Abstract
Immune checkpoint blockade (ICB) therapy has emerged as a pivotal advancement in cancer treatment, yet its efficacy varies among patients and resistance can develop. This study focuses on TIGIT, a newly identified immune checkpoint, to explore its expression, prognostic significance, and therapeutic potential in hematologic malignancies, particularly acute myeloid leukemia (AML). In this study, we found TIGIT highest expression levels in bone marrow and lymphoid tissues, with enrichment in immune cells such as NK-T cells and regulatory T cells (Tregs). A prognostic model incorporating TIGIT expression and other immune-related genes effectively stratified AML patients into high-risk and low-risk groups, with the former displaying significantly shorter overall survival times. Our model outperformed traditional prognostic factors, highlighting TIGIT’s potential as a superior predictive biomarker. Additionally, our in vitro and in vivo studies showed that combining tiragolumab with azacitidine (AZA) synergistically enhanced anti-tumor efficacy, reducing tumor burden and extending survival in a murine AML model. Our findings underscore TIGIT’s role in hematologic malignancies and its potential as a therapeutic target in AML. The combination of AZA with TIGIT inhibition offers a promising new approach for AML treatment, warranting further clinical evaluation.
Similar content being viewed by others
Introduction
Immune checkpoint blockade (ICB) therapy is currently a research hotspot in the field of cancer treatment. Breakthrough progress has been made in the research and drug development targeting immune checkpoints such as programmed death-1 (PD-1) and its ligand (programmed death-ligand 1, PD-L1), as well as cytotoxic T lymphocyte associated antigen-4 (CTLA-4)1,2,3. Although drugs targeting PD-1 and CTLA-4 have achieved success in clinical treatment of various tumors, their therapeutic effects remain unsatisfactory for some tumors4. Additionally, the use of single-target therapies in clinical settings can easily lead to drug resistance, prompting researchers to continue exploring new immune checkpoints5.
TIGIT, also known as WUCAM, Vstm3, or VSIG9, is a newly discovered immune checkpoint in recent years6. As a receptor of the immunoglobulin superfamily, it plays a crucial role in limiting adaptive and innate immunity7. Zhang et al. found that the deletion of TIGIT specifically in NK cells can inhibit tumor growth to a certain extent8. Blocking TIGIT with monoclonal antibodies can reverse the exhaustion of antitumor NK cells in various tumor models, thereby improving the overall survival rate of tumor-bearing animals9. The co-expression of multiple immune checkpoints on T cells suggests that tumor cells can evade the attack of the immune system through various pathways, which also implies that the combined use of drugs targeting these different pathways may exert a synergistic antitumor effect10,11. Research conducted on mouse models of melanoma and lung cancer metastasis has demonstrated that TIGIT antibodies, either alone or combined with PD-1 antibodies, can prevent tumor growth by enhancing the antitumor effects of CD8+ T cells12,13. In patients with head and neck squamous cell carcinoma and mouse models, overexpression of TIGIT has been observed on tumor-infiltrating CD8+ and CD4+ T cells, and this overexpression is associated with the expression of the immune checkpoint molecule PD-114. Indeed, blocking either TIGIT or PD-1 alone can enhance the effector function of tumor antigen-specific CD8+ T cells and more effectively suppress tumor growth15.
The novel immune checkpoint TIGIT exerts regulatory effects on various cells, including NK cells, effector T cells, and DC cells, and the mechanisms underlying these effects are diverse11. Although no drugs targeting the immune checkpoint TIGIT have been approved for market globally, more than 10 TIGIT antibodies are currently in clinical trials worldwide. In clinical trials for non-small cell lung cancer and extensive-stage small cell lung cancer, the survival rate of subjects in the tiragolumab and atezolizumab combination therapy group was significantly higher compared to the atezolizumab group16. In the study of MK-7684 combined with pembrolizumab for the treatment of metastatic solid tumors, both treatment tolerability and safety were good, and no dose-limiting toxicities were observed17.
TIGIT, as an underappreciated immune checkpoint in NK cells, is poised to play a significant role in future antitumor treatment. While most current research focuses on solid tumors, the role of TIGIT in hematologic malignancies remains largely unexplored. We focus on TIGIT, exploring its expression, distribution, prognostic impact, and therapeutic implications in hematologic cancers.
Results
Single-cell RNA expression level of TIGIT in tissues
TIGIT plays a crucial role in the immune regulation within the body. Our initial investigation focused on examining the expression of TIGIT across various normal tissue types. The results revealed that TIGIT displayed the highest expression levels in bone marrow and lymphoid tissues, followed by the kidney, urinary bladder, and gastrointestinal tract (Fig. 1a). To confirm these findings, we cross-referenced TIGIT RNA expression data from other databases. Both the Consensus and HPA databases indicated that TIGIT RNA tissue specificity was predominantly found in lymph nodes, tonsils, spleen, appendix, thymus, and bone marrow (Fig. 1b). Additionally, the FANTOM5 database demonstrated that TIGIT protein expression was also elevated in these tissues (Fig. 1c).
a Bar chart showing the tags per million (TPM) for different tissues, highlighting the relative abundance of RNA expression in each tissue type. b RNA Expression Overview from Consensus and HPA Databases:The top panel (Consensus Database) and bottom panel (HPA Database) display the normalized transcripts per million (nTPM) for various tissues, indicating the level of RNA expression across different tissues. c Bar chart depicting the tags per million (TPM) for specific cell types within a tissue, providing a detailed view of RNA expression at the cellular level. d Single Cell Type Specificity: Group Enhanced (T cells, NK cells, and Dendritic cells): This bar chart shows the nTPM for specific cell types within the groups of T cells, NK cells, and dendritic cells, highlighting the enhanced expression in these cell types. e UMAP (Uniform Manifold Approximation and Projection) plots for Bone Marrow: The left panel shows the clustering of different cell types in the bone marrow based on RNA expression profiles. The right panel displays a heatmap of gene expression levels for various genes across the identified cell types.
