Abstract
In (immune)oncology, virtual twins (VTs) offer patient-individual decision support. Nevertheless, current VTs do not incorporate the unique properties of engineered adoptive cellular immunotherapies (eACIs). Here, we outline the minimal design specifications for VTs for engineered ACIs (eACI-VTs) to model the complex interplay between cell product and patient physiology. We motivate utilizing VTs in eACIs to provide decision support and reflect on how eACI-VTs can support the widespread use of eACIs.
Similar content being viewed by others
Introduction
Adoptive cellular immunotherapy (ACI) is a novel therapy with the potential to revolutionize the treatment of cancer and other diseases1. Prominent examples include engineered ACIs (eACIs), such as chimeric antigen receptor (CAR) T cells and T cell receptor (TCR) engineered T cells, as well as non-eACI-like tumor infiltrating lymphocytes2, with CAR T cells being the most prevalent eACI in current clinical practice. CAR T cell therapy equips a patient’s own (autologous) or a healthy donor’s (allogenic) T cells with CARs, enabling them to recognize a defined target antigen on the surface of tumor cells3. Upon encountering tumor cells, CAR T cells become activated and are thereby enabled to kill the target cells. Six autologous CAR-based therapies are approved by the European Medicines Agency (EMA), and seven by the United States Food and Drug Administration (FDA), for hematological malignancies. With CAR T cells, an unprecedented proportion of patients experience long-lasting remission or even cure, with manageable adverse events4,5. However, not all patients respond to treatment5. Predicting which patients will benefit from eACI is crucial due to outcome uncertainty, high therapy costs, and limited manufacturing capacities, because the cell product is manufactured individually through a complex, multi-step process.
In healthcare, digital twins (DTs) and virtual twins (VTs) are computer-based models that digitally represent interacting biological systems across multiple scales. These models support monitoring disease prevention, diagnosis, treatment decision-making, and follow-up care, while also assisting both clinical and nonclinical research, thus accelerating the development of new medicines and medical devices. Although DTs/VTs are still in their early stages of development, their potential to transform precision medicine through individualized care is increasingly recognized6,7,8,9.
Definitions of the term DT vary in the literature and often lack a clear distinction from VTs. Viceconti et al. define a DT in healthcare as an application-specific virtual representation of a single organ of an individual patient, intended to guide patient-specific decisions and requiring integration with the patient’s personal data at least once during its life cycle10. Specifically, a digital representation of a real-world object qualifies as a DT if it includes: (i) a computational model of the object, (ii) a dataset describing changes in the object, and (iii) methods for continuously updating the computational model with data derived from its real-world counterpart11. Importantly, a DT is expected to evolve in parallel with its real-world counterpart. In healthcare, this real-world object may represent a patient, a clinical study participant (e.g., from a control arm), or any biological system, such as individual cells or organs. These definitions for DTs explicitly exclude population-based models because they lack continuous updates based on patient-specific data10,11. Expanding upon this, we define a VT in healthcare as an application-specific in silico system covering at least two single-organ DTs from the same patient to simulate multi-organ biomedical interplay. As such, VTs offer the complexity of different biological scales, dynamic adaptability, and different organs to guide treatment decisions. This makes VTs particularly well-suited for use in patients eligible for eACIs. Unlike conventional drugs, eACIs require modeling the medicinal product not only as a dynamic biological system of living cells but also in terms of its interplay with multiple organs of the patient. To motivate the development of VTs for patients eligible for eACIs (eACI-VTs), we propose minimum design specifications for such models. We use autologous CAR T cell therapy as a representative example, given its status as the most widely approved eACI. The main principle, however, readily extends to all classes of eACIs, such as allogenic CAR T cells, CAR NK cells, CAR macrophages, and TCR-engineered T cells, and can also be adapted to accommodate the distinct features of non-eACIs.
Engineered adoptive cellular immunotherapies
Current limitations for a broader application of engineered adoptive cellular immunotherapies
In 2017, Tisagenlecleucel (Kymriah®) was approved by the FDA as a first-in-class therapy, and in 2018 by the EMA, as a third-line treatment for acute lymphoblastic leukemia (ALL), follicular lymphoma, and diffuse large B-cell lymphoma (DLBCL). Since then, five additional products have entered the market in the European Union (EU) and six in the United States. These therapies target either Cluster of Differentiation 19 (CD19) in BCL and B-cell precursor ALL, or B-cell maturation antigen (BCMA) in multiple myeloma (MM).
Numerous studies aim to improve CAR T cell therapies (Box 1). Due to the unprecedentedly high therapy response rates in heavily pre-treated patients with hematological malignancies, efforts are underway to expand this therapy to earlier lines of therapy and additional indications, including solid cancers and beyond, particularly autoimmune and infectious diseases12. These advances coincide with a substantial increase in the number of treatment-eligible patients. Nevertheless, three main challenges already limit the availability of CAR immunotherapies: (i) the high cost of this treatment13, (ii) limited manufacturing capacity3, and (iii) the need for optimal patient stratification considering efficacy and safety14,15. It remains uncertain how significantly treatment costs will actually decrease with manufacturing automation, point-of-care production, or off-the-shelf allogenic cell products3, especially given the complex reimbursement landscape. Nevertheless, overcoming manufacturing limitations and standardizing production, along with improving treatment success, will critically depend on the development of individualized decision-support tools16,17,18. Specifically, patient-specific predictive models can provide objective criteria for the optimal timing of eACI therapy, helping to address critical manufacturing constraints through the timely allocation of manufacturing resources.
Personalized medicine approaches for engineered adoptive cellular immunotherapies
Current personalized medicine approaches for eACIs lack patient-specific modeling approaches. Available predictive models that support clinicians to assess individual risks and outcomes of CAR T cell patients are population-based models, as they do not continuously update model parameters with each patient’s individual data. For example, the CAR-HEMATOTOX Score enables risk assessment for hematologic toxicity, severe infection, and disease progression following anti-CD19 CAR T cell therapy in refractory/relapsed large B-cell lymphoma (R/R LBCL)17,18, and also holds prognostic value for response and toxicity in MM patients treated with BCMA-directed CAR T cells19. Additionally, the Endothelial Activation and Stress Index (EASIX) can be applied to R/R LBCL patients receiving anti-CD19 CAR T cell therapy to predict several CAR T-related toxicities20. In relapsed/refractory MM, the Myeloma CAR T Relapse (MyCARe) model provides an outcome prediction model for anti-BCMA CAR T cell therapy16. As these models are trained and validated for a population and lack periodic updates with individual patient data, they do not qualify as VTs. Nevertheless, as autologous CAR T cell therapies are patient-specific and biologically complex, an elaborate VT model that represents an individual’s (patho)biology, the molecular and cellular characteristics of the retrieved T cells, and the properties of the resulting CAR T cell product would be a favorable solution for improving personalized treatment planning.
Minimum design specification for virtual twins in engineered adoptive cellular immunotherapies
Virtual twins for patients eligible for engineered adoptive cellular immunotherapies
While the first DTs dedicated to CAR T cell manufacturing processes are under development3,21, no VT currently encompasses decision-making tasks across the entire patient journey. Tang et al. propose a five-level roadmap for human body DTs22. However, from level three onward, the design specifications only consider perturbations of the human system by conventional drugs, making them too simplistic for eACIs (Fig. 1). With the increasing application of eACIs, their fundamental differences from conventional drugs, and the requirement that VTs should be deliberately designed for specific contexts of use, it is vital to establish the minimally required design specifications for eACI-VTs. Following the definition of VT components23, we outline these specifications for eACIs.
