Abstract
This retrospective study evaluated the efficacy of large language models (LLMs) in improving the accuracy of Chinese ultrasound reports. Data from three hospitals (January-April 2024) including 400 reports with 243 errors across six categories were analyzed. Three GPT versions and Claude 3.5 Sonnet were tested in zero-shot settings, with the top two models further assessed in few-shot scenarios. Six radiologists of varying experience levels performed error detection on a randomly selected test set. In zero-shot setting, Claude 3.5 Sonnet and GPT-4o achieved the highest error detection rates (52.3% and 41.2%, respectively). In few-shot, Claude 3.5 Sonnet outperformed senior and resident radiologists, while GPT-4o excelled in spelling error detection. LLMs processed reports faster than the quickest radiologist (Claude 3.5 Sonnet: 13.2 s, GPT-4o: 15.0 s, radiologist: 42.0 s per report). This study demonstrates the potential of LLMs to enhance ultrasound report accuracy, outperforming human experts in certain aspects.
Similar content being viewed by others
Introduction
Accurate ultrasound reports are essential for effective patient management and treatment decisions. Mistakes, such as logical inconsistencies, omitted examination items, and spelling errors can cause misunderstandings and reduce diagnostic accuracy1,2. For example, in the liver assessment, the conclusion states, “No abnormalities detected,” while another section reports, “Multiple hepatic cysts identified.” Similarly, in a female patient, the conclusion states “right breast nodule” while the ultrasound description section reports “left breast nodule”. This inconsistency could result in a re-scan and/or re-assessment of the ultrasound images. Moreover, it can lead to missed diagnoses or incorrect treatment prescriptions, which could be potentially fatal for patients. However, the reality of radiologist shortages and overwork, combined with the high-pressure clinical environments, makes the occurrence of report errors inevitable3,4,5,6. Minimizing errors in reports is essential for maintaining diagnostic accuracy and patient safety, as well as enhancing healthcare efficiency.
Currently, most Western countries employ a double-reading system, where a senior physician reviews the reports to ensure certain accuracy standards are met. This laborious process not only reduces radiologists’ efficiency but also increases their workload7. In countries without a double-reading system, such as China, the accuracy of reports relies entirely on the radiologist conducting the ultrasound. This may result in variability in report quality and potential misdiagnoses8. Therefore, finding an efficient way to maintain the accuracy of ultrasound reports while maintaining a reasonable workload for radiologists has become a pressing clinical issue.
Large language models (LLMs), such as OpenAI’s ChatGPT and Anthropic’s Claude, have shown great potential in addressing challenging clinical problems9,10,11. In recent years, they have demonstrated significant capabilities in generating structured reports12,13,14 and improving report comprehensibility15,16,17,18. More importantly, a recent experimental study has confirmed GPT-4’s potential in detecting errors in radiology reports, suggesting it could enhance report accuracy19. However, this study did not take multi-center data into consideration. And their errors were synthetically generated, hence not representing the real-world error scenarios. Moreover, previous research has indicated that GPT’s performance in non-English environments is not as good as its performance in English settings for certain tasks20,21.
To this end, this study set out to evaluate the capabilities of Claude 3.5 Sonnet, GPT-4o, GPT-4, and GPT-3.5 in zero-shot learning as well as the impact of few-shot learning on model performance. Claude 3.5 Sonnet22 and GPT-4o23 represent the current state-of-the-art multimodal LLMs, demonstrating enhanced performance in non-English languages. To explore the potential of LLMs in improving the accuracy of Chinese ultrasound reports, we collected reports from three hospitals in China, which included both naturally and artificially introduced errors. We also compared the error detection performance of these models with those of six radiologists with varying levels of experience in ultrasound examinations.
Results
Zero-Shot Error Detection Performance
In the zero-shot error detection task for reports, Claude 3.5 Sonnet achieved the best results, with a detection rate of 52.3% (127 of 243). For error detection, the PPV, TPR, and F1 score were 76.5% (95% CI: 69.8%, 83.4%), 52.3% (95% CI: 46.0%, 58.8%), and 62.1% (95% CI: 56.2%, 68.0%), respectively. GPT-4o was the second-best performing model, with a detection rate of 41.2% (100 of 243). For error detection, the PPV, TPR, and F1 score were 88.5% (95% CI: 82.1%, 94.0%), 40.8% (95% CI: 34.0%, 48.0%), and 55.9% (95% CI: 48.8%, 62.3%), respectively. In contrast, GPT-3.5 had a detection rate of only 4.9% (12 out of 243), with PPV, TPR, and F1 scores of 17.9% (95% CI: 8.5%, 28.2%), 5.0% (95% CI: 2.3%, 8.3%), and 7.9% (95% CI: 3.3%, 12.6%), respectively. Additionally, GPT-4 achieved a detection rate of 26.7% (65 of 243), with PPV, TPR, and F1 scores of 84.4% (95% CI: 76.3%, 92.1%), 26.7% (95% CI: 20.9%, 32.8%), and 40.6% (95% CI: 33.6%, 47.4%), respectively (Table 1).