In order to delve deeper into TIGIT expression within specific tissue cell types, we analyzed the RNA single-cell type specificity of TIGIT. The data indicated that TIGIT expression was particularly enhanced in blood and immune cells (Fig. 1d). Utilizing single-cell sequencing, we investigated the subpopulation expression of TIGIT in these key tissues. The results demonstrated that immune cells, particularly T-cells in bone marrow, lymph nodes, and spleen, exhibited relatively high TIGIT expression levels (Figs. 1e and S1). In addition, single-cell sequencing data from GSE116256 indicated that TIGIT and its ligands are mainly expressed in NK cells and CD8+ T cells, which was consistent with our Human Atlas data (Fig. S2). According to relevant CCLE data, in hematological tumors, the expression of TIGIT in AML, ALL, and CML was lower than that of its ligands (Fig. S3). However, in pan-cancers, the expression of TIGIT in hematological tumors was higher than that in other types of tumors, while the expression of its ligands was lower than that in other types of tumors (Fig. S4). We further examined the relevant data of core cell types, such as endothelial cells, smooth muscle cells, fibroblasts, macrophages, neutrophils, mast cells, T-cells, and plasma cells. Our analysis revealed that T-cells in subcutaneous, visceral, breast, colon, heart muscle, and kidney tissues were closely associated with TIGIT expression (Fig. 2a). Moreover, we categorized T-cells based on their functional characteristics and examined TIGIT expression across these distinct subsets. Our findings indicated that TIGIT displayed elevated expression levels in NK-T cells and regulatory T cells (Tregs) (Fig. 2b). Through UMAP analysis, we explored the immune cell type specificities of hematopoietic tissues. Among all clusters identified, cluster 4, representing T-reg cells, exhibited the strongest association with TIGIT expression (Fig. 2c). Finally, as illustrated in Fig. 2d, we identified the top 10 genes that displayed the strongest positive correlation with Cluster 4.
a The distribution of different cell types in the samples. Various cell types, such as smooth muscle cells, fibroblasts, macrophages, neutrophils, mast cells, T cells, and plasma cells, are represented using different colored dots. b The expression levels of specific genes (T-reg, CD4+T, CD8+T, and NKT) in different cell types, bar charts indicate the expression levels of these genes. c UMAP analysis of TIGIT in immune cell type-specificities of hematopoietic tissues. d The top 10 genes that displayed the strongest positive correlation with Cluster 4. e–g Survival curves for different risk groups (high risk and low risk) over time. Each plot includes survival probability curves and risk tables showing the number of individuals at risk at various time points. h–j Multivariate Cox regression analysis, including risk scores, age, ELN2017, and other variables in different datasets. The significance levels and hazard ratios are provided for each variable. k–m ROC curves for different datasets (BeatAML, TCGA, GSE106291) to evaluate the performance of models in predicting 5-year survival. Sensitivity and specificity curves, along with AUC values, are shown.
Prognostic model development and validation
To further elucidate the prognostic significance of TIGIT in acute myeloid leukemia (AML), we harnessed a panel of 15 genes from cluster 4, identified through robust bioinformatic analyses as exhibiting strong correlations with regulatory T cells (Tregs) and TIGIT, to construct an innovative prognostic model. The BeatAML dataset was meticulously selected as our primary training cohort, whereas the TCGA and GSE37642 repositories served as rigorous validation platforms. Employing Kaplan-Meier survival analysis, we observed a marked disparity in overall survival (OS) durations, with high-risk patients demonstrating significantly shorter OS times compared to their low-risk counterparts across both training and validation cohorts (Fig. 2e–g). Utilizing multivariate Cox regression forest plots, we discerned that the risk score derived from our model not only exhibits robust prognostic capabilities but also stands as an independent predictor of OS, rivaling the predictive power of established clinical indices such as age, French-American-British (FAB) classification, and ELN2017 (Figs. 2h–j and S5). We dug into the relevant information in the database as deeply as possible. Multivariate analyses in the BeatAML and GSE106291 databases both indicated that, after being combined with the current available prognostic information based on molecular characterization, this riskscore model can still serve as an independent prognostic factor for poor prognosis (Figs. 2j and S6).
To substantiate the predictive accuracy of our prognostic model, we conducted time-dependent receiver operating characteristic (ROC) curve analyses to evaluate the area under the curve (AUC), which quantifies the balance between sensitivity and specificity. In the BeatAML dataset, the AUC for predicting 5-year OS was notably high at 0.832 [95% confidence interval (CI): 0.75–0.90] for the integrated risk score group, outperforming the risk score alone (0.637 [95% CI: 0.54–0.73]), age (0.742 [95% CI: 0.65–0.83]), WBC count (0.536 [95% CI: 0.44–0.63]), and the ELN2017 risk stratification system (0.691 [95% CI: 0.61–0.77]) (Fig. 2k). Similarly, in the TCGA dataset, the integrated risk score group achieved an exceptional AUC of 0.959 [95% CI: 0.92–1.0] for 5-year survival prediction, markedly surpassing the performance of the risk score alone (0.781 [95% CI: 0.63–0.93]), age (0.772 [95% CI: 0.62–0.92]), FAB classification (0.562 [95% CI: 0.41–0.71]), and WBC count (0.652 [95% CI: 0.47–0.84]) (Fig. 2l). In the GSE106291 dataset, the integrated risk score group exhibited an AUC of 0.757 [95% CI: 0.647–0.8663] for 5-year OS prediction, again demonstrating superior predictive accuracy compared to the risk score alone (0.649 [95% CI: 0.5255–0.7723]) and ELN2017 (0.723 [95% CI: 0.6194–0.8259]) (Fig. 2m). In the GSE37642 dataset, the integrated risk score group also exhibited a higher AUC of 0.745 [95% CI: 0.69-0.80] for 5-year OS prediction (Fig. S5). Collectively, these compelling results underscore that the incorporation of our novel risk score significantly enhances the precision of OS prediction in AML patients, outstripping the predictive capabilities of traditional prognostic factors.
