simulations for the forecast period, years \((Y+1)\) and \((Y+2)\), for \(Y=\) 2012–2020. Planned breeding controls, issues and relocations for each forecast period were parameterized within the model, allowing them to be incorporated into the forecasts.
Each forecast period was simulated 200,000 times, and quantile summaries of the numbers of adults were produced. Two r.m.s.e.Med and r.m.s.e.Min metrics were calculated, as before. The resulting 2-year forecasts are shown in Fig. 2.
For each 2-year forecast period, median model predictions are depicted by solid colored lines, 95% credible intervals by shaded regions and ranges by dotted lines. All quantiles were calculated from 200,000 model realizations. Initial observed values for each forecast period (circles filled white) were used as initial conditions for model simulations. The r.m.s.e.Med and r.m.s.e.Min values quantify the r.m.s.e. of the subsequent observations (circles filled black) with the median model forecast or the realization closest to the observed data, respectively.
The range of the model forecasts captures all observed data points for each forecast period, except for three outliers: February in forecast 7 and February and March in Forecast 8. In addition, individual realizations were identified that closely reflect the real data, indicating that 2-year periods are forecastable using past data and that the model effectively simulates the underlying dynamics over a 2-year period.
The r.m.s.e.Med values were slightly larger than those for the adult predictions in the verification tests (cf. Fig. 1). However, this is to be expected, owing to errors being calculated for 24 time points in this case rather than 12 previously. Nonetheless, the model demonstrates consistently reliable performance across the forecast periods. These results provide confidence that the model can be used to generate credible 2-year forecasts.
Discussion
We developed a model of the population dynamics of a laboratory colony of common marmosets that is stochastic, compartmental, based on the demographic structure of the colony and which accounts for breeding control, planned issues and relocations. The model works as intended and generates credible 2-year forecasts (Figs. 1 and 2). Model median predictions displayed consistently good performance. The model generates individual realizations that closely reflect the observed data (mean r.m.s.e.Min of 2-year forecasts: 3.21), providing assurance that the colony dynamics are forecastable and that the model simulates the dynamics in a credible manner. These results provide confidence in using the model to support colony management decision-making.
The R code for the version of the model presented here is freely available for reuse and modification, and is available on Github14. The mathematics underlying the model is straightforward and can be readily adapted to simulate the population dynamics of other laboratory animal colonies, even if structured differently. The first step in adapting the model would be to revise the colony schematic (Supplementary Fig. 1), to understand the demographic compartments and the movement of animals between them. The user may need to add additional compartments, depending on the colony structure. Next, the equations describing the monthly changes to the number of animals in each compartment (Algorithm 1, steps 14 and 15) can be modified accordingly. Any new compartments will require the addition of their own equations at this point in the algorithm. Success in the process of adapting the model will depend on an effective collaboration between colony managers and modelers.
The solid lines depict median predictions for adults and the dashed lines depict median predictions for offspring. All quantiles were calculated from 100,000 model realizations. In this illustrative example, it is assumed unacceptable for the number of offspring to fall below 50% of the initial value or for the number of adults to increase more than 25% above the initial value.
Whether the model needs modifying to represent a different colony or not, the availability of data is of crucial importance. A high-quality dataset is required, detailing the dates of birth, transfer and death for every animal, as well as a record of historical litter sizes. These data are essential for parameterizing the model (see the Methods for a greater description of data requirements).
The structure of the model is based on domain knowledge of the colony of interest, and it is completely parameterized using historical data. Consequently, the forecasts depend on the accuracy with which the colony is represented in the equations, as well as the trends and variance inherent in the past data. This presents three potential limitations for the model. First, if the model is adapted to a different colony but the structure is not represented correctly in the mathematical equations, then the forecasts should not be expected to be accurate. Second, if the colony structure is correctly represented in the equations, but there is insufficient data to parameterize the rates of transition between compartments, then this will present a problem. One solution would be to estimate unknown parameters by fitting the model to past data. This is a pragmatic solution, but it would introduce an additional uncertainty into the model predictions. Third, by design, variance in the past data is propagated through the model and is reflected in the uncertainty of the forecasts. Therefore, historical data with high variance will lead to forecasts with greater uncertainty. Individual model users will want to validate the forecasts of the adapted model against their own historical data to assess the level of confidence with which they can use the forecasts in their context.