Regarding the negative impact, the false positives generated by GPT-4o and GPT-4 were comparable. For example, the FPRR for GPT-4o was 3.3% (95% CI: 1.5%, 5.3%), compared to 3.0% (95% CI: 1.5%, 4.8%) for GPT-4 but with no statistically significant difference. In contrast, GPT-3.5 generated significantly more false positives, with an FPRR of 13.8% (95% CI: 10.0%, 17.5%). Claude 3.5 Sonnet had an intermediate FPRR of 9.8% (95% CI: 6.8%, 13.3%), which was higher than GPT-4o and GPT-4 but lower than GPT-3.5 (Table 1).
To further elucidate the models’ performance, we conducted a detailed analysis of their behavior across different error types (with 0 representing no error and 1-6 representing six different error types), as illustrated in Fig. 1. Claude 3.5 Sonnet demonstrated superior performance across all error categories. The model was particularly good at identifying contradictory conclusions (error type 2), correctly detecting 40 of 51 such errors, significantly outperforming the other models. It also performed well in detecting item omission errors (error type 1), identifying 23 out of 51 errors. However, this high sensitivity came at the expense of increased false positives, with the model misclassifying 30 non-error reports as contradictory conclusion errors. GPT-4o identified 24 of 51 item omission errors (error type 1), while also producing eight false positives for this error type. It performed well in detecting spelling errors (error type 5), identifying 23 out of 62 cases. GPT-4 correctly identified 18 of 51 contradictory conclusions errors (error type 2) and detected 19 of 46 descriptive errors (error type 3). It also showed moderate performance in identifying spelling errors, detecting 11 out of 62 errors. Compared to the other models, GPT-3.5 had lower detection rates across error types. It only detected 4 out of 51 contradictory conclusion errors (error type 2) and 5 out of 51 item omission errors (error type 1), and failed to identify any spelling errors. When detecting spelling errors (error type 5), Claude 3.5 Sonnet and GPT-4o performed similarly, identifying 22 and 23 out of 62 errors, respectively. This performance was notably better than that of GPT-4 and GPT-3.5 in this category. Regarding other error types, all models showed varying degrees of performance. For content repetitions (error type 4), Claude 3.5 Sonnet detected 10 out of 17 cases, outperforming other models. Other errors (error type 6) were generally less frequently detected across all models.
Specifically, the confusion matrices show the performance of GPT-3.5, GPT-4, GPT-4o and Claude 3.5 Sonnet in detecting specific error types in 400 reports. 0 = Error-free; 1 = Item omission; 2 = Contradictory conclusion; 3 = Descriptive error; 4 = Content repetition; 5 = Spelling error; 6 = Other error.
Few-Shot Error Detection Performance
In the test set analysis, both Claude 3.5 Sonnet and GPT-4o demonstrated higher error detection rates in the few-shot setting compared to the zero-shot setting, though these improvements were not statistically significant. Claude 3.5 Sonnet’s detection rate increased from 44.9% (57/127) to 50.4% (64/127), while GPT-4o’s rate rose from 37.0% (47/127) to 40.9% (52/127).
Claude 3.5 Sonnet exhibited significant improvements in the few-shot setting: PPV increased significantly from 75.0% to 91.4% (P < 0.05), F1 score improved from 56.2% to 65.0%, and TPR increased from 44.9% to 50.4%, though the latter two metrics lacked statistical significance. Notably, its FPRR significantly decreased from 9.5% to 3.0% (P > 0.05). In contrast, GPT-4o showed mixed results in the few-shot setting: F1 score and TPR improved marginally without statistical significance, while PPV decreased significantly from 87.0% to 70.3%. Additionally, FPRR increased from 3.5% to 11.0%, although this increase was not statistically significant.
These findings indicate that while both models showed increased error detection rates in the few-shot setting, Claude 3.5 Sonnet demonstrated more significant and comprehensive performance improvements, particularly in enhancing predictive accuracy and reducing false positives. Conversely, GPT-4o, despite slight improvements in detection rate, experienced a negative impact on overall report quality, showing no substantial advantage over its zero-shot performance. (Table 2; Fig. 2)
The bar charts show the performance evaluation indices of Claude 3.5 Sonnet, GPT-4o, GPT-4 and GPT-3.5 in terms of error detection. Higher values of Positive Predictive Value, True Positive Rate and F1 Score indicate better detection performance of the model, while a higher False Positive Report Rate value suggests poorer detection performance. Error bars represent 95% confidence interval.