Clinical features and prognosis of TIGIT
Accordingly, we endeavored to investigate the prognostic value of TIGIT in hematological malignancies. Notably, the forest plot prognostic analysis suggested that TIGIT expression could serve as a predictor of favorable overall survival (OS) rates across nearly all hematological malignancies (Fig. S7). We especially observed that TIGIT holds considerable prognostic significance in various acute myeloid leukemia (AML) datasets (Fig. 3a). Moreover, we examined the mutational status of TIGIT in AML. The mutational landscape of TIGIT revealed that patients exhibiting low TIGIT expression tend to possess a greater mutational load (Fig. 3b). Among prevalent AML mutation genes, FLT3 mutations were more frequently identified in individuals with low TIGIT expression, whereas TP53 and RUNX1 mutations were predominantly found in those with high TIGIT expression (Fig. 3b). We further explored the correlation between TIGIT expression and common molecular genetics in AML. Analyses using GSE13159, GSE147515, and BeatAML datasets revealed no significant differences in TIGIT expression among AML patients with different chromosomal abnormalities (Fig. S8). Additionally, there were no significant differences in TIGIT expression between patients with different prognostic risks (Fig. S9). This may suggest that the TIGIT-related prognostic model we constructed could be independent of currently used molecular genetic prognostic warning models and might serve as one of the independent prognostic factors.
a The survival probability curves for TIGIT in AML patients, comparing high and low TIGIT expression groups. The p-values (p = 0.0016 and p = 0.013) indicate statistically significant differences in survival between the groups. b The mutation count distribution across different sample groups (LowExp and HighExp) for various genes. The bar chart shows the percentage of mutations for each gene, highlighting the differences between the groups. c The UMAP plots represent the clustering of AML samples at different time points (D0, D14, D15, D18) and overall (Before and After). The plots show the distribution and changes in cell populations over time. d TIGIT expression levels across different clinical stages (Initial Diagnosis, Residual, Remission, Relapse, Unknown). The p-value (p = 0.018) indicates a significant difference in TIGIT expression between these stages. e The methylation patterns of genes related to the TIGIT group, showing variations in methylation levels across different samples. f The methylation status of the TIGIT gene promoter and gene body in remission and initial diagnosis samples. Green dots represent methylated regions, while empty dots represent unmethylated regions. g The methylation rates between remission and initial diagnosis samples, showing the distribution and differences in methylation levels.
Additionally, we utilized single-cell RNA sequencing data from AML patients at diagnosis and their matched samples post-chemotherapy to assess TIGIT expression across immune cell subtypes. Uniform Manifold Approximation and Projection (UMAP) analysis demonstrated that TIGIT expression levels in total bone marrow (BM) cells were significantly lower at diagnosis compared to those in matched samples following chemotherapy (Fig. 3c). This finding was further corroborated in additional beatAML datasets, where TIGIT expression was found to be diminished in patients presenting with high tumor burdens, encompassing both treatment-naive and relapsed cases (Fig. 3d). Furthermore, heatmap analyses unveiled substantial disparities in epigenetic-associated genes between the high and low TIGIT expression cohorts. In primary patient samples, TIGIT expression was elevated in patients attaining remission post-chemotherapy but reduced in treatment-naive individuals (Fig. 3e). Our analysis of the GSE106291 data suggests that there is no significant difference in the expression of TIGIT among ND-AML, s-AML, and t-AML (Fig. S10). Consistently, bisulfite sequencing PCR (BSP) results substantiated that the methylation status of both the promoter and gene body regions of the TIGIT gene was appreciably heightened in treatment-naive patients compared to those in remission, potentially constituting one of the underlying mechanisms driving differential TIGIT expression (Fig. 3f and g).
AZA enhances TIGIT expression and synergizes with TIGIT inhibitors in AML treatment
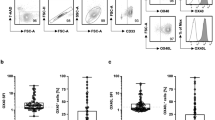
Given that Azacitidine (AZA) can regulate tumor-related gene expression by affecting DNA methylation levels, we explored the potential association between AZA and TIGIT. Initially, we measured TIGIT expression levels before and after AZA treatment in three AML cell lines and found that AZA significantly increased TIGIT mRNA expression in all three leukemia cell types (Fig. 4a). Moreover, TIGIT protein levels were also markedly elevated (Fig. 4b). Consequently, we further examined the methylation levels of CpG islands in the promoter and gene body regions of the TIGIT gene before and after AZA treatment. Intriguingly, AZA treatment resulted in a significant reduction in methylation levels of CpG islands in both the promoter and gene body regions, with the promoter region showing the most pronounced decrease (Fig. 4c).
The expression levels of TIGIT in Kasumi-1, OCI-AML3, and MOLM13 cell lines treated with DMSO or Azacitidine. The upper panel (a) displays the quantitative data, while the lower panel (b) shows the Western blot analysis. c The methylation status of the TIGIT promoter region in these three cell lines. Green circles indicate methylated regions, while white circles indicate unmethylated regions. d The cell viability of Kasumi-1, OCI-AML3, and MOLM13 cell lines under different treatment conditions, including mock, UCB-NK, UCB-NK with Azacitidine, UCB-NK with Tiragolumab, and UCB-NK with both Azacitidine and Tiragolumab. e The cell viability under conditions involving high and low NK cells, with and without treatments. The synergistic effect of tiragolumab plus azacitidine was calculated by CompuSyn software. f A schematic illustration of the experimental setup for the mouse model, showing the process of leukemia cell injection, infection, and treatment with different compounds. g Kaplan-Meier survival curve depicting the survival rates of mice over time after leukemia cell injection, comparing different treatment groups (vehicle, TIGIT, Azacitidine, and Azacitidine + TIGIT). h Photographs of spleens from treated mice, showing the physical changes due to different treatments. i The distribution of cells in peripheral blood (PB), bone marrow (BM), and spleen (SP) under different treatment conditions by Flow cytometry. j The percentage of leukemia cells in PB, BM, and SP under different treatment conditions.