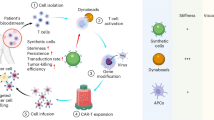
Upon a newly diagnosed disease or relapse, clinicians select the treatment option that is most likely to benefit the patient at that point in time. In the case of conventional drug treatment (left panel), all patients receive an identical drug that is pre-manufactured and precisely defined. By contrast, autologous engineered adoptive cellular immunotherapies (autologous eACIs) are manufactured individually from the patient’s own immune cells after the patient is found eligible for this option (middle panel). During manufacturing time, the patient receives a bridging therapy. Allogenic eACIs (right panel) are pre-manufactured, off-the-shelf products derived from healthy donors’ immune cells. Still, the products are complex, dynamic, biological systems that may be characterized well, but cannot be defined as precisely as conventional drugs. In both types of eACI treatment, the unique and complex cell product interacts with the patient, leading to dynamic, patient-specific interactions along various biological scales and targeted cells. This warrants that approaches towards virtual twins (VTs) for eACI-eligible patients comprise in silico models of multiple biological scales reflecting (CAR) immune cells before and after manufacturing, as well as models of the immune system and targeted cells throughout therapy. The underlying principle applies to all eACI classes and not only to chimeric antigen receptor (CAR) T cell therapies. The figure was created by the authors using Canva.com.
Minimum data categories required in observations of the real-world instance
The efficacy and toxicity of engineered T cells are influenced by factors spanning multiple biological scales14,15. To accurately model eACI-introduced changes in patient biology, it is essential to collect not only longitudinal high-level laboratory and clinical values, as is done for conventional drugs, but also longitudinal data on multiscale processes in (CAR) T cells and patient organs.
Therefore, we postulate three minimal required observation categories for an eACI-VT. First, longitudinal multiomics at the single-cell level are needed to measure intra- and intercellular processes influencing, for example, T cell activation, expansion, exhaustion, genotoxicity, on-target/off-tumor binding, immunosuppressive environment, or imbalances in (CAR) T cell clones24,25. In this context, the rapid advancement and growing adoption of (single-cell) multiomics technologies are highly promising26,27. Without such molecular and cellular data, eACI-VTs cannot infer decisions related to (CAR) T cells and their target cells. Second, longitudinal observations along the organ and body scale, such as CAR T cell expansion and persistence, response to treatment, comorbidities, and side effects, are required. They can be assessed through laboratory data, electronic health records (EHRs), imaging technologies, and sensors. For example, flow cytometry is routinely used to track CAR T cell numbers in peripheral blood, while medical imaging is valuable for monitoring tumor volume during CAR T cell therapy28, including extramedullary and minimal residual disease in MM29. Third, integration of patient-reported outcomes in combination with socio-economic factors like gender, income, education, and geographic location enhances eACI-VT simulations and validation, prevents bias, and increases predictive accuracy. This results in better-informed decisions and personalized treatment plans that reflect real-world diversity and individual health complexities.
As VTs are patient-specific in silico models aiming to accurately represent the patient under real-world conditions, they benefit from integrating these data categories as real-world data (RWD). For this, international standards for data models (Box 2) and data sharing across healthcare and research, including genomic and single-cell multiomics data, must be followed30,31. Importantly, clinical disease development is influenced not only by biological mechanisms, but also by medical decisions (e.g., prior lines of therapy, bridging therapy or follow-up treatment)32,33. Integrating longitudinal data that reflects this complexity is crucial. International and national patient registries for eACIs-eligible individuals are valuable sources of patient-specific RWD prior and during therapy. They allow the systematic connection of multiscale information, such as lab test results, patient phenotype, treatment history, clinical decision, treatment efficacy and safety, and (long-term) patient outcomes. Transparent access policies and the ability to interact with one another are a prerequisite for seamless integration of data from registries.
Minimum design specifications for patient-specific models of the virtual representation
For a VT to support clinical decisions during eACI, it must provide patient-specific model predictions or simulations for all treatment phases (Fig. 1). This includes initial eligibility, leukapheresis, bridging therapy, CAR T cell manufacturing, infusion, and long-term follow-up. Furthermore, the virtual representation of an eACI-VT must combine in silico models across biological scales relevant to the specific context of use (Fig. 2). Below, we highlight example models suitable for eACI-VTs.
CAR T cells are complex, patient-specific (autologous) or donor-specific (allogeneic) therapies. Therefore, it is beneficial to model a patient’s unique (patho)biology alongside the molecular and cellular characteristics of the retrieved T cells and the resulting CAR T cell product. There are four levels of context of uses of an eACI-VT (outer layer): (i) characterizing the patients’/donors’ status prior, the patients’ status during and after therapy, (ii) characterizing the cells during leukapheresis, manufacturing, and in the final cell product, (iii) characterizing the changes of the cell product due the interaction with the patient, and (iv) characterizing the changes of the target cells due to interaction with the CAR T cells. Therefore, the minimal set of in silico models required in an eACI-VT encompass multiple biological scales (middle layer): models for the whole body and for organs reflecting the system-wide status prior to treatment and the impact of treatment, tissue scale, and intercellular scale models for CAR T cell interaction with target cells and the target cells’ tissue during therapy, and cellular scale models that represent intracellular signaling of T cells at time of leukapheresis, (CAR) T cells during manufacturing, the CAR T cells in the medicinal cell product and their target cells. Appropriate in silico model categories that can be used to model events at the different biological scales include (inner layer) system biology models, knowledge-driven models, mechanistic cell models, stochastic models, statistical models (SL), machine learning (ML), and artificial intelligence (AI), ordinary or partial differential equations (ODEs, PDEs), as well as computational structural biology (3D protein structure and 3D/2D RNA structure models). These in silico models receive data generated on different devices and from various systems throughout patient care (central circle). In inpatient care, data from hospital information systems like the electronic health record, as well as lab data and molecular data, fuel the models. Outpatient care provides data via wearables, digital health devices, and the Internet of Things (IoT). During manufacturing, data are supplied via IoT and Cyber-Physical Systems (CPS). The figure was created by the authors using Canva.com.
Models for intracellular signaling in (CAR) T cells and their target cells
Ma and Gurkan-Cavusoglu34 compare different computational methods to model intracellular signaling and provide guidance on selecting the appropriate model for a specific task. Models of intracellular signaling of (engineered) T cells typically rely on existing knowledge of biochemical reaction mechanisms and are often built using continuous and discrete approaches35,36,37. Single-cell multiomics data supports the inference of gene regulation networks38. Recently, the advancement of large language models in natural language processing has led to their application in genomics and single-cell studies. The first pre-trained language models for intracellular biology are available. For instance, scBERT39 supports cell-type annotation, and scGPT40 is the first foundation model covering diverse tasks like cell-type annotation, multi-batch/multiomic integration, perturbation response prediction, and gene network inference. Modeling intracellular biomolecular networks in the context of CAR T cell therapy supports simulation of subcellular processes related to treatment responses by linking a patient’s genotype to their phenotype. A favorable functional status of immune cells is crucial for successful eACI response41. Models of this kind could thereby help to use the patient’s individual immune cell status and the status of individual subcellular factors responsible for long-term remission42. These simulations could support predictions of manufacturing success43 or therapeutic response25,41 (Fig. 2). Generative single-cell AI models are also emerging, enabling the creation of patient-specific in silico cells44 that may facilitate modeling of immune cell status in in silico clinical trials. Lastly, cellular modeling plays a growing role in DT technologies applications to enhance CAR T cell manufacturing processes45.
