Abstract
Pseudomonas aeruginosa is an emergent threat due to the antimicrobial resistance crisis. Bacteriophages (phages) are promising agents for phage therapy approaches against P. aeruginosa. It has been proposed that metazoans harbor phages on their mucosal surfaces, and this could be exploited for the rational design of prophylactic phage therapy. The goal of this study was to evaluate the potential of phage-mucus interaction to prevent infections caused by P. aeruginosa. We isolated two phages capable of infecting P. aeruginosa. Both are similar in morphology and closely related genetically. However, phage VAC3 is more efficient in replicating in mucin-exposed P. aeruginosa in vitro and is preferentially held in the respiratory tract of C57BL/6 mice. Pre-treatment with VAC3 phage protects mice from a lethal dose of P. aeruginosa while VAC1 does not. This shows that phages adapted to mucosal conditions have potential to be applied as prophylactic measures against an ESKAPE pathogen.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) is an emerging public health problem. It is estimated that 4.95 million deaths in 2019 could be associated with bacterial AMR1 and that by 2050 around 10 million people will die annually from infections caused by multidrug-resistant (MDR), extensively drug-resistant (XDR), or pandrug-resistant (PDR) bacteria2. Therefore, innovative strategies to control the spread of MDR/XDR/PDR microorganisms are needed. Phage therapy, the use of bacteriophages (phages) as antibacterial agents, is considered a potential therapeutic substitution or adjuvant therapy to antibiotics and currently holds immense relevance for scientific and technological development in controlling MDR/XDR/PDR infections3. Developed in the early 20th century, phage therapy was widespread until a shift towards antibiotic therapy took place in the Western World4.
The increase in bacterial resistance has renewed the interest in the therapeutic potential of bacteriophages. There are reports of successful cases in humans where phage therapy has been promising in eliminating infections caused by MDR microorganisms5,6,7,8, such as Acinetobacter baumannii, Mycobacterium abscessus, and Pseudomonas aeruginosa. Infections caused by resistant P. aeruginosa are considered extremely serious and one of the main contemporary problems regarding the AMR crisis9,10,11, especially in cystic fibrosis (CF) patients. Bacteriophages that infect the Pseudomonas genus were first discovered in the mid-20th century, and several species have been isolated and described to date7,12,13. P. aeruginosa was the subject of one of the largest modern phage therapy clinical trials ever conducted (Phagoburn)14. A retrospective analysis of 100 consecutive cases of modern personalized phage therapy in Belgium revealed that 45 cases had the involvement of P. aeruginosa, and that phage therapy led to clinical improvement in 37 of these cases15. Traditional phage therapy is based on curing an already established disease. The benefits of such an approach are indisputable. However, phage therapy also has the potential for being used as prophylaxis, improving its benefits in reducing mortality and morbidity from bacterial infections. The prophylactic use of bacteriophages has been recorded in laboratory conditions and on a large scale in human populations16,17,18, although a clear mechanism explaining their preventive efficacy had not yet been described until recently. Recent research using two different in vivo models based on fish pathogens hints that interactions between phages and mucosal layers could be the explanation behind the prophylactic potential of phages19,20. The connection between mucosal adherence and phage-based prophylaxis of bacterial infections has also been shown in a mice model using Escherichia coli and phage øPNJ-621.
Despite these pioneering studies, little attention has been given to the isolation and characterization of mucosal-adapted phages. A factor to consider in studying phages is their interaction with the environment, particularly with mucosal surfaces in various animals. These surfaces are habitats for many microorganisms, including bacteriophages and their hosts. Mucus is an essential component of the innate immune system and serves as a selective barrier between organisms and their environments19,22,23,24,25. It is known that many organisms host bacteriophages on their mucosal surfaces in a symbiotic relationship, where phages provide protection to hosts against bacterial infections, and the organism provides a means for phages to infect bacterial hosts24. Therefore, the bacteriophage adherence to mucus (BAM) model24 proposes that phages concentrate on mucosal surfaces through weak interactions with mucins, creating an immunity not derived from the host against bacterial invaders during the mucous colonization process. This phage-mucus interaction is primarily carried out by Ig-like phage structural proteins, which have been identified in about 25% of the species studied24. Recently, BAM model dynamics have been shown to apply to a group of four P. aeruginosa phages in vitro. Three of the phages had mucin-binding properties, while two were able to replicate more efficiently in mucin-exposed P. aeruginosa26. This shows that likely phage-mucin interactions can play a role in favoring phage-mediated prophylaxis in P. aeruginosa cases, but no in vivo data was presented.