AZA is one of the clinically approved therapeutic agents for AML18; however, its monotherapeutic efficacy is restricted, typically requiring combination with venetoclax or alternative targeted agents/chemotherapies to treat AML19. In this context, the prognostic risk model of TIGIT implicates it as a promising predictive biomarker for AML prognosis, leading us to hypothesize that inhibitors targeting the TIGIT pathway might synergize with targeted agents to eradicate AML tumor cells. To validate our hypothesis, we treated AML cell lines OCI-AML3, Kasumi-1, and MOLM13 with the TIGIT inhibitor tiragolumab in conjunction with azacitidine, followed by co-culture with umbilical cord blood-derived natural killer (UCB-NK) cells. Cellular viability assessments demonstrated that UCB-NK cells could synergize with tiragolumab and azacitidine to eliminate leukemic cells (Fig. 4d). Furthermore, we sought to substantiate these findings using mononuclear cells isolated from the bone marrow (BM) of newly diagnosed AML (ND-AML) patients. Specimens were stratified according to the NK cell proportion within the BM. Concordant with the in vitro studies, patient samples exhibiting low NK cell proportions treated with tiragolumab displayed diminished viability compared to those with high NK cell proportions (Fig. 4e). The synergistic effect of tiragolumab plus azacitidine was somewhat greater than that of the single drugs (Fig. 4e).
Tiragolumab synergizes with Azacitidine to Kill AML cells in Vivo
To ascertain the in vivo therapeutic efficacy of tiragolumab, we utilized lentiviral vectors encoding the MLL-AF9 oncogene to infect hematopoietic stem cells harvested from humanized TIGIT knock-in mice. Following engraftment via bone marrow transplantation, we successfully established a murine model of AML expressing human TIGIT (hTIGIT) (Fig. 4f). Survival analyses demonstrated that tiragolumab exerted significant antitumor activity in vivo, with concomitant administration of azacitidine resulting in marked prolongation of median survival time (Fig. 4g). Furthermore, quantitative assessments of spleen dimensions and mass revealed substantial reductions post-treatment, with imaging studies illustrating a pronounced decrease in spleen size towards normative values upon combined therapy (Fig. 4h). To evaluate the impact of treatment on tumor burden, we employed GFP fluorescence to monitor the presence of neoplastic cells within peripheral blood, bone marrow, and spleen compartments. Data analysis revealed a significant attenuation in GFP-positive tumor cell populations following combination therapy, thereby underscoring the potential efficacy of this synergistic approach for the treatment of AML (Fig. 4i, j).
Discussion
Immune checkpoint blockade therapy represents a significant advancement in cancer treatment by leveraging the immune system to combat tumor cells5. However, the effectiveness of current ICB therapies, such as those targeting PD-1 and CTLA-4, varies among patients and can lead to resistance in some cases4. This has prompted extensive research into alternative immune checkpoints like TIGIT, which hold promise as new therapeutic targets. Our study has delved into the expression patterns and prognostic significance of TIGIT in hematologic malignancies, with a particular focus on AML. We found that TIGIT expression is most pronounced in bone marrow and lymphoid tissues, aligning with its role in immune regulation. Single-cell RNA sequencing further revealed that TIGIT is abundantly expressed in immune cells such as NK-T cells and Tregs, highlighting its crucial role in modulating immune responses. Joint analysis of multiple databases shows that TIGIT has certain correlations with other immune regulatory molecules, but without regularity. For example, it is positively correlated with PD-L1 (CD274), CTLA4, and LAG3, but has no clear correlation with other immune molecules such as CD276, PVR, TGFB1, and ULBP1 (Fig. S11). To assess the prognostic value of TIGIT, we developed and validated a prognostic model incorporating TIGIT expression and other genes related to Tregs and immune regulation. Six genes including TIGIT were found to be associated with the risk score. They were FOXP3, PTTG1, TIGIT, RTKN2, BATF and BTG3 respectively. FOXP3 is a key transcription factor for Tregs and maintains immune tolerance20. Tumor cells inhibit effector T cells through TIGIT and simultaneously induce Treg differentiation. BATF+ Tfh promotes immunosuppressive antibodies, while TIGIT directly inhibits effector immune cells, forming a double barrier21. This model effectively stratified patients into high-risk and low-risk groups, with the former exhibiting significantly shorter overall survival times. Notably, our model’s risk score outperformed traditional prognostic factors like age, ELN2017, and white blood cell count, underscoring its potential as a superior predictive biomarker.
Clinical trial data further bolsters the potential of TIGIT as a therapeutic target22. For instance, a phase I/II clinical trial assessing the safety and efficacy of tiragolumab, an anti-TIGIT antibody, in combination with atezolizumab, an anti-PD-L1 antibody, in non-small cell lung cancer patients, reported a higher objective response rate (ORR) and progression-free survival (PFS) for the combination therapy compared to atezolizumab monotherapy23. Specifically, the ORR was 38.8% for the combination therapy versus 24.1% for atezolizumab alone, and the median PFS was 5.6 months compared to 3.9 months, respectively24. Similarly, another phase I/II trial evaluating the safety and efficacy of MK-7684, another anti-TIGIT antibody, in combination with pembrolizumab, an anti-PD-1 antibody, in patients with advanced solid tumors, demonstrated a favorable safety profile and promising antitumor activity [NCT02964013]25. The ORR for the combination therapy was 24%, with some patients experiencing durable responses.
IL-12 could promote NK cell polarization towards the Th1 phenotype, upregulate IFN-γ secretion, enhance anti-tumor activity, and simultaneously induce TIGIT expression26. IL-15, on the other hand, promotes NK cell proliferation and survival through IL-15Rα and enhances cytotoxicity by releasing perforin/granzyme B, but prolonged stimulation may lead to functional exhaustion27. Our research indicated that umbilical cord blood-derived NK cells, after being treated with appropriate levels of IL-12 and IL-15, could expand and proliferate more effectively, and in combination with a TIGIT inhibitor, could exert enhanced anti-tumor effects.