Figure 3 shows the results of the subgroup analysis. In the error-type-specific performance evaluation, Claude 3.5 Sonnet demonstrated superior performance in few-shot learning compared to zero-shot learning. It detected significantly more spelling errors (error type 5) (16 vs 8) and descriptive errors (error type 3) (17 vs 13). Notably, false positives for contradictory conclusions (error type 2) decreased substantially from 19 to 4. Conversely, the subgroup analysis of GPT-4o revealed mixed results. Few-shot learning outperformed zero-shot learning in detecting content repetition (error type 4) (6 vs 3) and spelling errors (error type 5) (19 vs 12). However, it demonstrated reduced efficacy in identifying item omission errors (error type 1) (1 vs 11) and generated more false positives for contradictory conclusions (error type 2) (12 vs 0).
Specifically, the confusion matrices show the performance of Claude 3.5 Sonnet and GPT-4o in zero-shot setting and few-shot setting in detecting specific error types in 200 reports. 0 = Error-free; 1 = Item omission; 2 = Contradictory conclusion; 3 = Descriptive error; 4 = Content repetition; 5 = Spelling error; 6 = Other error.
Radiologists study
In the test dataset (n = 127), we compared the performance of Claude 3.5 Sonnet and GPT-4o in both zero-shot and few-shot settings against radiologists of various experience levels. Among the human readers, only one attending radiologist (attending radiologist 2) achieved a higher error detection rate of 54.3% (69/127), slightly surpassing Claude 3.5 Sonnet’s few-shot performance. Two senior radiologists demonstrated error detection rates of 45.7% (58/127) and 41.7% (53/127), respectively, both outperforming GPT-4o’s few-shot performance. The other radiologists did not detect more errors than GPT-4o did, whether in the zero-shot or few-shot setting. The detection rate of attending radiologist 1 was 33.9% (43/127), while that of two resident radiologists was 29.1% (37/127) and 36.2% (46/127), respectively. Detailed performance metrics for both LLMs and radiologists across all experience levels are presented in Table 2. Statistical analysis revealed no significant differences between Claude 3.5 Sonnet in the few-shot setting and the average performance of radiologists across experience levels in terms of error detection and negative impact (all P ≥ 0.05). However, Claude 3.5 Sonnet in the zero-shot setting and GPT-4o in both settings exhibited lower positive predictive values (PPV) compared to radiologists, and lower F1 scores compared to senior and attending radiologists. Furthermore, senior and attending radiologists demonstrated higher average PPV, true positive rates (TPR), and F1 scores compared to residents. Notably, Resident 2 significantly outperformed Resident 1, highlighting considerable individual variability in performance.
Analysis of error detection in ultrasound reports across different physician seniority levels (Senior, Attending, and Resident) revealed complex patterns. All physician groups demonstrated high accuracy in identifying error-free reports (Type 0). However, detection capabilities for specific error types did not show a clear correlation with experience levels. Ranked by overall detection frequency across all physician groups, the most commonly identified errors were Type 3 (descriptive errors), Type 2 (contradictory conclusions), Type 5 (spelling errors), and Type 1 (item omission). Types 4 (content repetition) and 6 (other errors) were detected significantly less frequently than the other four categories. Notably, intra-level individual variability was observed, particularly in the detection of Type 1 (item omission) and Type 5 (spelling errors). These findings suggest that error detection capabilities in ultrasound reporting may be more closely associated with specific error characteristics and individual work habits rather than solely physician experience, highlighting potential areas for targeted training and quality improvement initiatives (Fig. 4).
Inter-observer variability
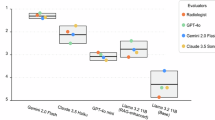
Figure 5 displayed the heatmap of Cohen’s Kappa coefficients among different readers, which measured their agreement in detecting various error types in the reports. The AI models demonstrated high internal consistency, with a correlation of 0.53 between Claude 3.5 Sonnet Few and Zero and 0.47 between GPT-4o Few and Zero. In contrast, agreement between human raters was generally low, with Kappa coefficients between radiology residents, attending radiologists, and senior radiologists all below 0.4, indicating significant differences between observers in reporting error detections. This scoring pattern revealed significant differences in judgments between different raters or systems, which may reflect the inherent complexity and ambiguity of the task of reporting error detection. In particular, the low agreement between human raters highlights the challenging nature of this task. In contrast, the AI system showed potential advantages in providing consistent judgments. Although the intraclass correlation coefficient between the six radiologists showed a moderate degree of agreement (0.45 [95%CI: 0.37, 0.54]), this level of agreement is still suboptimal given the criticality of the task, highlighting the importance of establishing uniform standards in error detection.
The figures quantify the consistency across readers with respect to the various error types in the test reports. -1: Completely inconsistent, 0: Occasional consistency; 0.0 ~ 0.20: Very low consistency; 0.21 ~ 0.40: Fair consistency; 0.41 ~ 0.60: Moderate consistency; 0.61 ~ 0.80: high consistency; 0.81 ~ 1: Almost perfect.