Models for intercellular signaling of CAR T cells and their target cells
Numerous models focus on cell–cell interactions without the details of molecular interactions, investigating the conditions for optimal CAR T treatment responses36,46,47. A more recent approach proposes using agent-based models (ABMs) to describe cell–cell interactions within a virtual environment via defined rules48. Each agent represents an individual cell, moving and interacting with other cells according to specified rules. Since the interaction of two cells may result in intercellular signaling events, each agent may also contain a mechanistic model, resulting in a hybrid modeling scheme49 for which predictions can be made. Integration of patient-specific bulk or single-cell data allows for individual predictions50. Furthermore, deep learning models from the AlphaFold family51 can predict 3D protein structure. An individual’s genomic sequence data may be used to map genetically observed patient-specific differences to antigens52. In turn, these models can be used to predict changes in tumor-associated antigen binding of therapeutics, including the CAR antigen interaction. However, this modeling task remains an area that still needs further improvement in the future53,54. Besides protein-protein interaction models, cell–cell communication networks inferred from patient-derived single-cell multiomics38,55, spatially resolved, if available, also inform intercellular signaling models.
In CAR T cell therapy, intercellular models simulate the dynamics of the interactions between tumor and CAR T cells47 or on-target/off-tumor binding effects56, allowing predictions of treatment responses or adverse effects. Deep learning models for protein structure prediction can help identify treatment failures due to individual differences in antigens52. In the future, cell-specific DTs combined with a multicellular VT parameterized with data from in vitro or in vivo experiments may support model-informed drug development by simulating treatment success for different CAR T cell designs alongside well-defined experiments. A first variant of this concept systematically explores the multidimensional CAR T cell engineering design space, allowing only the most promising CAR T cell designs to be tested in vitro or in vivo, which reduces the number of experiments conducted and makes the process more cost-effective and ethically favorable48.
Models for CAR T cell therapy beyond the intra- and intercellular scale
CAR T cell therapy, as a treatment with living cells, affects not only the target cells but also the target’s tissues, organs, and ultimately impacts the whole body14,15. For example, cytokine release syndrome, although manageable, is a serious side effect of CAR T cell treatment that leads to systemic inflammation. As such, an ACI-VT must comprise models that go beyond the intra- and intercellular scales. Knowledge-driven modeling methods, such as systems biology maps of immune-related adverse outcome pathways, help assess toxicity profiles57,58. As CAR T cells interact with the patients’ immune system, integrating a DT of the human immune system is valuable. Community-driven efforts that build immune system DTs for different human pathologies59,60 must therefore be integral to eACI-VTs development. Whole-body models are typically built using multimodal artificial intelligence/machine learning (AI/ML) and are data-driven22. Ferle et al. proposed a patient-specific model combining a long short-term memory network with a conditional restricted Boltzmann machine to predict individual blood values over patient trajectories61. Maura et al. recently published the first multi-state model for MM that combines genomic and clinical data for individualized prognosis62.
Applications of whole-body eACI-VTs include clinical decision-support software that combines guideline-based reasoning with probabilistic assessments of therapy-associated success factors based on real-world evidence63. These tools can guide optimal treatment sequences for patients. Additional applications include monitoring during critical and acute medical care shortly after CAR T cell therapy64. At later time points, remote monitoring of patients by combining VTs with wearables65 could improve outpatient care. First applications detect late cytokine release syndrome66 or predict patient-individual blood values61. Furthermore, eACI-VTs may also serve as educational platforms for clinicians and nurses, offering realistic and safe environments for learning about CAR T cell eligibility and patient care, as modeled in the digital pathology field67.
Towards credible virtual twins in engineered adoptive cellular immunotherapies
A VT collects data from observations of its real-world counterpart and processes the data to update the parameters of the virtual representation, which in turn derives decision support for a specific context of use in the real-world (Fig. 3). However, bidirectional interchange is subject to uncertainties in both directions, impacting the credibility, i.e., the trust in “the predictive capability”68,69, of a VT. Below, we discuss the main hurdles that likely lead to low eACI-VT credibility and must be addressed during eACI-VT implementation to quantify and control uncertainty arising from model design choices, imprecise parameter fitting, missing information, or biological variance.
A VT differs from population-based models by the bidirectional flow of data between the real-world instance and its virtual representation. Data from the observations of the real-world instance (upper panel) must be collected, processed, and prepared to update the parameters of the virtual representation. This comprises data from the path of the patient, the path of the product, as well as their interaction upon treatment. As different factors on multiple biological scales (patients’ and the cell products’ scales) influence patient trajectory, a virtual representation of interconnected models spanning the whole process of eACI is required (lower panel). This leads to a high need for digitalization and ensuring interoperability. In the virtual representation, parameters dynamically derived from harmonized real-world data covering multiple biological scales are fed into a carefully curated set of models. The updated virtual representation can thus in turn derive decisions with an impact in the real world. AI artificial intelligence, eACI engineered adoptive cellular immunotherapy, ML machine learning, ODEs ordinary differential equations, PDEs partial differential equations, SL statistical learning. The figure was created by the authors using Canva.com.
First, VT parameters derived from a patient population must build on a representative number of high-quality data entries stemming from the real-world instances. Challenges for eACI-VTs lie in the different data types required to parameterize the virtual representation. These range from electronic health records (EHRs) for patient-level data, patient-reported outcomes, and clinical lab test data—including clinical genomic data—to data derived from single-cell multiomics. While methods exist for integrating clinical genomic data into EHRs, thereby enabling eACI-VTs to seamlessly access data from different biological scales, such approaches have yet to be developed for single-cell multiomics data (Box 2). The systematic collection of data can be addressed by utilizing a decentralized, cloud-based federated learning network, allowing to incorporate datasets from multiple sources while maintaining controlled access to patient data. Initial solutions for integrating clinically derived single-cell multiomics data into a federated learning network have been developed70. Organizations participating in such cloud-based platforms should adhere to common data models to allow seamless data integration71 (Box 2).
Second, trust in model predictions diminishes when the uncertainty in the observations used for model training/parametrization is high. Single-cell multiomics data is pivotal for informing (multi-)cellular in silico models69,72, but using (single-cell) multiomics-derived observations poses a fundamental challenge in model training. Multiomics data is multimodal, often sparse, noisy, and expensive to generate, thus only available in a small number of biological replicates, and has a large signal-to-noise ratio73. Moreover, tracking changes of the molecular state of the same cell over time or spatially resolved is only now becoming feasible74,75. Therefore, datasets are often unpaired in terms of cell organization and time scale. Observations from “similar” cells need to be integrated to obtain a comprehensive view of a class of comparable cells. This uncertainty in defining a cell’s true ground state presents a significant challenge for data derived in silico models for individual cells or cell–cell interactions. This can be addressed by tailored solutions accounting for the uncertainty in single-cell clustering approaches76, expression quantification77,78, or cell-type annotation79. The problem of missing data can be managed through imputation methods using transfer learning with external reference data73,79, underpinning the need for external single-cell reference atlases of engineered immune cells. Additionally, variance in the dimension of the patients’ socio-cultural, economic, and ethical background, as well as sex and gender, influence data quality on all biological scales and should be documented in bioinformatic analyses.
Third, for cell models that are built on general biochemical rules, the causality of input-output relationships is at least approximately understood. In such cases, a mechanistic cell model can be considered trustworthy if intermediate steps and outputs can be inferred from a given input with a reasonably small error margin, e.g., not greater than negligible biological noise or measurement error. Parameter calibration and validation for mechanistic models can be supported by patient-specific in vitro models like patient-derived xenograft (PDX) models, allowing high-throughput analyses27,80,81. In cases where patient-specific data is sparse, parameter ranges could be defined and iteratively refined by repeated observations of the readout parameters of PDX models under different conditions. However, the impact on uncertainty quantification of such a nested approach is unknown and must be addressed.