The number of animals issued to experiments can differ from plans if projects experience changes in requirements, delays or cancellations. These uncertainties can create risks for colony managers, who seek to maintain the population within acceptable stocking densities. We can integrate these uncertain factors into our forecasts, providing previously unattainable insights to inform welfare-based colony management. To illustrate how this can be done, we revisited the forecast for 2021–2022 using data from the observation period 2020. Considering this as a baseline case, we explored the impact on the forecasts of incorporating usage uncertainty. For illustrative purposes, the planned number of animals to be issued to experiments during the forecast period was split across five projects. Each project was assigned two probability parameters. First, a probability of proceeding, to quantify expert opinion about the likelihood that each project would go ahead or not. For illustrative purposes, these probabilities were set to be 1, 1, 0.8, 0.5 and 1, respectively. Second, each project was assigned a discrete probability distribution over the potential number of months by which each project might be delayed, if it were to go ahead. The assumed maximum possible delays were 0, 0, 3, 6 and 1 months, respectively.
Using these illustrative parameters, 100,000 realizations of the model were performed, with each run randomly sampling from the distributions defined above, to generate 2-year forecasts incorporating usage uncertainty. The median and maximum predictions of the numbers of adults at the end of 2022 were 23.1% and 48.8% higher than in the baseline case, respectively. These figures reflect that when some projects do not proceed as planned, animal usage is less, hence a higher stock level at the end of the forecast period. These figures can be compared with housing capacity limitations to allow the potential risk of exceeding acceptable stocking densities to be assessed.
Continuing with the illustrative scenario, supposing that the increased numbers of adults at the end of 2022 present a risk of exceeding acceptable stocking densities, we explored how this risk could be mitigated by relocating a proportion of marmosets to alternative external primate facilities during the forecast period. We found that relocating 17% of the adult stock available in September 2022 during the final quarter of 2022 would be sufficient to reduce the median number of adults down to the baseline level, thus mitigating the risk.
Analysis of the illustrative scenario enables us to identify an unintended negative impact of this relocation plan, demonstrating a further benefit of using the model to support decision-making. The minimum model prediction of the number of adults remaining at the end of 2022 was 29.8% lower than in the baseline case due to the relocated animals being removed from the system. This represents the case where all projects proceed as planned (which occurs with a probability of 0.4 according to our assumed probabilities, assuming they are independent). Therefore, the decision to relocate animals to mitigate the risk of exceeding acceptable housing densities could unintentionally create a new risk of a shrinking population, should all projects proceed on time. Consideration of actual stock numbers enables the level of risk to be assessed in context, to help refine the relocation plan. For instance, the relocation process may only be initiated if projects do not proceed by a certain date.
We note that the usage uncertainty assumptions considered above are conservative. Often, the uncertainties in project likelihood and timing can be much greater, leading to larger potential impacts than the illustrative examples presented here.
The model can also be used to inform decision-making in the context of high-impact operational risks. For instance, we might want to assess the best breeding strategy in the event that all research projects are paused for an unspecified duration, for instance, due to a global pandemic, such as COVID-19. When laboratory work is paused, over time, unrestricted breeding could lead to maximum acceptable stocking densities being exceeded, so the goal would be to identify a strategy that maximizes breeding subject to the capacity constraints of animal housing. For illustrative purposes, suppose that the maximum acceptable stocking density is 25% higher than the number of adults at the beginning of the forecast period. Also, suppose that the minimum acceptable number of offspring is 50% of the initial number at the start of the same period. With these constraints in mind, we used the model to explore the impact of hypothetical breeding strategies that maintain 100%, 75%, 50%, 25% and 0% of the maximal breeding rate for a 2-year period, to identify which strategy would best maintain acceptable levels of offspring and adults while research projects are paused. Simulating each case 100,000 times, we calculated statistical summaries of the numbers of offspring and adults over time for each case (the median predictions are displayed in Fig. 3).