Time analysis
In the task of error detection for 200 reports, AI models demonstrated significant time efficiency advantages. Claude 3.5 Sonnet processed 200 reports in 0.7 and 1.0 hours under zero-shot and few-shot settings, respectively. For the same task, GPT-4o required 0.8 hours in the zero-shot setting and 1.1 hours in the few-shot setting. In contrast, human radiologists’ processing times ranged from 2.3 to 7.5 hours (Fig. 6a). For individual report processing, AI models exhibited even more pronounced speed: Claude 3.5 Sonnet required only 13.2 seconds per report in the zero-shot setting, while GPT-4o needed 15.0 seconds. The fastest human radiologist required 42.0 seconds per report (Fig. 6b). In the few-shot setting, AI models showed a slight increase in processing time but remained substantially faster than human experts.
Discussion
This study represents the first systematic evaluation of LLMs in Chinese ultrasound report quality assurance, providing innovative perspectives and methodologies for digital healthcare quality control. Our findings demonstrate the significant potential of LLMs, particularly Claude 3.5 Sonnet and GPT-4o, in detecting errors in ultrasound reports, which has important clinical implications for improving report accuracy and optimizing patient management.
While previous research by Gertz et al.19 explored GPT-4’s potential in detecting errors in radiology reports using synthetically generated errors across radiography, CT, and MRI, our study extends this work in several crucial ways. First, we specifically focus on ultrasound reports, an area that has not been explored previously in this context. Second, we collected reports from real clinical settings across three hospitals, covering a variety of anatomical regions and pathologies, including a variety of error types, to exclude misdiagnoses from images. This approach enhances the practical relevance of our findings and better reflects the complexity of real clinical settings. More importantly, our study is the first to explore the application of LLMs for error detection in Chinese ultrasound reports, which provides valuable insights for evaluating the performance of these models in non-English settings.
In addition, our study introduces a few-shot learning setting, providing examples for the models to potentially enhance their error detection capabilities. This approach yields interesting results, with Claude 3.5 Sonnet showing significant improvements in the few-shot setting, outperforming most radiologists on multiple metrics. In contrast, GPT-4o showed higher error detection rates, but also higher false positive rates, highlighting the subtle effects of few-shot learning on different models.
These findings not only confirm the potential of LLMs for medical report error detection shown in previous studies, but also reveal new insights into their performance in different learning paradigms and language environments. The outstanding performance of Claude 3.5 Sonnet in the few-shot setting and the ability of GPT-4o to surpass human performance in detecting spelling errors highlight the significant advances in the capabilities of LLMs. However, the increased false positive rate observed for GPT-4o in the few-shot setting also highlights the need for careful consideration when applying these models in clinical practice, especially in non-English settings. This exploration in the Chinese context provides an important reference for the future application of LLMs for medical report quality control in diverse language and cultural contexts.
In comparing the performance of LLMs with human experts, we observed significant advantages and potential limitations. The primary strengths of LLMs lie in their efficiency and consistency in processing large volumes of reports. Specifically, Claude 3.5 Sonnet and GPT-4o completed analysis in less than 20 seconds per report on average, whereas the best-performing radiologist required 135.6 seconds. Moreover, LLMs excelled in detecting spelling and logical errors, sometimes surpassing human experts. This efficiency position LLMs as potentially powerful tools for preliminary screening, augmenting the overall workflow efficiency of healthcare professionals. However, LLMs also exhibit a few key limitations. Firstly, they generate higher false-positive rates, possibly due to their inability to accurately interpret the correspondence between ultrasound terminology and findings. For instance, GPT-4o failed to correctly interpret the relationship between “anechoic nodule” and “cyst”, and between “fine liver echo” and “fatty liver” (Supplementary Fig. 1). And the occurrence of false positives will increase the workload of radiologists, leading to a decrease in efficiency. Secondly, the models lack the rich clinical experience and contextual understanding possessed by human experts. This was evident in a case with a logical error where the ultrasound description stated, “Scattered strong echoes seen in the right kidney, with the largest diameter about 6 mm; a 10*10 mm anechoic area seen in the left kidney,” while the conclusion read, “Left kidney stone; right kidney cyst.” All models failed to identify this error, whereas among the physicians, only one resident missed it.
Crucially, errors undetected by LLMs could lead to severe consequences, such as major localization errors potentially resulting in inappropriate treatment decisions24. Given these potential risks, we emphasize that LLMs should be viewed as auxiliary tools. It is imperative to establish rigorous quality control mechanisms, regularly evaluate and update LLM performance, and provide continuous training for healthcare professionals using these tools.