In this study, we isolated two phages from Brazilian environmental samples and showed that one of them replicates better in mucin-exposed hosts, persists in the respiratory tract of mice, and can be used as prophylaxis against P. aeruginosa infections in vivo. This shows that even phages infecting the same host might possess distinct lifestyles concerning mucosal interactions, and this can be exploited for rational prophylactic approaches against clinically relevant pathogens. Phage therapy based on mucosal-adapted phages holds the potential for the protection of mucosal membranes against infections. The administration of these phages preventively in susceptible patients could prevent bacterial colonization in these anatomical sites, as these phages remain in the body for a longer time after administration. Therefore, it is a potential relevance of phage-mucus interactions for designing phage treatments.
Results
VAC1 and VAC3 are two phages that infect P. aeruginosa
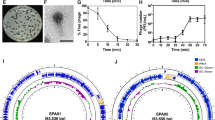
A bioprospection effort aimed at finding phages capable of infecting P. aeruginosa PA14 was made using environmental water samples collected from two southeastern cities from Minas Gerais and São Paulo states. Visible round plaques in double-layer agar overlay plates appeared following the enrichment cultures of two of our samples. These isolates were plaque-purified and further propagated. These two new phages, named P. aeruginosa phage VAC1 and P. aeruginosa phage VAC3, are able to reduce the proliferation of P. aeruginosa PA14 in planktonic cultures. A significant reduction in P. aeruginosa load was seen from 18 to 40 h post infection (p = 0.0002) (Fig. 1a). Transmission electron micrograph (TEM) showed that phages VAC1 and VAC3 have similar-tailed morphologies since both present an icosahedral head and a long tail. TEM images showed that phage VAC1 had a head of 58.82 ± 1.7 nm in diameter and a tail with a length of 183 ± 3.03 nm. Phage VAC3 had a head of 54.75 ± 6.66 nm in diameter and a tail of 202.23 ± 27.62 nm (Fig. 1b). At least 10 viral particles were measured to calculate these dimensions. To verify the influence of mucin in the replication of phages VAC1 and VAC3, P. aeruginosa PA14 strain was infected with either phage in the presence of 0.1% mucin and phage progeny was measured by titration over time. VAC1 titers in the presence and absence of mucin remained similar, while VAC3 titer at 24 h of infection was almost 3 logarithmic orders higher when the infection was done in the presence of mucin (p = 0.0286) (Fig. 1c). Both phages are resistant to different pH and temperatures, with VAC1 likely being more affected by temperature increase (Supplementary Fig. 1).
a Activity of the phages VAC1 and VAC3 against P. aeruginosa PA14. b Transmission electron micrograph of phages VAC1 and VAC3. Scale bar: 50 nm. c VAC1 and VAC3 titers in LB cultures supplemented with 0.1% (w/v) purified porcine mucin at the time of infection. Experiments were performed in three replicates, and the data is represented as the mean ± standard deviation range in a and in c. The phage concentration at the time of infection in (c) was 4 × 104 PFU/ml for all cultures. *p > 0.05 using unpaired t-student test for each individual time point.
Phages VAC1 and VAC3 are genetically similar but not identical
The phages used in this study have circular dsDNA genomes of 36601 (VAC1) and 37663 (VAC3) base pairs (bp) in size (Fig. 2a). Genome comparison of VAC1 and VAC3 phages demonstrate a conserved synteny with a high degree of identity (Fig. 2b), except for a unique region of 1062 bp present in the 5′ region of VAC3 phage. Phylogenetic analysis based on amino acid sequences of homologous major capsid proteins of VAC1 and VAC3 genes showed that they are highly similar and belong to the same lineage (Supplementary Fig. 2). VAC1 genome contains 56 predicted ORFs and a 64.1% G + C content while VAC3 genome contains 57 ORFs as well as a 64.2% G + C content (Supplementary Fig. 3 and Supplementary Data Set 1). The pangenome analysis identified that 55 ORFs are shared between the two phages (Fig. 2c). The annotation indicates that the only gene exclusive to VAC1 is an intermediate transcription factor (VAC1_ORF1) while the two genes exclusive to VAC3 codes for a LysR substrate-binding domain-containing protein (VAC3_ORF1) and a nucleoside-specific channel-forming protein (VAC3_ORF2). Neither phage codes for tRNAs, and both have one predicted anti-CRISPR gene. Regarding phylogeny, VAC1 and VAC3 have over 97% genomic similarity with Casadabanvirus JBD69 (NCBI Reference Sequence: NC_030908.1), as determined by the taxmyPHAGE taxonomy tool27. It is important to note that both phages have ORFs annotated as integrases (VAC1_ORF52 and VAC3_ORF53), meaning that despite being good models in this study, their therapeutical potential in clinics should be considered. However, no experimental evidence of lysogeny has been noted during the experiments shown here.