Under these circumstances, if breeding is unrestricted (that is, 100%), then the number of adults exceeds acceptable levels within 12 months of research projects being paused. Restricting breeding to 75% of maximum levels (for example, by administering chemical contraceptive intermittently to one-quarter of dams at any given time) also leads to unacceptably high numbers of adults and does not substantially limit stock levels in the 2-year timeframe. Restricting breeding to either 25% or 0% ensures that adult stock levels remain well within the acceptable range; however, in both these cases, the offspring population decreases below the acceptable level within 12 months. Restricting breeding to 50% ensures that the adult stock limit is not exceeded until the final month of the second year and the number of offspring remains above the minimum limit in this timeframe. Therefore, out of the options tested here, a 50% breeding strategy would be preferred.
As demonstrated by the illustrative scenarios described above, the model provides a means of analyzing the interactions among usage, relocations, breeding control, housing capacity and the associated risks of colony management decisions. To the best of our knowledge, this is the first model of its kind, based on the underlying structure of the colony, simulating life events and taking into account interventions.
This evidence-based approach to colony management supports the 3Rs objectives. It supports reduction by providing a quantitative approach to help mitigate overbreeding and advice to inform optimal stocking densities. It supports refinement by providing colony managers with forecasts that can influence decisions about future housing arrangements, helping to enhance welfare throughout the lifetimes of animals within the colony. Furthermore, this approach is repeatable, auditable and transparent, enhancing accountability to external regulatory bodies and stakeholders.
Methods
Model structure
The population is partitioned into three disjoint compartments, corresponding to broad demographic categories: offspring, breeders and nonbreeding adults, labeled \(\left\{{\mathrm{C,B,A}}\right\}\), respectively. Denoting by \({n}_{t}^{({\mathrm{C}})}\), \({n}_{t}^{({\mathrm{B}})}\) and \({n}_{t}^{({\mathrm{A}})}\) the number of animals alive in each compartment at time \(t\), the total population of the colony is equal to \(({n}_{t}^{{\mathrm{(C)}}}+{n}_{t}^{\mathrm{(B)}}+\,{n}_{t}^{{\mathrm{(A)}}})\), at time \(t\). The colony is described by the state vector \({{\mathbf{x}}}_{t}={({n}_{t}^{{\mathrm{(C)}}},{n}_{t}^{{\mathrm{(B)}}},{n}_{t}^{{\mathrm{(A)}}})}^{\mathrm{T}}\), where ‘T’ denotes the transpose. Time is a discrete variable with a default time-step of 1 month.
Animals move between compartments in a well-defined manner. The possible moves into or out of compartments are called transition events (or simply ‘events’). Each event has an associated transition rate governing the number of animals undergoing the transition in a fixed time interval. There are four events: births (new infants are born into the offspring compartment with an average rate \(\beta\) per month), transfers (mature offspring are transferred from family cages to adult cages with an average rate \(\omega\) per month), new breeders (preselected marmosets become breeders with an average rate \(\gamma\) per month) and deaths (animals in compartment \(X\) die with average rate \({\delta }^{(X)}\) per month, for \(X={\mathrm{C,B,A}}\)).
We also consider three interventions, where human decision-making affects the system: breeding controls (breeders partially or completely prevented from breeding at their maximal natural rate), issues (adult animals issued to experiments, according to a planned schedule) and relocations (adult animals relocated to external primate facilities, according to a planned schedule). A schematic of this compartmental system is shown in Supplementary Fig. 1.
The system can be simplified as follows. When a breeding animal dies, it is replaced by a previously selected animal to maintain a target number of breeders. Therefore, it can be assumed that for a given forecast period, the number of breeders is a constant, \(\widehat{B}\). Under these circumstances, the rates \(\gamma\) and \({\delta }^{(B)}\) are no longer required. The resulting simplified system is shown in Supplementary Fig. 2
The state vector can now be written as \({{\mathbf{x}}}_{t}={\left({n}_{t}^{{\mathrm{(C)}}},\hat{B},{n}_{t}^{{\mathrm{(A)}}}\right)}^{\mathrm{T}}\). Denoting the change in the numbers of offspring and adults between two time points \({t}_{1}\) and \({t}_{2}\) by \({\varDelta }^{{\mathrm{(C)}}}(t,{{\mathbf{x}}}_{t})\) and \({\varDelta }^{{\mathrm{(A)}}}(t,{{\mathbf{x}}}_{t})\), respectively, the change in state during this period is
Mathematical expressions for these changes are described in Algorithm 1 below.