Fourth, given that different factors on multiple biological scales influence the trajectory of patients eligible for CAR T cell therapy, a single model DT is insufficient. Instead, interconnected models are needed to span the whole process in eACIs (Fig. 3), including integration with DTs for the manufacturing process3,21. Interactions between models must be thoroughly investigated and validated. Consequently, interoperability across all four levels—technical, syntactic, semantic, and organizational—is essential82 but is often not seamlessly addressed by model developers83.
Fifth, biomarkers at multiple biological scales represented in the eACI-VT are an essential component for driving meaningful decision-making. In the case of new and innovative therapies like eACIs, validated biomarkers predicting patient outcome remain scarce. Also, due to the complex interaction between drug and host determining outcome on a single patient level, likely composite biomarker signatures allow a more trustworthy prediction84. Sensibly, biomarker development can be implemented and advanced in the process of building a VT. Structurally gathering multimodal data across various biological scales from many patients to train and validate the VT enables simultaneously identifying and confirming biomarker signatures from available multiomics data. These biomarkers can then, in turn, be applied in the VT to drive transparent decision-making.
Lastly, AI/ML models need to consider the temporal development of diseases over months and years, which is in contrast to the millisecond-scale of intercellular signaling. Integrating data-driven AI/ML models operating on clinical data with mechanistic models of intracellular signaling and cell–cell interactions remains an open challenge. Potentially, it is possible to extract features of ABM-based simulations, which could be employed as part of the data to train AI/ML models. Solutions to this challenge place high demands on hardware, data storage, and IT infrastructure for emerging digital healthcare technologies (Box 3).
The development of software as medical devices is a detailed process that, if stringently followed, can address some of the above-mentioned uncertainties85. Following the life cycle for medical software (IEC 62304), developers can begin with a comprehensive analysis of the technical, medical, ethical, legal, and societal requirements, which can then be refined throughout the development cycles. An incremental, iterative development process allows for the early release of a first prototype that can be tested in selected, relevant environments, enabling co-creation with stakeholders. This approach sets the basis for successfully translating an eACI-VT into a practical and impactful tool for use in both clinical and nonclinical settings. However, challenges that emerge in a hospital setting and with a larger patient population compared to academia, where the eACI-VT was developed, must be addressed during the co-creational process. This includes adhering to legal requirements and ethical considerations with regard to processing sensitive personal data and using AI (Boxes 4 and 5).
How virtual twins improve patient management prior and during treatment with engineered adoptive cellular immunotherapies
EACI-VTs are powerful tools designed to model the complex and unique interactions between administered cells and the patient. They support and educate clinicians and patients throughout the decision-making process, ultimately improving patient outcome and well-being over the entire CAR T cell therapy life cycle (use case of CAR T cell treatment of MM patients: Fig. 4). Of particular importance is the ability of eACI-VTs to integrate patient-specific longitudinal, real-world multimodal data on multiple biological scales. For instance, CD4+ CAR T cells can persist for years, keeping the patient in remission and potentially offering life-long therapeutic benefit4. EACI-VTs can anticipate and support future developments in the rapidly evolving field of eACIs, while accounting for time-dependent and dynamic variance in clinical care and diverse patient populations. EACI-VTs enhance knowledge about the mode and mechanism of action underlying these novel treatment options, also supporting the identification of biomarkers, especially those specific to eACI biology86, and enhancing post-authorization monitoring. In the development phase, eACI-VTs can simulate treatment response prior to first-in-human studies or aid in clinical trial planning, for example, by sampling synthetic patient populations87. Additionally, cases of off-label use and application of out-of-specification CAR T cell products can sensibly be monitored over longer periods of time. This is particularly relevant given recent reports on secondary T cell malignancies following CAR T cell therapy88,89. Currently, these cases are investigated by the FDA and the EMA for a possible link between the malignant transformation to insertion site mutagenesis during CAR T cell manufacturing. The FDA recommends life-long monitoring of treated patients90. Considering the high costs associated with eACI therapies, which currently limit access, the use of eACI-VTs for accurate patient stratification could have a significant impact on healthcare systems. Applications include cost-effectiveness analysis, which supports performance-based reimbursement models and strategies to reduce long-term costs of eACI treatment, ultimately achieving a more widespread use of this innovative therapy. Overall, VTs91, including eACI-VTs, are emerging as a key concept for advancing personalized, risk-adapted decision support in next-generation immunotherapy.
Throughout the journey of a multiple myeloma patient who is eligible for treatment with CAR T cells, an eACI-VT can support the decisions regarding optimal patient care for each step along the journey. This is facilitated by the frequent update of the eACI-VT with the relevant data on the patient, the product, and their interaction. These data are collected with various devices and systems in the monitoring periods of the patient journey. CAR chimeric antigen receptor, CPS Cyber-Physical System, eACI engineered adoptive cellular immunotherapy, eACI-VT virtual twin for engineered adoptive cellular immunotherapy, EHR electronic health record, GMP good manufacturing practice, ICU intensive care unit, MRD minimal residual disease, IoT internet of things, PRO patient-reported outcome, QoL quality of life. The figure was created by the authors using Canva.com.
How virtual twins improve model-informed drug development for engineered adoptive cellular immunotherapies
In addition to supporting clinical decision-making for treating patients, eACI-VTs used in model-informed drug development offer the potential to make eACI development and clinical trials more cost-effective and ethically responsible92,93. Clinical trials for regulatory approval of eACIs are typically lengthy94. One challenge lies in clinical trial design and recruitment, as access to eACIs is limited, often resulting in underrepresentation of patient diversity in clinical trials compared to post-approval product phases. In silico (clinical) trials could expedite eACIs’ availability by improving predictive accuracy of trials, reducing required sample size, and helping to predict the risk of side effects. These trials rely on synthetic patient populations simulated using computational models parametrized with retrospective RWD. A VT parametrized with RWD can serve as a prior to strengthen the control arm of a clinical trial93,95. Patient-specific pharmacokinetic/pharmacodynamic (PK/PD) models reveal dose-exposure-relationships96. Ordinary equations can simulate the dynamics of the interactions between tumor and CAR T cells47, helping to the reduction of trial sample sizes92. Mechanistic models may be used to refine trial designs by avoiding the inclusion of individuals at risk for side effects or by testing efficacy in patient subgroups with characteristics that are not available in the clinical trial population92,97. Initial approaches that help to reduce the risk of side effects in first-in-human studies of eACIs combine disease maps, immune-related adverse outcome pathways, and advanced nonclinical in vitro models58,98,99. Furthermore, generative AI approaches are expected to play a pivotal role in in silico clinical trials100, including models on multiple biological scales. An eACI-VT utilizing generative models can simulate potential clinically relevant outcomes in a study population, hence providing insights about eACI efficacy and safety101. Generative AI models for biological cells allow the generation of patient-individual in silico cells40,44, which is an essential biological scale for eACI-VTs to model, for example, immune-response upon ACI treatment. Before integrating eACI-VTs into clinical trial workflows, the credibility of each individual in silico model must be rigorously validated. Only after establishing confidence in individual components can the credibility of the overall VT be adequately demonstrated102.
Conclusion and outlook
VTs in healthcare aim to guide biomedical researchers and clinicians in optimizing therapies and treatment regimens tailored to individual patients. Additionally, they empower patients to better understand their unique disease trajectories.