a Genomic synteny analysis. Comparison of genome composition and organization. The schematic whole-genome-alignment diagram was obtained on the PhageScope platform using the sequence alignment analysis. Red boxes indicate asyntenic regions, and blue arrows indicate the position and orientation of the ORFs. b Dot plot showing genetic collinearity and identity between covered regions. c A network graph illustrating the components of the pangenome. The pangenome comprises a total of 113 genes grouped in 57 clusters, of which 54 are shared, constituting the core genome. Larger nodes represent the bacteriophages VAC1 (1) and VAC3 (3). Smaller nodes represent individual clusters, with each color indicating its functional category in the final annotation. The graph was generated using a force-based algorithm.
Phage VAC3 is preferentially retained in the respiratory tract of mice
To verify the possibility of VAC1 and VAC3 adhering and persisting in the primary mucosal surface of the respiratory tract, C57BL/6 mice were treated via the nasal route with either phage. After 24 h, mice were euthanized, and the presence of phages in the trachea and lungs was checked by collecting tissue and quantifying the phages by titration. The material collected was divided between the trachea, lungs (Fig. 3a), and nasal wash. Phage titers indicate that phage VAC1 and VAC3 have a differential ability to persist in the different regions of the respiratory tract of the mice. As shown in Fig. 3b, phage VAC3 was recovered from the nasal wash and lungs of 75% and in the trachea of all treated mice. VAC1 was not detected in the nasal wash and trachea of the treated mice, being found in only one mouse lung in low titers. No phage plaques appeared when samples from the control group (mice not treated with phages) were tested, indicating the absence of detectable endogenous phages.
a Representative figure showing the regions of the respiratory tract of mice used to quantify the bacteriophages. The respiratory tract was excised aseptically, and the trachea (T) and lungs (L) were used separately to quantify phages. The red line in A indicates the separation between the trachea and lungs. b Quantification of VAC1 (n = 4) and VAC3 (n = 4) in the nasal wash (pfu/mL), trachea (pfu/g), and lungs (pfu/g) of mice treated with these bacteriophages. The individual replicates are shown in (b), with the SD and mean marked in the graph. The limit of detection in b was 100 pfu/ml.
Pretreatment with phage VAC3 improves survival and clinical signs when followed by a lethal dose of P. aeruginosa
After verifying the persistence of phage VAC3 along the respiratory system of mice, we checked whether pre-exposure with our phages could protect C57BL/6 mice. Therefore, mice pretreated via the nasal route with phages were infected 24 h after the phage treatment with a lethal dose of P. aeruginosa PA14, using the same route. During the course of infection, clinical parameters were followed. No significative difference was seen regarding the weight of mice in the different tested conditions (Fig. 4a), despite a drop in weight for the mice infected with the bacterium without phage pretreatment. However, mice in the groups pretreated with phages and then infected had fewer clinical signs compared to the non-pretreated mice, with a stronger reduction seen for the VAC3 pretreated group (p = 0.0025 in 24 h and p = 0.0147 in 48 h) (Fig. 4b). Both groups pretreated with phages had a higher survival rate when compared to the group not treated with phages (Fig. 4c). Of note, survival was higher and significant for the VAC3 pretreated group when compared to the non-treated group (p = 0.0122). One mouse survived the lethal dose of P. aeruginosa in the VAC1 pretreated group, while three survived in the VAC3 pretreated group. In addition, none of the mice treated with phages alone (no P. aeruginosa challenge) had recorded clinical symptoms or mortality, demonstrating that treatment with phages did not cause significant systemic changes. The quantification of bacteria in the lungs of mice shows that only treatment with phage VAC3 was able to reduce the number of colony-forming units (CFU) per milligram of tissue compared to the group of infected mice (p = 0.047) (Fig. 4d).