We use the model to forecast the future state of the system, starting at an initial time \({t}_{0}\) for a period of \(N\) months, that is, for a forecast period \(\left({t}_{0},\,{t}_{0}+N\right)\). The months during this forecast period will be indexed \(i=0,\,\cdots ,{N}\), where \(i=0\) corresponds to \({t}_{0}\). To generate forecasts, the model is parameterized using real data extracted from the colony record-keeping system for a prior period of \(M\) months, referred to as the observation period, \(({t}_{0}-M,\,{t}_{0})\).
Data requirements
The following data items for the observation period are required to parameterize the model: the distribution of litter sizes (\({{\mathcal{L}}}_{0}\)), the number of transfer events each month (\({r}_{0,\,j}\)), the median number of offspring and adults alive each month (\({\bar{n}}_{0,\,j}^{\mathrm{C}}\) and \({\bar{n}}_{0,\,j}^{\mathrm{A}}\)), and the numbers of offspring and adults dying each month (\({d}_{0,\,j}^{\mathrm{C}}\) and \({d}_{0,\,j}^{\mathrm{A}}\)), where the index \(j\) denotes the \(j\) th month of the observation period.
The following data items for the forecast period are also required to allow the model to make predictions: the planned number of dams per month (\({D}_{i}\)), the planned proportion of dams receiving contraception per month (\({p}_{i}\)), the efficacy of the contraception used (\({c}_{\mathrm{eff}}\)), the planned number of issues per month (\({u}_{i}\)) and the planned number of relocations per month (\({v}_{i}\)), where the index i denotes the i th month of the forecast period.
Transition rates
To implement the simplified model depicted in Supplementary Fig. 2, we quantify the transition rates using data from the observation period. First, we derive a formula for the rate of births.
Let \({D}_{i}\) denote the target number of dams for month i of the forecast period. Breeding control interventions (that is, chemical contraception) are modeled as follows. Let \({p}_{i}\) be the proportion of dams receiving contraception during month i and let \({c}_{\mathrm{eff}}\) denote the efficacy of the contraceptive, expressed as a proportion (that is, \({c}_{\mathrm{eff}}=1\) corresponds to 100% efficacy and so on). Then, the fraction of maximal breeding that is permitted during month i, denoted \({f}_{i}\), is
Using equation (1), breeding control interventions to be implemented during a forecast period are fully specified by a time series \({\mathbf{f}}=(\,{f}_{1},\,{f}_{2},\cdots ,{f}_{N})\). Then, the number of dams able to give birth to a litter of marmosets during month i of the forecast period can be approximated by \({\widehat{D}}_{i}=\lfloor\;{f}_{i}{D}_{i}\rceil\), where the asymmetric brackets indicate rounding to the nearest integer.
Next, we model the inter-litter interval. The average gestation period for marmosets is 143–144 days5. Postpartum ovulation within 10–20 days of litter delivery is a feature of the marmoset reproductive cycle, leading to reported median inter-litter intervals of 162 days15, 154 days16 and 158 days17. Therefore, we assume that the average inter-litter time is between 153 and 164 days, or between approximately 5.0 and 5.4 months. Let \({t}_{{m}}^{l}\) denote the time from the start of the forecast period (in months) at which the \(m\) th dam gives birth to its \(l\) th litter during that period. Each breeding dam is assumed to produce its first litter of the forecast period between 0 and 5.4 months after the start of the forecast period. This means that the time of the first litter can be simulated by sampling from a uniform distribution for each of the \(m\) dams, \({t}_{m}^{1} \sim {{\mathrm{{Uniform}}}}(0,\,5.4)\). Then, since the inter-litter interval is between 5.0 and 5.4 months, it can be assumed that the times of subsequent litters are calculated by the formula
for \(l > 1\), where \(\tau \sim {\mathrm{Uniform}}(5,\,5.4)\). Litter times greater than \({(t}_{0}+N\)) are discarded, since they are beyond the end of the forecast period.