Although this study is the first to investigate the role of LLMs in detecting errors in ultrasound reports, it also has shortcomings. First, the average error detection rate of all six radiologists was 40.3% which appeared to be lower than that reported by Gerz et al.19. This could be attributed to visual fatigue and decreased attention from prolonged reading of extensive text sections. This challenge in long-term text interpretation can potentially affect their efficiency and accuracy. It may also be related to the fact that the reports came from different hospitals, and there were differences in the structured standard report templates of each hospital. Additionally, while LLMs reached levels comparable to radiologists in some cases, its performance in Chinese settings was not as outstanding as in English settings, which is consistent with model’s performance in other non-English environments25,26. Furthermore, while our study included state-of-the-art models like GPT versions and Claude 3.5 Sonnet, it did not encompass the full range of available large language models. Although no significant differences were observed in LLMs’ performance between PDF and plain text formats, direct text input is recommended for clinical applications to avoid potential PDF parsing issues. Finally, our study focused on logical and spelling errors, rather than misdiagnoses and missed diagnoses caused by misinterpretation of the ultrasound images. Detecting image errors is crucial for diagnostic accuracy, as both diagnostic conclusion errors and feature description errors significantly affect report accuracy27. We tested a sample image, but the results were unsatisfactory. Therefore, we have reasons to believe that the current model will require further extension to detect errors in image interpretation.
This multi-center study provided the first systematic evaluation of LLMs for error detection in ultrasonography reports. In few-shot learning settings, LLMs such as Claude 3.5 Sonnet demonstrated superior error detection capabilities compared to most radiologists, confirming their potential as adjunctive tools in radiological workflows. However, the discrepancy in LLM performance between non-English and English environments underscores the challenges in cross-lingual applications. Future research should focus on optimizing the use of LLMs in multilingual medical contexts, particularly in enhancing their understanding of complex medical concepts, while exploring their synergistic potential with medical image analysis. These advancements promise not only to improve the quality of ultrasonography reports but also to pioneer new avenues for AI-assisted quality control in healthcare.
Building on these findings, we have identified several specific strategies for future research. We hypothesize that a more promising approach might involve specialized training of one or multiple models to better address specific challenges. For instance, we are exploring strategies such as 1) Employing segmentation and classification models for feature extraction, and 2) Utilizing LLMs for feature analysis and result interpretation. Furthermore, we are actively investigating the potential of LLMs to address the interpretability challenges inherent in existing deep-learning models. Additionally, future research should focus on optimizing LLMs for multilingual medical contexts and exploring synergies with medical image analysis to improve their understanding of complex medical concepts in diverse linguistic contexts.
Methods
This retrospective study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Review Board of Zhejiang Cancer Hospital (IRB-2024-494), the Ethics Review Board of Dongyang People’s Hospital (IRB-2024-097) and the Ethics Review Board of Taizhou Cancer Hospital (IRB-2024-049). Due to its retrospective design, informed consent has been waived. None of the patient identification information was provided to GPT-3.5, GPT-4, GPT-4o, or Claude 3.5 Sonnet.
Data Set and Error Categories
To increase the diversity of the dataset, 300 error-free ultrasound diagnostic reports that passed quality control and 100 erroneous reports that did not pass quality control were collected from three hospitals: Zhejiang Cancer Hospital (ZJCH), Dongyang People’s Hospital (DPH), and Taizhou Cancer Hospital (TZCH). Table 3 shows the baseline characteristics of the study sample. These reports cover a wide range of pathological abnormalities across various regions, including superficial, abdominal, and pelvic areas. Out of the 300 error-free reports in our dataset, an algorithm randomly selected 100 reports for error insertion, then human experts in ultrasound diagnostics were then tasked with inserting errors into these 100 selected reports. The experts randomly determined the types of errors to be introduced and their locations within each report. Consequently, the dataset now consists of 200 reports without any errors, 100 reports with real-world errors, and 100 reports with artificially inserted errors. These reports were further divided into non-test and testing datasets with 200 cases in each group. The test dataset comprises of 100 correct reports, 50 with real-world errors and 50 with artificial errors. Each report included 1 to 3 errors. From the non-test set reports, seven reports, with six of them representing each of the six error types and one representing an error-free report, were randomly selected to serve as few-shot examples for the best model (Fig. 7a).
a Description of the datasets. b Stage one involved collecting a total of 400 US reports, including both error-free and erroneous reports, from three hospitals. These reports were used to evaluate the performance of different models in detecting errors in ultrasound reports under a zero-shot setting, with performance assessed at both the error and report levels. The two best-performing models were selected to proceed to the next stage. In stage two, a test set consisting of 200 reports (50% error-free and 50% erroneous) was randomly selected to test the error detection performance of the two best models in a few-shot setting, thereby assessing the model's rapid learning capability. In stage three, the test set was used to evaluate the error detection performance of six radiologists with varying levels of experience, providing a comparison with the model's performance. ZJCH= Zhejiang Cancer Hospital, DPH= Dongyang People's Hospital, TZCH= Taizhou Cancer Hospital, PPV = Positive Predictive Value, TPR = True Positive Rate, FPRR = False Positive Report Rate.