a Weight loss of mice during the first 24 h of pretreatment with phages and the following 48 h after infection with P. aeruginosa PA14 strain. The red arrow indicates the moment in which P. aeruginosa PA14 was administered to the mice. b Clinical sign scores in bacteriophages-treated mice after infection with P. aeruginosa PA14 strain 24- and 48-h post-infection with P. aeruginosa. Note that the 24 h and 48 h time points are counted after the administration of P. aeruginosa PA14. c Survival curve of phage-treated mice after infection with P. aeruginosa PA14 strain. The mice were monitored for 48 h after infection or until reaching a clinical score of 15 or death. The numbers of deaths and survivors were recorded. Note that the VAC1 and VAC3 curves are superposed (no death in both groups). d Quantification of colony forming units in the lungs of phage-treated mice after infection with P. aeruginosa PA14 strain. ND not detected. The graph shows the average and SD of each group comprised of four mice. The individual replicates plus SD are shown in (b) and (d). **Significance of p < 0.01 compared to PA14 group. *Significance of p < 0.05 compared to PA14 group. All groups had 4 mice (n = 4). The results were analyzed using Kruskal–Wallis non-parametric ANOVA for (a, b and d). Survival curves c were analyzed using Log-rank (Mantel–Cox) test.
Histopathology confirms the efficacy of VAC3 in diminishing P. aeruginosa damage
Biological material collected from the infection experiment shown in Fig. 4 was used for histopathology and P. aeruginosa colony counting. Histopathological analysis of lung sections showed areas of intense inflammation in all groups of mice infected with PA14 when compared to non-infected ones. However, lung injury was more intense in the PBS and VAC1 pretreated group compared to the VAC3 pretreated group (p = 0.0003 and p = 0.0018, respectively) (Fig. 5a, b). Infection with PA14 induced a greater loss of the alveolar architecture in the lung in all groups, with diminished effect in VAC3-treated mice. Thickening of the alveolar septum, foci of vascular congestion, an intense interstitial inflammatory infiltrate, and the presence of exudates in the PBS and VAC1 treated group after infection were observed. In contrast, VAC3 pretreated mice showed a similar lung architecture to non-infected mice (Fig. 5a). The lung injury histopathological score was similar between PBS and VAC1 groups after infection with PA14 (Fig. 5b). In contrast, mice treated with VAC3 showed a reduction of all inflammatory parameters after infection demonstrating a lower interstitial inflammatory infiltrate, little alveolar septum thickening and some few vascular congestion (Fig. 5a, b). The presence of phages in the lungs of infected mice was also determined. Phage titration from the lung indicated that only one mouse from VAC1 and one from VAC3 treated groups (not infected with P. aeruginosa) were positive for phage detection after 48 h of phage administration dose (Fig. 5c). When the same analysis was done in mice that were pretreated with phages and then infected with P. aeruginosa, phage VAC1 was detected in only one individual while all mice were positive for VAC3 phage (Fig. 5c).
a Histopathological analysis of the lungs of C57BL/6 mice that were pre-treated with bacteriophages VAC1 and VAC3 before P. aeruginosa infection. Representative photomicrographs of the lung region in a 20× objective. Staining with hematoxylin and eosin (HE); magnification at 20×; scale bar: 100 μm. b Histopathology scores of lungs from C57BL/6 mice pre-treated with bacteriophages VAC1 and VAC3 before P. aeruginosa infection. c Quantification of VAC1 and VAC3 by titration in the lungs (pfu/g) of mice treated with both phages. NI is non-infected with P. aeruginosa. The individual replicates plus SD are shown in (b) and (c). *** Significance of p < 0.0001 compared to the non-infected group. The results were analyzed using an unpaired t-student test.
Discussion
P. aeruginosa is an opportunistic pathogen responsible for a wide range of acute and chronic infections with significant levels of morbidity and mortality. This bacterium is frequently found infecting patients with cystic fibrosis, cancer, severe burns, and also immunocompromised patients10,11,28,29,30. A complicating factor of the infection by P. aeruginosa is the presence of MDR and XDR isolates, which in turn compromise the antimicrobial therapy and increase the mortality and morbidity rates of the populations affected by this infection31,32. In vitro and in vivo studies have already demonstrated that bacteriophages are promising agents in the elimination or inhibition of P. aeruginosa MDR when used with a curative approach7,12,13,21,30,33,34,35,36,37,38,39,40,41,42.