In the context of AML, our study discovered that azacitidine, an FDA-approved therapeutic agent for AML, significantly increases TIGIT expression by decreasing methylation levels in its promoter and gene body regions. This finding implies a potential synergistic effect between AZA and TIGIT inhibitors. We analyzed and compared the RNA expression and flow cytometry expression results of TIGIT in tumor cells from 10 AML patients. Correlation analysis indicated that in AML, RNA expression correlates with flow cytometric expression of TIGIT (Fig. S12). This suggests that FCM evaluation of TIGIT in Tregs might guide the choice of chemotherapy. Our in vitro and in vivo studies indeed showed that combining the TIGIT inhibitor tiragolumab with AZA enhances anti-tumor efficacy, resulting in substantial reductions in tumor burden and extended survival in a murine model of AML. The success of this combination therapy can be attributed to AZA’s dual role in upregulating TIGIT expression and exerting direct anti-tumor effects, while tiragolumab blocks the inhibitory signals mediated by TIGIT, thereby bolstering the overall immune response against the tumor. This synergistic approach not only improves treatment efficacy but also potentially mitigates the risk of resistance development by targeting multiple pathways simultaneously. Clinical trial data on the combination of AZA with other immunotherapies further support this strategy28,29. For example, a phase II study evaluating the safety and efficacy of azacitidine in combination with nivolumab, an anti-PD-1 antibody, in relapsed/refractory AML patients reported an ORR of 33% and a median overall survival of 10.3 months [NCT02534303]30. Although this trial did not specifically target TIGIT, it illustrates the potential benefits of combining DNA methylation inhibitors with immune checkpoint inhibitors in AML treatment.
This study still has the following limitations: (1) Current mainstream bioinformatics databases (including TCGA, BeatAML, and GEO) lack complete datasets of commonly used prognostic indicators; (2) Due to data limitations, we could only perform limited-dimensional feature mining based on existing data. Regarding the aforementioned deficiencies, we will further analyze the relationship between this prognostic risk factor and currently routine prognostic-related factors in a multi-center study. (3) As there is no murine TIGIT inhibitor at present, we chose humanized TIGIT mice for our animal model. However, these mice have a deficiency in the integrity of the immune cell lineage, specifically manifested as incomplete coverage of T cell subsets and the absence of immune memory cells, among other issues. (4) Indeed, due to the substantial individual differences among patients, there are certain variations in the proportion of intrinsic NK cells. Some reports have indicated that AML patients with a high level of intrinsic NK cells tend to have a better prognosis, while those with a low level of intrinsic NK cells have a poorer prognosis31. From our research results, it can be observed that patients with a high NK group have better killing effects either when Tiragolumab is used alone or in combination with AZA. Our current study cannot rule out whether this is related to the presence of intrinsic NK cells.
In summary, our study highlights the importance of TIGIT in hematologic malignancies, particularly in AML. TIGIT expression serves as a valuable prognostic biomarker, and its upregulation by AZA, combined with TIGIT inhibition, presents a novel and potentially highly effective therapeutic strategy. The clinical trial data discussed above provide robust evidence supporting the efficacy and safety of this combinatorial approach. Future clinical trials should evaluate the safety and efficacy of this combinatorial approach in AML patients, with the potential to revolutionize the treatment landscape of this disease. Additional research should explore the broader implications of TIGIT in other hematologic malignancies and its potential role in other cancer types. Gaining a deeper understanding of the molecular mechanisms governing TIGIT regulation and its interplay with other immune checkpoints will be essential for developing more effective and personalized immunotherapies.
Methods
Primary samples and cell culture
Mononuclear cells from the bone marrow of patients with initial onset and those who achieved remission after chemotherapy in Guangzhou First People’s Hospital were extracted and cultured in DMEM medium with 10% fetal bovine serum and cytokines. All participants provided written informed consent in accordance with the regulations of the Institutional Review Boards of the Hospitals in agreement with the Declaration of Helsinki. This work has been approved by the Ethnics Committee of Guangzhou First People’s Hospital (S-2024-136). The AML cell lines such as Ontario Cancer Institute-Acute Myeloid Leukemia-3 (OCI-AML3, this cell line is derived from the peripheral blood of a 57-year-old male patient diagnosed with AML-M4 in 1987, carrying an NPM1 gene mutation (type A) and a DNMT3A R882C mutation), Kasumi-1 (this cell line is derived from the peripheral blood of an Asian male AML patient, characterized by a translocation between chromosomes 8 and 21, resulting in the fusion of the AML1 gene with the ETO gene to form the AML1-ETO protein), and MOLM13 (Established in 1997 from the peripheral blood of a 52-year-old female patient with AML-M5 type. It carries an NPM1 gene mutation (type A, i.e., NPM1c) and an FLT3-ITD mutation) were obtained from the Yu Shan-he’s laboratory.
Data acquisition
The transcript level expression data of TIGIT in different tissues and different cancers types were obtained from Human Altas32. The clinicoprognosis and clinicopathologic information and gene expression data of patients with hematological malignancies, including B-cell lymphoma (GSE10846, GSE34171), AML (GSE37642, GSE106291, GSE116256, GSE13159 and GSE147515), T-cell lymphoma (GSE58445, GSE90579), and myeloma (GSE4204, GSE2658), are derived from the GEO database. In addition, the analysis of AML patients also utilized relevant data from TCGA and BeatAML, respectively. The expression profiles were normalized by Log2 transformation.
Single-cell expression and prognostic analysis
Single-cell RNA sequencing (scRNA-seq) analysis was conducted on various tissues33. Initially, cells intended for scRNA-seq underwent quality control to ensure they were single-suspension cells without prior enrichment. Subsequently, the sequencing data required a minimum of 4000 cells and at least 20 million read counts. Ultimately, TIGIT expression in tissues was visualized using UMAP. Moreover, scRNA-seq data from AML patients at diagnosis and their matched samples post-chemotherapy, obtained from GSE116256, were utilized for immune cell subtype analysis34. UMAP analysis revealed the discernible TIGIT expression in immune cells within the total BM cells of AML samples at diagnosis and corresponding samples after chemotherapy.
Quantitative Real-Time PCR (qRT-PCR)
TIGIT expression levels were quantified using qRT-PCR. For this purpose, we employed the commercially available assay (ESscience; QP002) and adhered strictly to the manufacturer’s instructions. The relative expression data were analyzed employing the widely accepted 2−ΔΔCt method, which allows for the comparison of gene expression between different samples. The specific primers utilized for amplifying the TIGIT gene were as follows: Forward Primer: TCTGCATCTATCACACCTACCC; Reverse Primer: CCACCACGATGACTGCTGT. These primers were meticulously designed using the Primerbank database, ensuring high specificity and efficiency in amplifying the target sequence.