The gold standards for error detection were established through a rigorous three-phase process (Fig. 8). First, true errors were identified based on the Chinese 2022 Ultrasonography Quality Control Guidelines28, with reports classified as non-compliant if they lacked required examination content, had discrepancies between descriptions and conclusions, or contained clear errors such as organ misidentification or orientation mistakes. Senior physicians with over 15 years of experience reviewed and categorized these errors, ensuring consistency through expert panel discussions. Second, artificial errors were introduced into 100 randomly selected reports from a pool of 300 high-quality, error-free reports. The types and locations of these errors were determined by the expert panel, with the distribution aligned to reflect real-world error frequencies while ensuring experimental controllability. Finally, a total of 400 reports—comprising true errors, artificially inserted errors, and error-free reports—underwent a final review by three senior radiologists, who verified that each report contained only the intended errors. This comprehensive process ensured that the gold standards accurately reflected both real and artificial errors, providing a reliable benchmark for error detection. The list of the three-member final review panel and the senior radiologists’ expert panel responsible for routine ultrasound report quality control, hence establishing the gold-standards for the real-world errors, along with their years of experience, can be found in Supplementary Note 1. Furthermore, supplementary Fig. 2 depicts the distribution of the six error types for both real and artificially errors.
Identification of True Errors: True errors were determined according to the quality control standards outlined in the Chinese 2022 Ultrasonography Quality Control Guidelines, which define non-compliant reports as those that fail to include requested examination content, have discrepancies between descriptions and conclusions, or contain clear errors (e.g., missing organs described as normal, orientation errors, incorrect units or data, or unedited template text). Senior radiologists with over 15 years of experience conducted detailed reviews of ultrasonography reports to identify errors based on these criteria. Errors were further categorized and confirmed through collective discussions among an expert panel to ensure consistency and accuracy. Creation of Artificially Inserted Errors: Artificial errors were introduced into 100 reports selected randomly from a pool of 300 high-quality reports that passed routine quality checks. Reports were randomized using Excel’s random number generator, and the types and locations of errors were determined by an expert panel. Artificial errors were distributed based on the observed frequency of error types in true error samples and adjusted for controllability. Final Review Process: A total of 400 reports, including true and artificially inserted errors, underwent a rigorous final review by three senior radiologists with extensive diagnostic experience. This review involved in-depth discussions to confirm that each report contained only the intended errors without additional, unintended ones.
Based on the severity of errors, the categories include: 1. Item omission: Missing ultrasound descriptions for examination items listed on the ultrasound prescription form. 2. Contradictory conclusion: Discrepancies between ultrasound descriptions and the conclusions provided, including incorrect descriptions of the examined organ, location, and disease orientation (e.g. left/right, up/down, front/back). 3. Descriptive error: Errors in the units or data associated with the examined organ, location, or disease in the report, such as excessively high values. 4. Content repetition: Failure to remove ambiguous template text from the ultrasound report, resulting in irrelevant or duplicated content. 5. Spelling error: Chinese spelling errors may occur when radiologists quickly type using pinyin input methods (e.g., “回声“ misspelt as “会声“, “集合“ misspelt as “几何“). 6. Other error: errors that did not fit into the above categories, including incorrect use of punctuation marks. Supplementary Fig. 3c shows examples of various errors.
Study Design
This study was conducted in three stages to evaluate the capabilities of different LLMs and radiologists in detecting errors in ultrasound reports (Fig. 7b).
Stage one, using the full set of reports (400 cases), the capabilities of Claude 3.5 sonnet, GPT-4o, GPT-4, and GPT-3.5 were assessed in a zero-shot setting to detect errors in ultrasound reports, with the aim of determining the two best-performing models.
Stage two, a test set (200 cases) was composed by randomly selecting 100 correct reports, 50 real-error subgroup reports, and 50 artificial-error subgroup reports. This phase explores explored whether the error detection capabilities of the optimal model improve in a few-shot setting.
Stage three, employing the same test set as Stage Two, the error detection performance and time taken by radiologists of different experience levels (senior radiologists, attending radiologists, and residents) were assessed via a customized online survey platform (https://www.wjx.cn/).
Performance Evaluation
Six radiologists with varying levels of experience in ultrasound examination, including two senior radiologists (L.Z. with 18 years and K.W. with 15 years), two attending radiologists (Y.D. with ten years and J.Y. with five years), and two residents (T.J. with two years and Y.Z. with one year) were tasked to detect errors in 200 cases. Each radiologist independently evaluated each ultrasound report to identify potential errors using the customized online survey platform. We also measured the total time taken by each radiologist to review the test set on this platform.