A factor to be considered in the study of phages is their interaction with the environment, especially with the mucosal surfaces of various animals. These surfaces are habitats for microorganisms, including bacteriophages19,22,23,24,25. The BAM model suggests that phages would concentrate on the mucosal surfaces through weak interactions with mucins, creating a ubiquitous non-host-derived immunity against bacterial invaders during the mucus colonization process19. Additionally, some bacteria were already shown to have increased susceptibility to mucosal-adapted phages when exposed to mucin. This has been shown directly for Flavobacterium columnare, Aeromonas sp, Streptococcus mutans, A. veronii and P. aeruginosa phages19,20,26,43 and indirectly for Clostridioides difficile and E. coli phages44,45. This points out that likely phages associated with mucosal layers are effective predators of bacteria in their mucosal-invasive physiological state in nature. Phage therapy based on phages that bind to mucus has the potential to act as a preventive approach to protect the mucosa against infections that affect these sites19. The impact of phage-mucosa dynamics for phage-based prophylaxis of bacterial infections has already been shown in vivo for three distinct bacterial pathogens: F. columnare19, A. veronii20, and E. coli21.
In this work, we isolated two P. aeruginosa PA14 infecting phages from environmental water samples collected in Brazil. Despite being genetically and morphologically similar to each other, phage VAC3 was more efficient in proliferating in mucin-exposed P. aeruginosa in vitro, whereas VAC1 was not. Differential response of similar phages to mucin was also recently recorded in a different collection of P. aeruginosa phages from the Eliava Institute in Georgia26. When tested in vivo, VAC3 was preferentially held in the respiratory system of mice, while VAC1 was not. VAC3 retention was higher in the trachea, an organ that has areas containing large clouds of mucus along the respiratory apparatus46. The absence of VAC1 phage along the trachea and nasal region of the treated mice could be associated with the passage of the virus to the lungs without adherence to areas containing primary mucus. The mechanism behind VAC3 attachment to the upper respiratory tract is still not understood, and it is important to note that this phage lacks the canonical HOC protein shown to mediate phage-mucin interactions in T4 phage14. Differential stability between VAC1 and VAC3 is likely not an issue, given that both phages are similar and survived equally under different temperature and pH treatments. Pretreatment of mice with VAC3 resulted in a higher survival rate, fewer clinical signs, and lower bacterial counts in the lungs of the mice. This protection could be associated with the fact that the presence of VAC3 phages in the nasal cavity and trachea before infection could favor the infection of bacteria by this phage at these two anatomical sites. This could, at the same time, reduce live P. aeruginosa cells in the lungs and increase the production of a new viral progeny of VAC3 phage in the infected tissues. The lower degree of tissue injury and inflammatory parameters in mice pre-treated with VAC3 phages in comparison to VAC1, together with the high detection rate of VAC3 phage titers in the lungs of all mice after infection, strengthen this hypothesis.
So far, the rational prophylactic use of phages based on mucosal interactions in vivo has been shown in two different fish disease models19,26 and against E. coli in BALB/c mice21. To our knowledge, this is the first time the BAM model dynamics were applied to prevent P. aeruginosa infections in vivo, pointing out to the potential relevance of such an approach against ESKAPE pathogens. The success of the use of phage therapy against P. aeruginosa in vitro and in vivo models is already well established30,40,41,42. The main difference between those studies and our study is the use and validation of preventive phage therapy based on a phage that remains attached to the upper respiratory tract and replicates more efficiently in the presence of mucin. This approach could be important to prevent P. aeruginosa infections in patients with CF. CF is a genetic-based disease, characterized by higher concentrations of mucins in the lungs of affected individuals. This scenario contributes to the establishment of a chronic inflammatory response in the lungs and also airway obstruction47. Together with this hypersecretion mucus phenotype, there is a chronic bacterial infection that could, in turn, exacerbate lung inflammation48. One of the major bacterial pathogens infecting CF patients is P. aeruginosa49. Therefore, the high frequency of antibiotic consumption by CF patients contributes to an increased rate of MDR pathogens in the lungs, including P. aeruginosa50. A multi-center clinical trial showed that phage therapy in CF patients infected with P. aeruginosa showed that the phage cocktail was well tolerated and also reduced bacterial load in the lungs of participants51.