Having simulated a set of litter times for each breeding dam during the forecast period, \(\left\{{t}_{m}^{l}\right\}\), the model proceeds month-by-month to evaluate the number of infant offspring born in each case. For each i, dams that give birth during the i th month are identified. This is achieved by determining the subset of dams, \({\mu }_{i}\), that satisfy the following condition:
where \(m=1,\ldots ,\,\max ({\widehat{D}}_{i})\) indexes the breeding dams, and the half-square brackets indicate taking the floor of the number inside. This form of rounding (taking the floor and including the \(+1\) term) ensures that simulated litter times are allocated to the correct ordinal month number. For instance, randomly generated litter times between 0 and 1 months after the beginning of the forecast period are allocated to the first month (that is, \(i=1\), January), litter times that are between 1 and 2 months after the start of the forecast period are allocated to the second month (that is, \(i=2\), February) and so on.
Using this notation, \(\left|{\mu }_{i}\right|\) denotes the number of litters birthed in month i. It remains to simulate the number of infant offspring born into each litter using historical data. Let \({{\mathcal{L}}}_{0}\) denote the empirical distribution of litter sizes (that is, the number of infant offspring per litter) from the observation period. Then, for each month i, a set of \(\left|{\mu }_{i}\right|\) litter sizes is sampled
where \({s}_{i}^{{k}_{i}} \sim {{\mathcal{L}}}_{0}\), for \({k}_{i}=1,\cdots ,\left|{\mu }_{i}\right|\).
The total number of new infant offspring born during month i is then the sum of these samples,
Repeating this process sequentially through months \(i=1,\,\cdots ,{N}\) yields a time series of births for the \(N\)-month forecast period, \({\mathbf{b}}=\left({b}_{1},\,{b}_{2},\,\cdots ,\,{b}_{N}\right)\), generated according to the empirical inter-litter intervals and litter sizes.
Next, we consider the rate of transfer events, where mature offspring are moved from their family cages and pair-bonded with opposite sex partners in adult cages (males in this category will have previously been vasectomized to prevent subsequent breeding). The transfer rate for month i of the forecast period can be simulated using the empirical transfer rate for the prior observation period, together with the number of offspring alive during the previous month, \({n}_{(i-1)}^{\mathrm{C}}\), where \({n}_{0}^{\mathrm{C}}\) is the initial number of offspring at the start of the forecast period. Let \({r}_{0,\,j}\) denote the number of transfer events in the \(j\) th month of the observation period and let \({\bar{n}}_{0,\,j}^{\mathrm{C}}\) be the median number of offspring alive in the same month. Then, the mean monthly rate of transfers for the observation period is calculated as
where \(M\) is the number of months in the observation period. Equation (5) expresses the rate of transfers as the number of transfers per month per offspring. Then, the number of transfer events during the i th month of the forecast period, \({w}_{i}\), can be simulated using a Poisson random variable with rate proportional to \({\omega }_{0}\),
where \({\omega }_{0}\) is defined by equation (5) and \({n}_{(i-1)}^{\mathrm{C}}\) is the number of offspring in month \((i-1)\) or the forecast period.
The final transition rates to quantify are the average monthly death rates for offspring and adults. These are estimated using the respective rates observed during the observation period, similarly to transfers. Denote by \({d}_{0,\,j}^{\mathrm{C}}\) and \({d}_{0,\,j}^{\mathrm{A}}\) the numbers of offspring and adults dying during month \(j\) of the observation period, and let \({\bar{n}}_{0,\,j}^{\mathrm{C}}\) and \({\bar{n}}_{0,\,j}^{\mathrm{A}}\) be the median numbers of offspring and adults alive during the \(j\) th month. The mean monthly per capita death rates for offspring and adults during the observation period are then
The model assumes that deaths are calculated at the end of each month, so that the death rates apply not just to the \({n}_{(i-1)}^{\mathrm{C}}\) and \({n}_{(i-1)}^{\mathrm{A}}\) offspring and adults alive at the start of month i, but also to the \({b}_{i}\) infant offspring born and the \({w}_{i}\) mature offspring transferred during the month. Under this assumption, the numbers of deaths during month i of the forecast period can be simulated by Poisson-distributed variables with rates determined by equations (7) and (8),
The death rate of breeders is not considered owing to the simplifying assumption that the number of breeders remains constant (\(\widehat{B}\)) during the forecast period.