EACIs are “living drugs” comprising individual cells that have complex and dynamic interactions with the host over time, which fundamentally distinguishes them from conventional drugs. Although currently described VT frameworks in healthcare incorporate multimodal data on all relevant biological scales to create single-organ DTs or complex whole-body VTs, they all lack the integration of virtual representations that model the drug and its interaction with target cells that evolve over the course of treatment, spanning decades. Here, we outline the minimum design specifications necessary to adapt VTs for use with “living drugs,” such as CAR T cell therapy. With these minimum specifications, we are pioneering the establishment of a foundational framework for developing eACI-VTs that can accurately simulate the individual pathophysiology of patients eligible for or undergoing eACI. As this complex and potentially revolutionary therapy option expands into earlier lines of therapy and other indications, possibly impacting a broader patient population, we envision a parallel development of eACI-VTs as a tool to support the implementation of true precision health.
Data availability
No datasets were generated or analyzed during the current study.
References
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Finck, A. V., Blanchard, T., Roselle, C. P., Golinelli, G. & June, C. H. Engineered cellular immunotherapies in cancer and beyond. Nat. Med. 28, 678–689 (2022).
Blache, U., Popp, G., Dünkel, A., Koehl, U. & Fricke, S. Potential solutions for manufacture of CAR T cells in cancer immunotherapy. Nat. Commun. 13, 5225 (2022).
Melenhorst, J. J. et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 602, 503–509 (2022).
Cappell, K. M. & Kochenderfer, J. N. Long-term outcomes following CAR T cell therapy: what we know so far. Nat. Rev. Clin. Oncol. 20, 359–371 (2023).
Kamel Boulos, M. N. & Zhang, P. Digital twins: from personalised medicine to precision public health. J. Pers. Med. 11, 745 (2021).
Fischer, R.-P., Volpert, A., Antonino, P. & Ahrens, T. D. Digital patient twins for personalized therapeutics and pharmaceutical manufacturing. Front. Digit. Health 5, https://doi.org/10.3389/fdgth.2023.1302338 (2024).
Katsoulakis, E. et al. Digital twins for health: a scoping review. npj Digit. Med. 7, 77 (2024).
Sun, T., He, X. & Li, Z. Digital twin in healthcare: recent updates and challenges. Digit. health 9, 20552076221149651 (2023).
Viceconti, M., Vos, M., de, Mellone, S. & Geris, L. Position paper From the digital twins in healthcare to the Virtual Human Twin: a moon-shot project for digital health research. IEEE J. Biomed. Health Inform. 28, 491–501 (2023).
Wright, L. & Davidson, S. How to tell the difference between a model and a digital twin. Adv. Model. and Simul. Eng. Sci. 7, https://doi.org/10.1186/s40323-020-00147-4 (2020).
Baker, D. J., Arany, Z., Baur, J. A., Epstein, J. A. & June, C. H. CAR T therapy beyond cancer: the evolution of a living drug. Nature 619, 707–715 (2023).
Karampampa, K., Zhang, W., Venkatachalam, M., Cotte, F.-E. & Dhanda, D. Cost-effectiveness of idecabtagene vicleucel compared with conventional care in triple-class exposed relapsed/refractory multiple myeloma patients in Canada and France. J. Med. Econ. 26, 243–253 (2023).
Donnadieu, E. et al. Time to evolve: predicting engineered T cell-associated toxicity with next-generation models. J. Immunother. Cancer 10, e003486 (2022).
Guedan, S. et al. Time 2EVOLVE: predicting efficacy of engineered T-cells—How far is the bench from the bedside?. J. Immunother. Cancer 10, e003487 (2022).
Gagelmann, N. et al. Development and validation of a prediction model of outcome after B-cell maturation antigen-directed chimeric antigen receptor T-cell therapy in relapsed/refractory multiple myeloma. J. Clin. Oncol.42, 1665–1675 (2024).
Rejeski, K. et al. The CAR-HEMATOTOX risk-stratifies patients for severe infections and disease progression after CD19 CAR-T in R/R LBCL. J. Immunother. Cancer 10, e004475 (2022).
Rejeski, K. et al. CAR-HEMATOTOX: a model for CAR T-cell-related hematologic toxicity in relapsed/refractory large B-cell lymphoma. Blood 138, 2499–2513 (2021).
Rejeski, K. et al. The CAR-HEMATOTOX score as a prognostic model of toxicity and response in patients receiving BCMA-directed CAR-T for relapsed/refractory multiple myeloma. J. Hematol. Oncol. 16, 7506–7508 (2023).
Boer et al. Population-based external validation of the EASIX scores to predict CAR T-cell-related toxicities. Cancers 15, 5443 (2023).
Hort, S. et al. Toward rapid, widely available autologous CAR-T cell therapy—artificial intelligence and automation enabling the smart manufacturing hospital. Front. Med. 9, 913287 (2022).
Tang, C. et al. A roadmap for the development of human body digital twins. Nat. Rev. Electr. Eng. 1, 199–207 (2024).
National Academies of Sciences, Engineering, and Medicine. Foundational Research Gaps and Future Directions for Digital Twins (National Academies Press, 2023).
Yang, J., Chen, Y., Jing, Y., Green, M. R. & Han, L. Advancing CAR T cell therapy through the use of multidimensional omics data. Nat. Rev. Clin. Oncol. 20, 211–228 (2023).
Rade, M. et al. Single-cell multiomic dissection of response and resistance to chimeric antigen receptor T cells against BCMA in relapsed multiple myeloma. Nat. Cancer 5, 1318–1333 (2024).
Irmisch, A. et al. The Tumor Profiler Study: integrated, multi-omic, functional tumor profiling for clinical decision support. Cancer Cell 39, 288–293 (2021).
Kropivsek, K. et al. Ex vivo drug response heterogeneity reveals personalized therapeutic strategies for patients with multiple myeloma. Nat. Cancer 4, 734–753 (2023).
Vercellino, L. et al. Current and future role of medical imaging in guiding the management of patients with relapsed and refractory non-hodgkin lymphoma treated with CAR T-cell therapy. Front. Oncol. 11, 664688 (2021).
Zamagni, E., Tacchetti, P. & Cavo, M. Imaging in multiple myeloma: How? When?. Blood 133, 644–651 (2019).
Amit, I. et al. The commitment of the human cell atlas to humanity. Nat. Commun. 15, 10019 (2024).
Rehm, H. L. et al. GA4GH: International policies and standards for data sharing across genomic research and healthcare. Cell Genom. 1, 100029 (2021).
O’Rourke, K. ASCO releases guideline on CAR T-cell therapy: a multidisciplinary team’s recommendations help in the recognition, workup, evaluation, and management of the most common chimeric antigen receptor (CAR) T-cell-related toxicities. Cancer 128, 429–430 (2022).
Kröger, N., Gribben, J., Chabannon, C., Yakoub-Agha, I. & Einsele, H. (eds) The EBMT/EHA CAR-T Cell Handbook (2022).
Ma, C. & Gurkan-Cavusoglu, E. A comprehensive review of computational cell cycle models in guiding cancer treatment strategies. npj Syst. Biol. Appl. 10, 71 (2024).
Ildefonso, G. V. & Finley, S. D. A data-driven Boolean model explains memory subsets and evolution in CD8+ T cell exhaustion. npj Syst. Biol. Appl. 9, 36 (2023).
Cess, C. G. & Finley, S. D. Data-driven analysis of a mechanistic model of CAR T cell signaling predicts effects of cell-to-cell heterogeneity. J. Theor. Biol. 489, 110125 (2020).
Kondratova, M., Barillot, E., Zinovyev, A. & Calzone, L. Modelling of immune checkpoint network explains synergistic effects of combined immune checkpoint inhibitor therapy and the impact of cytokines in patient response. Cancers 12, 3600 (2020).
Badia-I-Mompel, P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754 (2023).
Yang, F. et al. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 4, 852–866 (2022).