Besides a high degree of identity, the VAC3 phage genome is 1062 base pairs longer than the VAC1 phage genome. Pangenome analysis showed that VAC1 has one unique ORF (VTF3S_VAR67 Intermediate transcription factor 3 small subunit). In general, Viral Intermediate transcription factors (VIF) are proteins that can act with host RNA polymerase to initiate transcription from viral intermediate gene promoters or inhibit host cell transcription52. VAC3 genome has two unique ORFs, a LysR substrate-binding domain-containing protein and a Nucleoside-specific channel-forming protein tsx. The LysR substrate-binding domain-containing protein is a transcription factor widely expressed in bacteria. When a LysR protein recognizes a specific substrate, there are conformational changes that modulate the protein’s DNA-binding affinity and regulatory activity53. The other unique ORF in VAC3 codifies a potential nucleoside-specific channel-forming protein tsx. This protein is a porin expressed in the outer membrane of some Gram-negative bacteria and is described as a substrate-specific channel for nucleosides and deoxynucleosides. It also could act as a receptor for colicin K and bacteriophage T654,55. It was also demonstrated that mutations in the Tsx porin gene result in bacterial resistance to phage infection56,57. Therefore, the presence of these ORFs does not correlate directly with the differential mucosal retention of VAC1 and VAC3, since these ORFs are not identified as structural proteins. However, we could not exclude that these ORFs could be indirectly related to mucin or mucus response in phage-host interaction. VTF3S_VAR67 Intermediate transcription factor 3 small subunit and LysR substrate-binding domain-containing protein are involved in transcription regulation, and the activity of these transcription factors could be modulated by mucin exposure of the host. Additionally, the nucleoside-specific channel-forming protein tsx expressed by VAC3 could positively influence the infection of P. aeruginosa by this phage since once this protein channel is expressed in the outer membrane of infected bacteria, nucleoside transport across the bacterial is facilitated, and therefore precursors for viral nucleic acid synthesis. Future studies should be done to verify the real role of these ORFs in the differential mucin-binding phenotype, if any.
In conclusion, here we show the differential ability of two newly isolated P. aeruginosa phages to respond to mucin, persist in mucus-producing tissues, and protect mice against a lethal dose of the bacterium. Phage therapy based on mucosal-adapted phages has a protective potential against infections that affect mucosal surfaces, as the administration of these phages preventively could prevent bacterial colonization. Mucosal-adapted phages could be used for the development of preventive technologies (e.g., nasal sprays, sanitizing solutions for environmental use, or use in surgical equipment) that could act as preventive methods against infection in people linked to the hospital environment, enabling the protection of patients at risk as well as the protection of visitors/companions and health professionals.
Methods
Pseudomonas aeruginosa
P. aeruginosa strain PA14 was cultured in lysogeny broth (LB)58 at 37 °C and 150 rpm. In some experiments, cultures were made in LB supplemented with 0.1% of mucin (w/v). Mucin stocks were prepared by the autoclavation of a 2% mucin solution in water. The mucin used was purified porcine mucin (Sigma), shown previously to be a relevant model for studying phage–mucin interactions in the concentration used19,26.
Bacteriophage isolation and propagation
Bacteriophages that infect the P. aeruginosa PA14 strain were isolated from environmental water samples using the enrichment method and a double-layer agar overlay58. Environmental water samples were collected from two cities in southeast Brazil (Alfenas, Minas Gerais State—21° 26′ 0″ South, 45° 57′ 0″ West and Mococa, São Paulo State- 21° 28′ 0″ South, 47° 1′ 0″ West). Samples were filtered through 0.45 μm pore size filters, and 3 mL of the filtrates were mixed with 1 mL of 5× LB and 1 mL of a 48-h culture of P. aeruginosa for preparing the enrichment cultures. After 24 h incubation at 37 °C and 150 rpm, the cultures were centrifuged at 4000g for 30 min at 25 °C, and the supernatants were filtered through a 0.45 μm pore size filter. Dilutions (10−1 to 10−5) of enriched samples were plated by the overlay agar method containing PA14 strain. Plaques with differential morphology were picked and suspended in TM buffer (50 mM Tris–HCl, 10 mM MgCl2, pH 7.4). This suspension was further 10-fold serially diluted and used for plaque re-isolation. To ensure purity, each plaque was re-isolated three times before the experiments.