To include planned issues and relocations in model forecasts, time series detailing the number of animals required per month of the forecast period are specified. We denote these by \({\mathbf{u}}=({u}_{1},{u}_{2},\cdots ,{u}_{N})\) and \({\mathbf{v}}=({v}_{1},{v}_{2},\cdots ,{v}_{N})\), respectively. The first time series captures the planned schedule of (total) marmoset usage across a portfolio of research projects and the second captures the planned relocation schedule. In practice, many components of \({\mathbf{u}}\) and \({\mathbf{v}}\) may equal zero, representing no usage or relocations during those months. Since there may be some uncertainty in the numbers of animals that will be issued to experiments (for example, due to uncertainties in project likelihood, timing and requirements) or relocated, each component \({u}_{i}\) and \({v}_{i}\) can be expressed as a probability distribution, enabling uncertainty to be propagated through the model via simulation.
Simulating population dynamics
Let \({{\mathbf{x}}}_{{t}_{0}}\) denote the initial state of the system at the beginning of the forecast period. A single realization of the system is simulated using the pseudo-code in Algorithm 1.
Algorithm 1. Pseudo-code for simulating one stochastic realization of the model
1 Initialize \(i=1\) and an empty matrix \({{{X}}}_{(N+1)\times 3}.\)
2 \({{{X}}}_{[1,]}={{\mathbf{x}}}_{{t}_{0}}\)
3 \({\widehat{D}}_{i}=\lfloor\;{f}_{i}{D}_{i}\rceil\)
4 \({t}_{m}^{1} \sim {\mathrm{Uniform}}(0,\,5.4)\), for \(m=1,\,\cdots ,\,{\widehat{D}}_{1}\)
5 WHILE \(i\le N\)
6 \({\mu }_{i}=\left\{m:\,\left\lfloor {t}_{m}^{l}+1\right\rfloor =i\right\}\)
7 IF \(\left|{\mu }_{i}\right| > 0\)
8 \({{\mathbf{s}}}_{i}{{=}}\left\{{s}_{i}^{1},{s}_{i}^{2}\,\cdots ,\,{s}_{i}^{\left|{\mu }_{i}\right|}\right\}\), where \({s}_{i}^{{k}_{i}} \sim {{\mathcal{L}}}_{0}\), for \({k}_{i}=1,\,\cdots ,\,\left|{\mu }_{i}\right|\)
9 \({t}_{{k}_{i}}^{l+1}={t}_{{k}_{i}}^{l}+{\mathrm{Uniform}}(5,\,5.4)\), for \({k}_{i}=1,\,\cdots ,\,\left|{\mu }_{i}\right|\)
10 \({b}_{i}=\sum {{\boldsymbol{s}}}_{i}\)
11 ELSE
12 \({b}_{i}=0\)
13 Sample \({{w}}_{i}\), \({d}_{i}^{\mathrm{C}}\) and \({d}_{i}^{\mathrm{A}}\) according to equations (6), (9) and (10) (sample \({u}_{i}\) and \({v}_{i}\), if they are specified as probability distributions)
14 \({\varDelta }_{i}^{\mathrm{C}}={b}_{i}-{d}_{i}^{\mathrm{C}}-{w}_{i}\)
15 \({\varDelta }_{i}^{\mathrm{A}}={w}_{i}-{d}_{i}^{\mathrm{A}}-{u}_{i}-{v}_{i}\)
16 \({{\mathbf{x}}}_{{t}_{0}+i}={{\mathbf{x}}}_{{t}_{0}+\left(i-1\right)}+\left({\begin{array}{c}{{\varDelta }_{i}^{\mathrm{C}}} \\ 0\\ {\varDelta }_{i}^{\mathrm{A}}\end{array}}\right)\)
17 \({({{\mathbf{x}}}_{{t}_{0}+i})}_{a}=\max (0,{({{\mathbf{x}}}_{{t}_{0}+i})}_{{{a}}})\), for \(a=1,\,3\).
18 \({{{X}}}_{[1+i,]}={{\mathbf{x}}}_{{t}_{0}+i}\)
19 \(i=i+1\)
20 END WHILE
In step 13, the random variables are sampled from the distributions defined above. If \({\boldsymbol{u}}\) and \({\boldsymbol{v}}\) are specified as time series of probability distributions, then these are also sampled. The \(\max (0,\,\bullet )\) function in step 17 prevents negative values in the state vector for the \({\mathrm{C}}\) and \({\mathrm{A}}\) compartments. Negative values of animals, while obviously impossible in reality, could be numerically possible in the forecasts due to the random sampling step if, for instance, the user were to input parameter values sufficient to create a drastic decline in stock levels.