To investigate the ability of four LLMs in a zero-shot setting, we needed to modify the format of the ultrasound reports based on the model requirements: text format for GPT-3.529 and PDF format for GPT-430, GPT-4o23 and Claude 3.5 sonnet. Readers are referred to the supplementary materials for Chinese (Supplementary Fig. 3a) and English ultrasound report templates (Supplementary Fig. 3b).
The prompts for all four releases were the same and are shown as follows: “In the following content, I will provide you with an ultrasound report divided into sections: ‘Examination Items,’ ‘Ultrasound Description,’ and ‘Ultrasound Indications.’ Please identify any of the following errors if present:
-
1.
Missing examination items compared to the prescription, which is also listed at the top of the ultrasound report.
-
2.
Inconsistencies between the ultrasound description and the ultrasound indications, such as discrepancies in lesion location (left/right, up/down, front/back).
-
3.
Errors in units or texts describing organs and diseases.
-
4.
Irrelevant or duplicated template text that has not been removed.
-
5.
Spelling error.
-
6.
Any other error.”
To assess the performance of the two best models in few-shot learning scenarios, the following prompt was provided to the models: ‘I am going to provide you with seven example reports: one error-free report and six reports containing the various categories of errors listed. The sole purpose of these example reports is to improve your comprehension and to help you recognize the various error categories mentioned in the subsequent tasks.’ We then entered each report and its corresponding error description and categorization in order. For example, one example report contained ‘contradictory findings’, specifically, the ultrasound description showed a hypoechoic nodule in the left breast, whereas the ultrasound impression described a cyst in the right breast. At the end of the learning phase containing all 7 example reports, we entered ‘provided example reports completed’ to indicate the end of the examples. We then followed the prompts mentioned in the zero-shot setting to begin error detection on the test set reports.
The time required for Claude 3.5 sonnet, GPT-4o, GPT-4, and GPT-3.5 to correct each ultrasound report was evaluated by measuring the duration from submitting the prompt to receiving the final response. For each model, this assessment was conducted on a randomly selected sample of 20 ultrasound reports of varying lengths.
Statistical Analysis
All analyses were performed using R software (version 4.2.3). The performance of GPT models and radiologists in error detection was evaluated using Positive Predictive Value (PPV), True Positive Rate (TPR), and F1 Score. The negative impact of false positives on the overall performance of GPT models and radiologists in report evaluation was investigated using the False Positive Report Rate (FPRR). Bootstrap methods were utilized to generate 95% confidence intervals (CIs).
To compare performance metrics between radiologists and LLMs under both zero- and few-setting, the Wald χ² test was used to analyze differences in error detection performance (PPV, TPR, F1 Score) and the impact of false positives (FPRR). Bonferroni correction was applied to adjust for multiple comparisons, and a two-sided P value of less than 0.05 was considered statistically significant. Cohen’s kappa coefficient was used to evaluate the consistency between the model and each individual radiologist, while the Intraclass Correlation Coefficient was employed to assess the consistency among radiologists. The definitions of each of the metrics are provided in Supplementary Note 2.
Data availability
Individual participant data can be made available upon request, directed to corresponding author (D.X.). Once approved by the Institutional Review Board/Ethics Committee of all participating hospitals, deidentified data can be made available through a secured online file transfer system for research purpose only.
Code availability
All pre- and post-processing code employed in this study were standard code which can be accessed via Microsoft Excel and statistical software R. No customized code was written for this work.
References
Zeng, A. et al. Development of Quality Indicators for the Ultrasound Department through a Modified Delphi Method. Diagnostics 13, 3678 (2023).
G. E. Healthcare. Technology Solutions Can Help Achieve Accurate Ultrasound Reports. https://www.gehealthcare.com/insights/article/technology-solutions-can-help-achieve-accurate-ultrasound-reports (2023).
Meng, F., Zhan, L., Liu, S. & Zhang, H. The Growing Problem of Radiologist Shortage: China’s Perspective. Korean J. Radio. 24, 1046 (2023).
Jeganathan, S. The Growing Problem of Radiologist Shortages: Australia and New Zealand’s Perspective. Korean J. Radio. 24, 1043 (2023).
Do, K.-H., Beck, K. S. & Lee, J. M. The Growing Problem of Radiologist Shortages: Korean Perspective. Korean J. Radio. 24, 1173 (2023).
Stephenson, J. US Surgeon General Sounds Alarm on Health Worker Burnout. JAMA Health Forum 3, e222299 (2022).
Geijer, H. & Geijer, M. Added value of double reading in diagnostic radiology,a systematic review. Insights Imaging 9, 287–301 (2018).
Tao, X. et al. A National Quality Improvement Program on Ultrasound Department in China: A Controlled Cohort Study of 1297 Public Hospitals. IJERPH 20, 397 (2022).
Han, T. et al. Comparative Analysis of Multimodal Large Language Model Performance on Clinical Vignette Questions. JAMA 331, 1320 (2024).