TIGIT-related prognosis model
TIGIT-related genes (TRGs) were delineated based on Human Altas analysis. The BeatAML cohort was adopted as the training dataset, whereas the TCGA and GSE37642 cohorts were selected as external validation datasets to substantiate the extensibility of our conclusions. To screen for prognostic pertinent genes, we implemented the log-rank analysis, which evaluates the disparity in survival profiles between two cohorts based on their optimal cutoff expression values, with a stringent significance threshold of P < 0.05 established to solely retain statistically significant correlations. Furthermore, we devised a prognostic TRGs signature through the employment of the least absolute shrinkage and selection operator (LASSO) regression analysis. This sophisticated statistical approach utilizes penalized maximum likelihood estimators to precisely identify a subset of genes exhibiting the most substantial predictive capacity for patient outcomes while concurrently mitigating the risk of overfitting. The model development process incorporated tenfold cross-validation, a robust resampling technique that partitions the dataset into ten equal subdivisions and iteratively employs nine-tenths for training purposes and the remaining one-tenth for validation, thereby ensuring a comprehensive and reliable assessment of model performance. The TRGs riskscore (RS) for each AML patient was calculated by using the following formula:
Patients were classified into high- and low-risk categories based on the optimal RS cutoff. Kaplan-Meier and time-dependent ROC analyses were conducted to assess the prognostic accuracy and reliability of the TRGs signature.
Mutation profiling and methylation analysis
KEGG pathway enrichment analysis was conducted to delineate the underlying physiological functionalities across the high-risk and low-risk cohorts. A threshold of a normalized p-value < 0.05 was established as the criterion for statistical significance. Furthermore, the immune cell infiltration scores for a panel of 22 distinct immune cell types were determined utilizing the ESTIMATE deconvolution algorithm35. Extract 1 µg of total genomic DNA; perform bisulfite conversion utilizing the EZ DNA Methylation-Gold Kit (Zymo) in accordance with the manufacturer’s protocol; design methylation-specific primers employing the QUMA software36; amplify the target sequences post-bisulfite conversion via Bland-Tag-Plus PCR; and assess the CpG methylation status of the amplified target regions using the QUMA analytical tool.
Reagent
TIGIT inhibitor (Tiragolumab, A2028) and Azacitidine (S1782) were obtained from Selleck.
Cell viability assay
Cell viability was examined using the Cell Counting Kit-8 assay (CCK8) (Dojindo) according to the manufacturer’s instructions. AML cell lines and primary samples were plated (5000~50,000 cells/well) in a 96-well plate and incubated in a humidified cell incubated with an atmosphere of 5% CO2 at 37 ◦C. Following cell exposure to Tiragolumab and Azacitidine for 48 h, the CCK-8 reagent was added, and incubation was continued for another 2 h. Optical density was detected by a microplate reader at a wavelength of 450 nm.
Flow cytometric analysis
Murine peripheral blood (PB), bone marrow (BM), and spleen (SP) cells were prepared in FACS buffer (PBS containing 1% FBS) and incubated with fluorochrome-labeled antibodies. Data acquisition was performed on an LSR II flow cytometer (Becton Dickinson), followed by analysis using FlowJo software (TreeStar).
Umbilical cord blood-derived natural killer cells (UCB-NK)
The UCB-NK samples originated from the Shandong affiliate of the China Cord Blood Bank. Incrementally cultivated UCB-NK cells were grown in RPMI-1640 medium (Gibco, New York) supplemented with 10% FBS (Biochrom AG, Berlin, Germany). Moreover, the cultures were treated with 50 ng/mL human interleukin-12 (IL-12, PeproTech) and 10 ng/mL interleukin-15 (IL-15, PeproTech)37. Low concentrations of IL-12 (10 ng/mL) might tend to maintain activity, while high concentrations (50 ng/mL) can enhance proliferation but also require attention to toxicity. Generally, when used in combination with IL-15, it could synergistically promote NK cell expansion and enhance function. These cells were subsequently co-cultured with OCI-AML3, Kasumi-1, and MOLM13 cell lines in 96-well plates.
Western blotting
Antibodies were purchased from FineTest (anti-TIGIT, FNab10904), and Santa Cruz (anti-GAPDH).
Animal models of TIGIT
The MSCV-MLL/AF9-IRES-GFP ecopack plasmid, generously provided by Dr. Yu Shanhe’s laboratory at the Shanghai Institute of Hematology, Ruijin Hospital affiliated with Shanghai Jiao Tong University, was utilized to transfect 293 T cells via Lipofectamine 2000-mediated transfection. Viral supernatants were harvested at 48 hours and 72 hours post-transfection, respectively, followed by centrifugation, filtration, and storage at −80 °C for future utilization.
BALB/c-hTIGIT mice were procured from Nanjing Bioway Company Limited and housed under specific pathogen-free (SPF) conditions at the Animal Center of South China University of Technology. At the onset of experimentation, mice were aged between 6 to 8 weeks. Each experimental cohort comprised a minimum of six animals, which were randomly allocated to distinct treatment groups. All experimental procedures strictly adhered to the National Institutes of Health (NIH) guidelines for the ethical care and use of laboratory animals.
Eight-week-old female BALB/c-hTIGIT mice received an intraperitoneal injection of 5-FU (5 mg/mouse), and five days post-injection, bone marrow cells were harvested and cultured in Cocktail medium (DMEM supplemented with 20% FBS, 20 ng/ml IL-3, 20 ng/ml IL-6, 50 ng/ml SCF, 20% WEHI-3 cell supernatant, and antibiotics) containing viral supernatant for 48 hours. Cells were subsequently centrifuged at 650 × g for 2 hours, resuspended in fresh culture medium, and incubated overnight. On the subsequent day, the infection process was repeated once. Cells were then resuspended in PBS and injected into eight-week-old female BALB/c mice (which had been irradiated with a total of 900 Gy in two fractions, 4 hours apart, with each fraction delivering a lethal dose of 450 Gy) via the tail vein (200 μl containing 4 × 105 cells). Following the development of MLL-AF9 leukemia, bone marrow and spleen cells from GFP-positive (MLL-AF9/h-TIGIT) primary cells were cryopreserved in liquid nitrogen.