The output of Algorithm 1 is a matrix X with \((N+1)\) rows containing the state vector for each month of the forecast period. The three columns of X are the time series for the compartments \({\mathrm{C}},{\mathrm{B}},{\mathrm{A}}\), respectively. The matrix \(X\) is a unique stochastic trajectory of the model through its state space. A distribution of trajectories is generated by performing many realizations using a Monte Carlo approach. The resultant distribution represents the range of possible outcomes for the colony during the forecast period. Statistical quantities (for example, range and quantiles) are calculated to summarize the realizations, allowing the many possible outcomes to be expressed probabilistically with uncertainty. Note that no parameter fitting is required, all parameters are derived from real data extracted from the colony record-keeping system.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Code availability
A set of scripts containing R code for the model are available for reuse and modification via GitHub at https://github.com/dstl/colony-forecast-model (ref. 14).
References
Prescott, M. & Lidster, K. Improving quality of science through better animal welfare: the NC3Rs strategy. Lab. Anim. 46, 152–156 (2017).
Hosey, G., Melfi, V. & Pankhurst, S. Zoo Animals: Behaviour, Management and Welfare (Oxford Univ. Press, 2009).
Wallace, P. Y., Asa, C. S., Agnew, M. & Cheyne, S. M. A review of population control methods in captive-housed primates. Anim. Welf. 26, 7–20 (2016).
Munson, L., Moresco, A. & Calle, P. P. in Wildlife Contraception: Issues, Methods and Applications (eds Asa, C. S. & Porton I. J.) 66–82 (John Hopkins Univ. Press, 2005).
Elliott, J. J. et al. in Management of Animal Care and Use Programs in Research, Education, and Testing 2nd edn (eds Weichbrod, R. H. et al.) Ch. 29 (CRC/Taylor & Francis, 2018); https://www.ncbi.nlm.nih.gov/books/NBK500423/
Bacaër, N. A Short History of Mathematical Population Dynamics Vol. 618 (Springer, 2011).
Boyce M. S. Population viability analysis. Ann. Rev. Ecol. Syst. 23, 481–506 (1992).
Lacy, R. C. VORTEX: a computer simulation model for population viability analysis. Wildl. Res. 20, 45–65 (1993).
Lacy, R. C. Structure of the VORTEX simulation model for population viability analysis. Ecol. Bull. 48, 191–203 (2000).
Ha, J. C. in Nonhuman Primates in Biomedical Research 2nd edn (eds Abee, C. R. et al.) 287–292 (Academic, 2012).
Akkoç, C. C. & Williams, L. E. Population modeling for a captive squirrel monkey colony. Am. J. Primatol. 65, 239–254 (2005).
NC3Rs Guidelines: Non-human Primate Accommodation, Care and Use 2nd edn (National Centre for the Replacement Refinement & Reduction of Animals in Research, 2017); https://nc3rs.org.uk/3rs-resources/non-human-primate-accommodation-care-and-use-guidelines
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2021); https://www.R-project.org/
Gillard, J. Colony-forecast-model. GitHub https://github.com/dstl/colony-forecast-model (2025).
Tardif, S. D. et al. Reproduction in captive common marmosets (Callithrix jacchus). Comp. Med. 53, 364–368 (2003).
Poole, T. B. & Evans, R. G. Reproduction, infant survival and productivity of a colony of common marmosets (Callithrix jacchus jacchus). Lab. Anim. 16, 88–97 (1982).
Box, H. O. & Hubrecht, R. C. Long-term data on the reproduction and maintenance of a colony of common marmosets (Callithrix jacchus jacchus) 1972–1983. Lab. Anim. 21, 249–260 (1987).
Acknowledgements
Many thanks to the numerous Dstl colleagues who offered advice and expertise in the preparation of this work. This work was funded by the UK Ministry of Defence.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Peer review
Peer review information
Lab Animal thanks Fernando Melleu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gillard, J.J. A computational model to support the welfare-based management of a laboratory colony of common marmosets. Lab Anim 54, 120–125 (2025). https://doi.org/10.1038/s41684-025-01518-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41684-025-01518-3