Transmission electron microscopy
Ten microlitres of a phage lysate solution (1 × 1010 pfu/mL) were added on a carbon film over the copper grid for negative staining with 2% phosphotungstic acid (pH 6.7). The grids were imaged in a Tecnai G2-12—FEI Spirit Biotwin 120 keV transmission electron microscope.
Effect of pH and temperature on phage stability
One hundred microliters of a phage suspension (1 × 1011 pfu/mL) was added to tubes containing 0.9 mL of sterile buffered saline with pH levels adjusted between 3 and 9 using 0.1 M HCl or NaOH. The phages were then incubated at 37 °C for 1 h, followed by the determination of phage titers using the double-layer agar method. Similarly, 100 μl of phage suspension was added to tubes containing 0.9 mL of sterile buffered saline and allowed to incubate in a heat block ranging from 30 °C to 60 °C for 1 h. Phage titers were determined using the double-layer agar method after the incubation period.
Sequencing, assembly, and annotation
Phage DNA was extracted from 1 mL of high-titer phage lysates (1 × 1010 pfu/mL) utilizing the ZnCl2 method as described by Santos et al.59. Following filtration, DNase (10 µg/mL) (Thermo Fischer) and RNase (10 µg/mL) (Thermo Fischer) were added to the lysates and incubated at 37 °C for 1 h. Subsequently, the particles were concentrated using 0.2 M filtered ZnCl2 (Sigma) and treated with 1 mg/mL of Proteinase K. Finally, DNA extraction was performed using a DNA extraction kit (NucleoSpin Blood Kit, Macherey-Nagel).
For DNA Library preparation, the Ready-To-Go™ GenomiPhi™ V3 DNA system (Cytiva®) was used following the manufacturer’s instructions to amplify DNA. The concentration of each sample was determined using the Qubit™ dsDNA HS Assay Kit (Invitrogen®), with initial concentrations of 4.73 ng/μL for VAC1 and 8.3 ng/μL for VAC3 genomic DNA. Each sample was diluted in nuclease-free water to achieve an input of approximately 10 ng in a 10 μL volume. After amplification, the concentrations were 357 ng/μL for VAC1 and 378 ng/μL for VAC3, respectively. The NGS library was prepared using the COVIDSeq kit (Illumina®), following the manufacturer’s procedural guide. The process began with the Tagment PCR Amplicons step, using 357 ng for VAC1 and 378 ng for VAC3 in a total volume of 20 μL per sample, adjusted with nuclease-free water as needed. The final library concentrations were 29.6 ng/μL for VAC1 and 26.9 ng/μL for VAC3, with an average fragment size of 434 bp for both. Sequencing was performed on a MiSeq system using a 600-cycle V3 flow cell.
The genomes were assembled de novo using Spades 3.12 software with the default parameters60,61. Synteny analysis was performed on PhageScope62. Open reading frames (ORFs) were predicted using Prodigal63. All of the predicted ORFs were annotated manually using BLASTp (considering a cutoff of <10−3), HHpred (considering only hits with probabilities greater than 80% and E value greater than or equal to 1 found in the PDB_mmCIF70_8_Mar, SCOPe70_2.08 and UniProt-SwissProt-viral70_3Nov_2021 databases) and InterPro, during March of 202464,65,66. The pangenome was constructed using the OrthoFinder 2.5.4 software with an MCL inflation parameter equal to 467. To visualize the constructed pangenome, a network was built using Gephi68. The layout was generated using a force-based algorithm.
For phylogenetic analysis, amino acid sequences of homologous major capsid proteins were obtained from the National Center for Biotechnology Information database using the BLASTp (<10−5) and aligned using the MUSCLE algorithm. After aligning these sequences, the best-fit substitution models were selected by the ModelFinder algorithm implemented in IQtree69. The phylogenetic tree was reconstructed using IQtree software (version 1.6.12)70, using the maximum-likelihood method and 1000 bootstrap replicates. Finally, the phylogenetic tree was visualized and edited using iTOL71.