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Tao, Z. et al. Impact of T cell characteristics on CAR-T cell therapy in hematological malignancies. Blood Cancer J. 14, 213 (2024).
Bai, Z. et al. Single-cell CAR T atlas reveals type 2 function in 8-year leukaemia remission. Nature 634, 702–711 (2024).
Baguet, C., Larghero, J. & Mebarki, M. Early predictive factors of failure in autologous CAR T-cell manufacturing and/or efficacy in hematologic malignancies. Blood Adv. 8, 337–342 (2024).
Rood, J. E. et al. The Human Cell Atlas from a cell census to a unified foundation model. Nature 637, 1065–1071 (2025).
Shoshi, A. et al. A flexible digital twin framework for ATMP production—towards an efficient CAR T cell manufacturing. Procedia CIRP 125, 124–129 (2024).
Barros, L. R. C. et al. CARTmath—a mathematical model of CAR-T immunotherapy in preclinical studies of hematological cancers. Cancers 13, 2941 (2021).
Liu, L. et al. Computational model of CAR T-cell immunotherapy dissects and predicts leukemia patient responses at remission, resistance, and relapse. J. Immunother. Cancer 10, e005360 (2022).
Prybutok, A. N., Yu, J. S., Leonard, J. N. & Bagheri, N. Mapping CAR T-cell design space using agent-based models. Front. Mol. Biosci. 9, 849363 (2022).
Ruscone, M. et al. Multiscale model of the different modes of cancer cell invasion. Bioinformatics 39, btad374 (2023).
Béal, J., Montagud, A., Traynard, P., Barillot, E. & Calzone, L. Personalization of logical models with multi-omics data allows clinical stratification of patients. Front. Physiol. 9, https://doi.org/10.3389/fphys.2018.01965 (2019).
Thompson, B. & Petrić Howe, N. Alphafold 3.0: the AI protein predictor gets an upgrade. Nature https://doi.org/10.1038/d41586-024-01385-x (2024).
Moth, C. W. et al. VUStruct: a compute pipeline for high throughput and personalized structural biology. Preprint at bioRxiv https://doi.org/10.1101/2024.08.06.606224 (2024).
Hitawala, F. N. & Gray, J. J. What does AlphaFold3 learn about antigen and nanobody docking, and what remains unsolved? Preprint at bioRxiv https://doi.org/10.1101/2024.09.21.614257 (2025).
Eshak, F. & Goupil-Lamy, A. Advancements in nanobody epitope prediction: a comparative study of AlphaFold2Multimer vs AlphaFold3. J. Chem. Inf. Model. 65, 1782–1797 (2025).
Dimitrov, D. et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 13, 3224 (2022).
Santurio, D. S. & Barros, L. R. C. A mathematical model for on-target off-tumor effect of CAR-T cells on gliomas. Front. Syst. Biol. 2, 923085 (2022).
Mazein, A. et al. A guide for developing comprehensive systems biology maps of disease mechanisms: planning, construction and maintenance. Front. Bioinform. 3, 1197310 (2023).
Mazein, A. et al. Using interactive platforms to encode, manage and explore immune-related adverse outcome pathways. J. Immunotoxicol. 21, S5–S12 (2024).
Laubenbacher, R. et al. Building digital twins of the human immune system: toward a roadmap. npj Digit. Med. 5, 64 (2022).
Immune Digital Twins Working Group. Building an international and interdisciplinary community to develop immune digital twins for complex human pathologies. https://zenodo.org/records/10783684 (2024).
Ferle, M. et al. Predicting progression events in multiple myeloma from routine blood work.npj Digit Med 8, 231 (2025).
Maura, F. et al. Genomic classification and individualized prognosis in multiple myeloma. J. Clin. Oncol. 42, 1229–1240 (2024).
Oeser, A. et al. KAIT—knowledge-driven and artificial intelligence-based platform for therapy decision support in hematology. White Paper. https://kait.health/KAIT_White_Paper.pdf (2021).
Halpern, G. A., Nemet, M., Gowda, D. M., Kilickaya, O. & Lal, A. Advances and utility of digital twins in critical care and acute care medicine: a narrative review. J. Yeungnam Med. Sci. 42, 9 (2025).
Cloß, K. et al. Application of wearables for remote monitoring of oncology patients: a scoping review. Digit. Health 10, 20552076241233998 (2024).
Rajeeve, S. et al. Early and consistent CRS detection using wearable device for remote patient monitoring following CAR-T therapy in relapsed/refractory multiple myeloma (RRMM): early results of an investigator-initiated trial. Blood 142, 1007 (2023).
Hanna, M. G. et al. Future of artificial intelligence-machine learning trends in pathology and medicine. Mod. Pathol. 38, 100705 (2025).
Center for Devices and Radiological Health. Assessing the Credibility of Computational Modeling and Simulation in Medical Device Submissions. Guidance for Industry and Food and Drug Administration Staff. (FDA, 2023).
Galappaththige, S., Gray, R. A., Costa, C. M., Niederer, S. & Pathmanathan, P. Credibility assessment of patient-specific computational modeling using patient-specific cardiac modeling as an exemplar. PLoS Comput. Biol. 18, e1010541 (2022).
Saldanha, O. L. et al. SwarmMAP: swarm learning for decentralized cell type annotation in single cell sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2025.01.13.632775 (2025).
Vorisek, C. N. et al. Towards an interoperability landscape for a national research data infrastructure for personal health data. Sci. Data 11, 772 (2024).
Polychronidou, M. et al. Single-cell biology: What does the future hold?. Mol. Syst. Biol. 19, e11799 (2023).
Lähnemann, D. et al. Eleven grand challenges in single-cell data science. Genome Biol. 21, 31 (2020).
Chen, W. et al. Live-seq enables temporal transcriptomic recording of single cells. Nature 608, 733–740 (2022).
Polanski, K. et al. Bin2cell reconstructs cells from high resolution Visium HD data. Bioinformatics 40, btae546 (2024).
Grabski, I. N., Street, K. & Irizarry, R. A. Significance analysis for clustering with single-cell RNA-sequencing data. Nat. Methods 20, 1196–1202 (2023).
Baldoni, P. L. et al. Dividing out quantification uncertainty allows efficient assessment of differential transcript expression with edgeR. Nucleic Acids Res. 52, e13 (2024).
van Buren, S. et al. Compression of quantification uncertainty for scRNA-seq counts. Bioinformatics 37, 1699–1707 (2021).
Engelmann, J. et al. Uncertainty quantification for atlas-level cell type transfer. Preprint at arxiv https://doi.org/10.48550/arXiv.2211.03793 (2022).
Alieva, M. et al. BEHAV3D: a 3D live imaging platform for comprehensive analysis of engineered T cell behavior and tumor response. Nat. Protoc. 19, 2052–2084 (2024).
Hernández-López, P. et al. Dual targeting of cancer metabolome and stress antigens affects transcriptomic heterogeneity and efficacy of engineered T cells. Nat. Immunol. 25, 88–101 (2024).
Lehne, M., Sass, J., Essenwanger, A., Schepers, J. & Thun, S. Why digital medicine depends on interoperability. npj Digit. Med. 2, 79 (2019).
Niarakis, A. et al. Addressing barriers in comprehensiveness, accessibility, reusability, interoperability and reproducibility of computational models in systems biology. Brief. Bioinform. 23, bbac212 (2022).
Levstek, L., Janžič, L., Ihan, A. & Kopitar, A. N. Biomarkers for prediction of CAR T therapy outcomes: current and future perspectives. Front. Immunol. 15, 1378944 (2024).
Markowetz, F. All models are wrong and yours are useless: making clinical prediction models impactful for patients. npj Precis. Oncol. 8, 54 (2024).