Primary cells isolated from MLL-AF9/h-TIGIT mice were administered via tail vein injection into eight-week-old female BALB/c-hTIGIT mice. Three days following injection, animals were randomly assigned to treatment cohorts. The Vehicle group received intravenous injections of saline (200 μl) for a duration of ten consecutive days, the Azacitidine group was treated with intravenous injections of 200 μl (100 mg/kg Ara-C, HY-13605, MCE), and the h-TIGIT group received intravenous injections of 200 μl every three days (10 mg/kg Tiragolumab, Selleck). Peripheral blood samples were collected from control mice at day 12 to assess GFP positivity and monitor leukemia progression. Mice were euthanized 14 days post-transplantation, and bone marrow and spleen GFP positivity were analyzed via flow cytometry to evaluate treatment efficacy. All animal studies were approved by the Animal Ethics Committee at Guangzhou Medical University (Guangzhou, P.R. China).
Combination Index Analysis
The CompuSyn software (version 1.0; ComboSyn, Inc., Paramus, NJ, USA) was employed to calculate the combination indexes based on the average fraction of viable cells in the cytotoxicity assays38. The combinatorial effects were categorized into four groups: strong synergism (CI = 0.1–0.3), distinct synergism (CI = 0.3–0.7), mild synergism (CI = 0.7–0.9), and additive (CI = 0.9–1.1).
Statistical analysis
Continuous variables were subjected to statistical analysis employing either the Student’s t-test or the Wilcoxon signed-rank test, depending on the normality of the data distribution. Comparative assessments of overall survival (OS) between distinct groups were accomplished utilizing the log-rank test, a widely accepted method for evaluating differences in survival distributions. The time-dependent Receiver Operating Characteristic (ROC) package was deployed to construct ROC curves and ascertain the Area Under the Curve (AUC) values, which serve as a measure of predictive accuracy. Independent prognostic indicators were derived through rigorous application of univariate and multivariate Cox proportional hazards regression analyses, facilitating the identification of factors significantly associated with patient outcomes. All statistical analyses were conducted using specialized software packages, specifically R (version 4.0.4) and SPSS (version 25.0), both of which are widely recognized for their robust statistical capabilities. Statistical significance was determined by a two-sided P-value threshold of less than 0.05. In the context of drug combination studies, the CompuSyn software (Inc., Paramus, NJ, USA) was utilized to analyze combination indices (CI) based on viability data obtained from cytotoxicity assays39. The interpretation of CI values followed established conventions: CI values less than 1 indicated synergistic interactions, whereas values greater than 1 implied antagonistic effects. Notably, lower CI values correlated with enhanced synergistic efficacy, underscoring the potential benefits of specific drug combinations. *p < 0.05, **p < 0.01, **p < 0.001.
Data availability
No datasets were generated or analysed during the current study.
Abbreviations
- ICB:
-
Immune checkpoint blockade;
- PD-1:
-
Programmed death-1;
- PD-L1:
-
Programmed death-ligand 1;
- CTLA-4:
-
Cytotoxic T lymphocyte associated antigen-4;
- scRNA-seq:
-
Single-cell RNA sequencing;
- TRGs:
-
TIGIT-related genes;
- LASSO:
-
Least absolute shrinkage and selection operator;
- RS:
-
Riskscore;
- CCK8:
-
Cell Counting Kit-8 assay;
- PB:
-
Peripheral blood;
- SP:
-
Spleen;
- SPF:
-
Specific pathogen-free;
- NIH:
-
National Institutes of Health;
- OS:
-
Overall survival;
- ROC:
-
Receiver Operating Characteristic;
- AUC:
-
Area Under the Curve;
- CI:
-
Combination indices;
- Tregs:
-
Regulatory T cells;
- AML:
-
Acute myeloid leukemia;
- WBC:
-
White blood cell;
- FAB:
-
French-American-British;
- UMAP:
-
Uniform Manifold Approximation and Projection;
- BM:
-
Bone marrow;
- BSP:
-
Bisulfite sequencing PCR;
- AZA:
-
Azacitidine;
- ND-AML:
-
Newly diagnosed AML;
- hTIGIT:
-
human TIGIT;
- GFP:
-
Green Fluorescent Protein;
- UCB-NK:
-
Umbilical cord blood-derived natural killer cells.
References
Liu, Z. et al. Progenitor-like exhausted SPRY1(+)CD8(+) T cells potentiate responsiveness to neoadjuvant PD-1 blockade in esophageal squamous cell carcinoma. Cancer Cell 41, 1852–1870.e9 (2023).
van Gulijk, M. et al. PD-L1 checkpoint blockade promotes regulatory T cell activity that underlies therapy resistance. Sci. Immunol. 8, eabn6173 (2023).
Auslander, N. et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 24, 1545–1549 (2018).
Morad, G. et al. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021).
Sun, Q. et al. Immune checkpoint therapy for solid tumours: clinical dilemmas and future trends. Signal Transduct. Target Ther. 8, 320 (2023).
Harjunpaa, H. & Guillerey, C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 200, 108–119 (2020).
Ge, Z. et al. TIGIT, the Next Step Towards Successful Combination Immune Checkpoint Therapy in Cancer. Front Immunol. 12, 699895 (2021).
Zhang, P. et al. Targeting TIGIT for cancer immunotherapy: recent advances and future directions. Biomark. Res 12, 7 (2024).
Chu, X. et al. Co-inhibition of TIGIT and PD-1/PD-L1 in cancer immunotherapy: mechanisms and clinical trials. Mol. Cancer 22, 93 (2023).
Ochoa, M. C. et al. Antibody-dependent cell cytotoxicity: immunotherapy strategies enhancing effector NK cells. Immunol. Cell Biol. 95, 347–355 (2017).