Li, J. et al. Integrated image-based deep learning and language models for primary diabetes care. Nat Med. https://doi.org/10.1038/s41591-024-03139-8 (2024).
Kresevic, S. et al. Optimization of hepatological clinical guidelines interpretation by large language models: a retrieval augmented generation-based framework. npj Digit. Med. 7, 1–9 (2024).
Huang, J. et al. A critical assessment of using ChatGPT for extracting structured data from clinical notes. npj Digit. Med. 7, 106 (2024).
Adams, L. C. et al. Leveraging GPT-4 for Post Hoc Transformation of Free-text Radiology Reports into Structured Reporting: A Multilingual Feasibility Study. Radiology 307, e230725 (2023).
Hasani, A. M. et al. Evaluating the performance of Generative Pre-trained Transformer-4 (GPT-4) in standardizing radiology reports. Eur Radiol https://doi.org/10.1007/s00330-023-10384-x (2023).
Doshi, R. et al. Quantitative Evaluation of Large Language Models to Streamline Radiology Report Impressions: A Multimodal Retrospective Analysis. Radiology 310, e231593 (2024).
Jeblick, K. et al. ChatGPT makes medicine easy to swallow: an exploratory case study on simplified radiology reports. Eur Radiol https://doi.org/10.1007/s00330-023-10213-1 (2023).
Lyu, Q. et al. Translating radiology reports into plain language using ChatGPT and GPT-4 with prompt learning: results, limitations, and potential. Vis. Comput. Ind. Biomed. Art. 6, 9 (2023).
Ali, R. et al. Bridging the literacy gap for surgical consents: an AI-human expert collaborative approach. npj Digit. Med. 7, 63 (2024).
Gertz, R. J. et al. Potential of GPT-4 for Detecting Errors in Radiology Reports: Implications for Reporting Accuracy. Radiology 311, e232714 (2024).
Cozzi, A. et al. BI-RADS Category Assignments by GPT-3.5, GPT-4, and Google Bard: A Multilanguage Study. Radiology 311, e232133 (2024).
Lai, V. D. et al. ChatGPT Beyond Engl.: Towards a Compr. Evaluation Large Lang. Models Multiling. Learn. https://doi.org/10.48550/arXiv.2304.05613 (2023).
Anthropic. Claude 3.5 Sonnet. https://www.anthropic.com/news/claude-3-5-sonnet (2024).
OpenAI. Hello GPT-4o. https://openai.com/index/hello-gpt-4o (2024).
OpenAI. GPT-3.5. https://chatgpt.com (2021).
OpenAI. GPT-4. https://openai.com/gpt-4 (2023).
Benjamens, S., Dhunnoo, P. & Meskó, B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. npj Digit. Med. 3, 118 (2020).
Jiao, W. et al. Is ChatGPT A Good Translator? Yes With GPT-4 As The Engine. https://doi.org/10.48550/arXiv.2301.08745 (2023).
Department of Medical Administration and Medical Management. Notice of the General Office of the National Health Commission on Issuing the Medical Quality Control Indicators for Five Specialized Fields, including Ultrasound Diagnosis (2022 Edition). http://www.nhc.gov.cn/yzygj/s7657/202205/56765f0f512f4f058efc4169a0e1c639.shtml.
Kao, Y.-S., Chuang, W.-K. & Yang, J. Use of ChatGPT on Taiwan’s Examination for Medical Doctors. Ann. Biomed. Eng. 52, 455–457 (2024).
Zhang, L. et al. Diagnostic error and bias in the department of radiology: a pictorial essay. Insights Imaging 14, 163 (2023).
Acknowledgements
This work was supported by the Pioneer and Leading Goose R&D Program of Zhejiang (2023C04039). The funder had no roles in study design, data collection and analysis, publication decisions, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
Y.Y., K.W. and B.F. conceived the study. B.F., Y.Z., L.S. and X.C. performed the literature search and data analysis. T.J., Z.J., Y.Z., C.C., Y.D., J.Y., Q.P., L.Z. designed the clinical study. All authors critically read and reviewed the manuscript. Y.Y., V.Y.W. and J.Y. were major contributors in writing the manuscript. Y.Y., K.W. and B.F. contributed equally in this study. L.Z., V.Y.W., P.L. and D.X. are co-corresponding authors and contributed equally.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, Y., Wang, K., Feng, B. et al. The use of large language models in detecting Chinese ultrasound report errors. npj Digit. Med. 8, 66 (2025). https://doi.org/10.1038/s41746-025-01468-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-025-01468-7
This article is cited by
-
Exploring the use of large language models for classification, clinical interpretation, and treatment recommendation in breast tumor patient records
Scientific Reports (2025)
-
An Exploratory Comparison of AI Models for Preoperative Anesthesia Planning: Assessing ChatGPT-4o, Claude 3.5 Sonnet, and ChatGPT-o1 in Clinical Scenario Analysis
Journal of Medical Systems (2025)