Biasco, L. et al. Clonal expansion of T memory stem cells determines early anti-leukemic responses and long-term CAR T cell persistence in patients. Nat. Cancer 2, 629–642 (2021).
Moore, J. H. et al. SynTwin: a graph-based approach for predicting clinical outcomes using digital twins derived from synthetic patients. Pac. Symp. Biocomput. 29, 96–107 (2024).
Braun, T., Kuschel, F., Reiche, K., Merz, M. & Herling, M. Emerging T-cell lymphomas after CAR T-cell therapy. Leukemia https://doi.org/10.1038/s41375-025-02574-x (2025).
Braun, T. et al. Multiomic profiling of T cell lymphoma after therapy with anti-BCMA CAR T cells and GPRC5D-directed bispecific antibody. Nat. Med. 31, 1145–1153 (2025).
Levine, B. L. et al. Unanswered questions following reports of secondary malignancies after CAR-T cell therapy. Nat. Med. 30, 338–341 (2024).
Topol, E. J. Medical forecasting. Science 384, eadp7977 (2024).
Arsène, S. et al. In silico clinical trials: Is it possible?. Methods Mol. Biol. 2716, 51–99 (2024).
Barrett, J. S., Nicholas, T., Azer, K. & Corrigan, B. W. Role of disease progression models in drug development. Pharm. Res. 39, 1803–1815 (2022).
Derraz, B. et al. New regulatory thinking is needed for AI-based personalised drug and cell therapies in precision oncology. npj Precis. Oncol. 8, 23 (2024).
Terranova, N. & Venkatakrishnan, K. Machine learning in modeling disease trajectory and treatment outcomes: an emerging enabler for model-informed precision medicine. Clin. Pharmacol. Ther. 115, 720–726 (2024).
Singh, A. P. et al. Development of a quantitative relationship between CAR-affinity, antigen abundance, tumor cell depletion and CAR-T cell expansion using a multiscale systems PK-PD model. mAbs 12, 1688616 (2020).
Viceconti, M. et al. Possible contexts of use for in silico trials methodologies: a consensus-based review. IEEE J. Biomed. Health Inform. 25, 3977–3982 (2021).
Alb, M. et al. Novel strategies to assess cytokine release mediated by chimeric antigen receptor T cells based on the adverse outcome pathway concept. J. Immunotoxicol. 21, 13–28 (2024).
Mazein, A. et al. An explorable model of an adverse outcome pathway of cytokine release syndrome related to the administration of immunomodulatory biotherapeutics and cellular therapies. Res. Sq. https://doi.org/10.21203/rs.3.rs-5163108/v1 (2024).
Bordukova, M., Makarov, N., Rodriguez-Esteban, R., Schmich, F. & Menden, M. P. Generative artificial intelligence empowers digital twins in drug discovery and clinical trials. Expert Opin. Drug Discov. 19, 33–42 (2024).
Moingeon, P., Chenel, M., Rousseau, C., Voisin, E. & Guedj, M. Virtual patients, digital twins and causal disease models: paving the ground for in silico clinical trials. Drug Discov. Today 28, 103605 (2023).
Aycock, K. I. et al. Toward trustworthy medical device in silico clinical trials: a hierarchical framework for establishing credibility and strategies for overcoming key challenges. Front. Med. 11, 1433372 (2024).
Bagley, S. J. et al. Intrathecal bivalent CAR T cells targeting EGFR and IL13Rα2 in recurrent glioblastoma: phase 1 trial interim results. Nat. Med. 30, 1320–1329 (2024).
Choi, B. D. et al. Intraventricular CARv3-TEAM-E T cells in recurrent glioblastoma. N. Engl. J. Med. 390, 1290–1298 (2024).
Blache, U., Tretbar, S., Koehl, U., Mougiakakos, D. & Fricke, S. CAR T cells for treating autoimmune diseases. RMD Open 9, e002907 (2023).
Müller, F. et al. CD19 CAR T-cell therapy in autoimmune disease—a case series with follow-up. N. Engl. J. Med. 390, 687–700 (2024).
Morte-Romea, E. et al. CAR Immunotherapy for the treatment of infectious diseases: a systematic review. Front. Immunol. 15, 1289303 (2024).
Carvalho, T. First two patients receive CAR T cell therapy for HIV. Nat. Med. 29, 1290–1291 (2023).
Labanieh, L. & Mackall, C. L. CAR immune cells: design principles, resistance and the next generation. Nature 614, 635–648 (2023).
Shabaneh, T. B. et al. Systemically administered low-affinity HER2 CAR T cells mediate antitumor efficacy without toxicity. J. Immunother. Cancer 12, e008566 (2024).
Bachmann, M. The UniCAR system: a modular CAR T cell approach to improve the safety of CAR T cells. Immunol. Lett. 211, 13–22 (2019).
Schlegel, L. S., Werbrouck, C., Boettcher, M. & Schlegel, P. Universal CAR 2.0 to overcome current limitations in CAR therapy. Front. Immunol. 15, 1383894 (2024).
Turtle, C. J. et al. CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J. Clin. Investig. 126, 2123–2138 (2016).
Mensurado, S., Blanco-Domínguez, R. & Silva-Santos, B. The emerging roles of γδ T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol. 20, 178–191 (2023).
Golubovskaya, V. & Wu, L. Different Subsets of T cells, memory, effector functions, and CAR-T immunotherapy. Cancers 8, 36 (2016).
Wang, W. et al. Breakthrough of solid tumor treatment: CAR-NK immunotherapy. Cell Death Discov. 10, 40 (2024).
Abdin, S. M., Paasch, D. & Lachmann, N. CAR macrophages on a fast track to solid tumor therapy. Nat. Immunol. 25, 11–12 (2024).
Caldwell, K. J., Gottschalk, S. & Talleur, A. C. Allogeneic CAR cell therapy-more than a pipe dream. Front. Immunol. 11, 618427 (2020).
Hamieh, M., Mansilla-Soto, J., Rivière, I. & Sadelain, M. Programming CAR T cell tumor recognition: tuned antigen sensing and logic gating. Cancer Discov. 13, 829–843 (2023).
Flugel, C. L. et al. Overcoming on-target, off-tumour toxicity of CAR T cell therapy for solid tumours. Nat. Rev. Clin. Oncol. 20, 49–62 (2023).
Tousley, A. M. et al. Co-opting signalling molecules enables logic-gated control of CAR T cells. Nature 615, 507–516 (2023).
Prommersberger, S. et al. Generation of CAR-T cells with sleeping beauty transposon gene transfer. Methods Mol. Biol. 2521, 41–66 (2022).
Metanat, Y. et al. The paths toward non-viral CAR-T cell manufacturing: a comprehensive review of state-of-the-art methods. Life Sci. 348, 122683 (2024).
Zhang, J. et al. Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature 609, 369–374 (2022).
Auw et al. Comparison of two lab-scale protocols for enhanced mRNA-based CAR-T cell generation and functionality. Sci. Rep. 13, 18160 (2023).
Fraessle, S. P. et al. Activation-inducible CAR expression enables precise control over engineered CAR T cell function. Commun. Biol. 6, 604 (2023).
Prommersberger, S. et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. 28, 560–571 (2021).
Parayath, N. N., Stephan, S. B., Koehne, A. L., Nelson, P. S. & Stephan, M. T. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells in vivo. Nat. Commun. 11, 6080 (2020).
Nicolai, C. J. et al. In vivo CAR T-cell generation in non-human primates using lentiviral vectors displaying a multi-domain fusion ligand. Blood 144, 977–987 (2024).