Annese, T., Tamma, R. & Ribatti, D. Update in TIGIT immune-checkpoint role in cancer. Front Oncol. 12, 871085 (2022).
Ziogas, D. C. et al. Beyond CTLA-4 and PD-1 inhibition: novel immune checkpoint molecules for melanoma treatment. Cancers. 15, 2718 (2023).
Patel, A. J. & Middleton, G. W. TIGIT-based immunotherapeutics in lung cancer. Immunother. Adv. 3, ltad009 (2023).
Mao, L. et al. TIGIT/CD155 blockade enhances anti-PD-L1 therapy in head and neck squamous cell carcinoma by targeting myeloid-derived suppressor cells. Oral. Oncol. 121, 105472 (2021).
Pawlowska, A. et al. The Dual Blockade of the TIGIT and PD-1/PD-L1 pathway as a new hope for ovarian cancer patients. Cancers. 14, 5757 (2022).
Brazel, D., Ou, S. I. & Nagasaka, M. Tiragolumab (Anti-TIGIT) in SCLC: Skyscraper-02, a Towering Inferno. Lung Cancer 14, 1–9 (2023).
Rutkowski, P. & Czarnecka, A. M. Pembrolizumab for the adjuvant treatment of IIB or IIC melanoma. Expert Rev. Anticancer Ther. 23, 897–902 (2023).
Tenti, E. et al. Efficacy of Azacitidine in the treatment of adult patients aged 65 years or older with AML. Expert Opin. Pharmacother. 17, 2479–2486 (2016).
Ma, C. et al. Azacitidine combined with venetoclax alleviates AML-MR with TP53 mutation in SDS: a case report and literature review. Anticancer Drugs 35, 548–555 (2024).
Zhang, Z. & Zhou, X. Foxp3 Instability Helps tTregs Distinguish Self and Non-self. Front Immunol. 10, 2226 (2019).
Lahmann, A. et al. Bach2 Controls T Follicular Helper Cells by Direct Repression of Bcl-6. J. Immunol. 202, 2229–2239 (2019).
Liu, Y., Liu, W. & Wu, T. TIGIT: Will it be the next star therapeutic target like PD-1 in hematological malignancies?. Crit. Rev. Oncol. Hematol. 204, 104495 (2024).
Park, S. et al. Phase III, randomized study of Atezolizumab Plus Bevacizumab and chemotherapy in patients with EGFR- or ALK-mutated non-small-cell lung cancer (ATTLAS, KCSG-LU19-04). J. Clin. Oncol. 42, 1241–1251 (2024).
Cho, B. C. et al. Tiragolumab plus atezolizumab versus placebo plus atezolizumab as a first-line treatment for PD-L1-selected non-small-cell lung cancer (CITYSCAPE): primary and follow-up analyses of a randomised, double-blind, phase 2 study. Lancet Oncol. 23, 781–792 (2022).
Niu, J. et al. First-in-human phase 1 study of the anti-TIGIT antibody vibostolimab as monotherapy or with pembrolizumab for advanced solid tumors, including non-small-cell lung cancer. Ann. Oncol. 33, 169–180 (2022).
Romagnani, S. Biology of human TH1 and TH2 cells. J. Clin. Immunol. 15, 121–129 (1995).
Marquardt, N. et al. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat. Commun. 10, 3841 (2019).
Verma, A. et al. Nivolumab Plus 5-Azacitidine in pediatric relapsed/refractory Acute Myeloid Leukemia (AML): Phase I/II trial results from the Therapeutic Advances in Childhood Leukemia and Lymphoma (TACL) Consortium. Cancers. 16, 496 (2024).
Ricard, L. et al. A combination of 5-azacytidine and nivolumab is a potentially effective rescue therapy in relapsed/refractory AITL. Front. Immunol. 15, 1410638 (2024).
Daver, N. et al. Efficacy, safety, and biomarkers of response to Azacitidine and Nivolumab in relapsed/refractory acute myeloid leukemia: a nonrandomized, open-label, Phase II study. Cancer Discov. 9, 370–383 (2019).
Dai, Y. J. et al. Bone marrow infiltrated natural killer cells predicted the anti-leukemia activity of MCL1 or BCL2 inhibitors in acute myeloid leukemia. Mol. Cancer 20, 8 (2021).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 7, 1275 (2021).
van Galen, P. et al. Single-cell RNA-Seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Kumaki, Y., Oda, M. & Okano, M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 36, W170–W175 (2008).
Alnabhan, R., Madrigal, A. & Saudemont, A. Differential activation of cord blood and peripheral blood natural killer cells by cytokines. Cytotherapy 17, 73–85 (2015).
Chou, T. C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70, 440–446 (2010).
Khushalani, N. I. et al. Breaking the barriers in cancer care: The next generation of herpes simplex virus-based oncolytic immunotherapies for cancer treatment. Mol. Ther. Oncolytics 31, 100729 (2023).
Acknowledgements
We especially thank Si-Yuan He (The University of Texas MD Anderson Cancer Center) for helping perform the analysis of samples. This work was supported by the National Natural Science Foundation of China (82000144 and 82400204), Guangzhou Basic and Applied Basic Research Foundation (202201011458), Guangzhou Health Technology Project (20231A011012), Guangzhou Municipal Science and Technology Project (2024A03J1022 and 2023A04J0616) and GuangDong Basic and Applied Basic Research Foundation (2023A15115110992).
Author information
Authors and Affiliations
Contributions
Y.Y.Z. and J.J.H. performed basic and murine experiments; Y.L.L., R.N.S., and K.H.B. performed bioinformatics analyses; T.G., X.P.L. interpreted data; Y.L.L. and S.Y.H. performed single-cell sequencing analysis and interpreted data; X.P.L. and P.H.W., edited the paper; Y.J.D. conceived and oversaw the study, interpreted data, and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, YY., He, JJ., Liu, YL. et al. Unlocking the potential of TIGIT in enhancing therapeutic strategies for acute myeloid leukemia through combined azacitidine therapy. npj Precis. Onc. 9, 142 (2025). https://doi.org/10.1038/s41698-025-00933-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00933-6