Reps, J. M., Schuemie, M. J., Suchard, M. A., Ryan, P. B. & Rijnbeek, P. R. Design and implementation of a standardized framework to generate and evaluate patient-level prediction models using observational healthcare data. J. Am. Med. Inform. Assoc. 25, 969–975 (2018).
Warner, J. L. et al. HemOnc: a new standard vocabulary for chemotherapy regimen representation in the OMOP common data model. J. Biomed. Inform. 96, 103239 (2019).
Belenkaya, R. et al. Extending the OMOP common data model and standardized vocabularies to support observational cancer research. JCO Clin. Cancer Inform. 5, 12–20 (2021).
OHDSI. OMOP Common Data Model Oncology Extension. OMOP Common Data Model Oncology Extension. https://ohdsi.github.io/CommonDataModel/oncology.html (2024).
Shin, S. J. et al. Genomic common data model for seamless interoperation of biomedical data in clinical practice: retrospective study. J. Med. Internet Res. 21, e13249 (2019).
ISO/TS 20428:2024. Genomics informatics—data elements and their metadata for describing structured clinical genomic sequence information in electronic health records. International Organization for Standardization (2024).
Danis, D. et al. Phenopacket-tools: Building and validating GA4GH Phenopackets. PLoS ONE 18, e0285433 (2023).
ISO 4454:2022. Genomics informatics—phenopackets: a format for phenotypic data exchange. International Organization for Standardization (2022).
Hrovatin, K. et al. Considerations for building and using integrated single-cell atlases. Nat. Methods 22, 41–57 (2025).
Alterovitz, G. et al. FHIR Genomics: enabling standardization for precision medicine use cases. npj Genom. Med. 5, 13 (2020).
Lau-Min, K. S. et al. Real-world integration of genomic data into the electronic health record: the PennChart Genomics Initiative. Genet. Med. 23, 603–605 (2021).
Matschinske, J. et al. The FeatureCloud platform for federated learning in biomedicine: unified approach. J. Med. Internet Res. 25, e42621 (2023).
King, C. H. S. et al. Communicating regulatory high-throughput sequencing data using BioCompute Objects. Drug Discov. Today 27, 1108–1114 (2022).
Banerjee, S., Jesubalan, N. G., Kulkarni, A., Agarwal, A. & Rathore, A. S. Developing cyber-physical system and digital twin for smart manufacturing: Methodology and case study of continuous clarification. J. Ind. Inf. Integr. 38, 100577 (2024).
Elayan, H., Aloqaily, M. & Guizani, M. Digital twin for intelligent context-aware iot healthcare systems. IEEE Internet Things J. 8, 16749–16757 (2021).
Oks, S. J. et al. Cyber-physical systems in the context of Industry 4.0: a review, categorization and outlook. Inf. Syst. Front. 26, 1731–1772 (2024).
Vanhellemont, A. et al. Remote patient monitoring for early detection of cytokine release syndrome in myeloma patients: a comparative study between standard care and remote monitoring. Blood 144, 3664 (2024).
Article 29 Data Protection Working Party. Opinion 05/2014 on Anonymisation Techniques. https://ec.europa.eu/justice/article-29/documentation/opinion-recommendation/files/2014/wp216_en.pdf, 10 (2014).
Gobert, A., Knockaert, M., Martin, R. & Van Gyseghem, J.-M. La donnée à caractère personnel et sa réutilisation.J. Trib 8, 124–127 (2024).
Zenker, S. et al. Data protection-compliant broad consent for secondary use of health care data and human biosamples for (bio)medical research: Towards a new German national standard. J. Biomed. Inform. 131, 104096 (2022).
ISO/TS 9491-1:2023. Biotechnology—predictive computational models in personalized medicine research. International Organization for Standardization (2023).
FDA. Artificial intelligence and machine learning in software as a medical device. https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device#regulation (2024).
Busch, F. et al. Navigating the European Union artificial intelligence act for healthcare. npj Digit. Med. 7, 210 (2024).
Acknowledgements
The CERTAINTY project is funded by the European Union (Grant Agreement 101136379). However, views and opinions expressed are those of the author(s) only and do not necessarily reflect those of the European Union or the Health and Digital Executive Agency. Neither the European Union nor the granting authority can be held responsible for them.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Consortia
Contributions
U.W. and K.R. wrote the main manuscript. S.B. and U.W. prepared fig. 3, U.W. and K.R. prepared figure 1 and 2, D.F. and U.W. prepared fig. 4. U.W., M.Kr., C.B., M.A., M.Q., L.C., S.O.-R., S.B., L.F., S.C., M.Ma., M.F., M.Ra., G.A., J.R.H., J.J., D.S., Z.S., J.K., G.P., C.A., M.Kn., C.T.S., D.F., C.Sa., V.V., N.G., M.Rü., O.P., S.Fri., A.S., C.W., C.St., J.V.G., A.N., L.G., M.H., T.N., U.P., U.K., R.D., A.K., S.Fra., H.F., M.Me., K.R. reviewed, revised and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
G.A. received speaker honoraria from Janssen Pharmaceutica NV. H.F. received funding from UCB Pharma and AbbVie for research purposes. S.Fri. received speaker's fees from Novartis Pharma GmbH, Janssen-Cilag GmbH, Vertex Pharmaceuticals (Germany) GmbH, Kite/Gilead Sciences GmbH, MSGO GmbH, Bristol-Myers Squibb GmbH & Co. KGaA and art tempi Communications. L.G.: Janssen, BMS, Sanofi, Pfizer. M.H. is an inventor on patent applications and has been granted patents related to CAR technology, licensed in part to industry. M.H. is a cofounder and equity owner of T-CURX GmbH, Würzburg. M.H. receives speaker honoraria from BMS, Janssen, Kite/Gilead, and Novartis and research support from BMS. J.R.H receives research funding from Adaptive Biotechnologies, BioLinRx, Sanofi, GlaxoSmithKline, Regeneron, Pfizer, Johnson and Johnson Innovative Medicine, and Takeda Oncology. U.K. has received consultant and/or speaker fees from AstraZeneca, Affimed, Glycostem, GammaDelta, Zelluna, CGT manufacturing: Miltenyi Biotec and Novartis Pharma GmbH, Bristol-Myers Squibb GmbH & Co. J.K. is shareholder on follow up companies of Gadeta, inventor on multiple patents dealing with genetic engineering of immune cells, received research support from Novartis, Miltenyi Biotech, and Gadeta. Maximilian Merz: Advisory Boards/Honoraria/Research support: Amgen, BMS, Celgene, Gilead, Janssen, Stemline, Springworks, Takeda, and Roche/Genentech. A.N. collaborates with SANOFI-AVENTIS R&D via a public–private partnership grant CIFRE contract, n° 2020/0766. O.P. has received grants and personal fees from Incyte, Neovii, Gilead, Merck Sharpe and Dohme, Omeros, Sobi, Takeda, and Jazz Pharmaceuticals. U.O. has received honoraria and research support from BMS, Johnson-Johnson, Gilead. A.S. is a shareholder and managing director of Springboard Health Angels, Schmidt Deng Ventures and their respective venture funds and portfolio companies. D.S. is shareholder at Masaryk University spinoff: Institute of Biostatistics and Analyses. Z.S. is inventor on multiple patents dealing with genetic engineering of immune cells. K.R. received honoraria from Novartis Pharma GmbH. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Weirauch, U., Kreuz, M., Birkenbihl, C. et al. Design specifications for biomedical virtual twins in engineered adoptive cellular immunotherapies. npj Digit. Med. 8, 493 (2025). https://doi.org/10.1038/s41746-025-01809-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41746-025-01809-6