Phage infections
To analyze the effect of our phages in liquid cultures of P. aeruginosa PA14, overnight cell cultures were adjusted to 2 × 108 cells/mL in sterile saline solution (0.9% NaCl). A 100 μl of cell suspension was inoculated in 5 mL of LB medium followed by incubation at 37 °C with regular shaking (120 rpm). The culture was infected with phages at the multiplicity of infection (MOI) of 0.01. After the desired times of incubation, samples were taken to quantify the number of cells by spectrophotometry at an optical density of 600 nm. To evaluate the influence of mucin supplementation in phage multiplication, overnight cultures were diluted to 1:10,000 and inoculated to LB supplemented with 0.1% mucin. Control cultures consisted of an LB without mucin. All samples were incubated at 37 °C for 4 h and after they were infected with phages at MOI of 0.01 After 4, 24, and 48 h of infection, samples were collected (100 µl each), mixed with 10 μl of chloroform, and titrated as described above.
In vivo binding of phages throughout the respiratory system of mice
To verify the ability of the phages to persist in the respiratory system, mice anesthetized intraperitoneally with xylazine (10 mg/kg) were treated with 10 μL of bacteriophages diluted in SM buffer (200 mM NaCl2 10 mM MgSO4 50 mM Tris-HCl, pH 7.5) via nasal route in each nostril (2 × 108 PFU/nostril). After 24 h, all mice were euthanized through a lethal dose of anesthetics (ketamine at 300 mg/kg and xylazine at 30 mg/kg). The respiratory system of each individual was externalized and divided into tracheal and pulmonary portions. The organs (trachea and lungs) were weighted and homogenized in 500 μl of sterile PBS. The homogenates were centrifuged at 10,000 rpm for 5 min, and the supernatant was used for bacteriophage titration. The nasal wash was collected using sterile PBS as described72. Briefly, after extracting the respiratory system, a syringe was inserted into the larynx region, and a sterile 1.5 mL tube was inserted in the nostril. Lavages were performed with 500 μl of sterile PBS, which was flushed through the larynx and collected through the nares in a 1.5 mL microtube. The collected sample was centrifuged as described above, and a supernatant was used for bacteriophage titration.
Prophylactic effect of phages against a P. aeruginosa challenge in vivo
Mice were treated with phages as described in the topic above. Twenty-four hours after the phage treatment, mice were anesthetized with xylazine (10 mg/kg) and challenged with 10 μL of P. aeruginosa PA14 in LB medium via the nasal route in each nostril (4 × 107 colony forming units/nostril). Mice were monitored every 8 h post infection, and survival was assessed until 48 h after infection. Clinical parameters were evaluated using the clinical parameters described by Cigana et al.73. Based on these parameters, a final total score of up to 15 points was calculated as a global indicator of the clinical symptoms (Supplementary Table 1). The right lungs of mice that died or reached an endpoint clinical score of 15 in this period were collected to quantify the number of colony-forming units (CFU) of P. aeruginosa per milligram of tissue. The left lungs of mice were preserved for histopathological analysis. For mice that survived beyond 48 h, euthanasia was performed through a lethal dose of anesthetics, and both right and left lungs were acquired for analysis, following the previously outlined procedure.
Lung microbiological and Histological analysis
The right lung from each mouse was excised and homogenized in 1 mL of sterile PBS. Subsequently, 100 μl of serially diluted lung homogenate samples were inoculated to LB agar. The culture plates underwent incubation at 37 °C, and the plates were examined for the presence of P. aeruginosa colonies after 24 h. For phage quantification, 100 μl of lung homogenates were mixed with 10 μl of chloroform and titrated as described above. The left lung from each specimen was preserved using 10% formaldehyde in PBS at room temperature. Tissues were submitted to dehydration in ethanol and embedded in paraffin. Semi-serial sections, measuring 5 μm in thickness, were stained with hematoxylin and eosin, and tissue slides were examined utilizing a Carl Zeiss Axio ScopeA1 microscope equipped with a Canon G10 Power Shot digital microscope camera. For both histopathological and morphometric examinations, 10 randomly selected histological images were acquired for analysis. The sections were evaluated in a double-blind way concerning lung injury was assessed using a modified scoring system, which comprised four categories: edema, hemorrhage, leukocyte infiltration, and vasocongestion. Each category was assigned a score ranging from 0 to 3. The total lung injury score was determined by adding together the scores from each category. Thus, the maximum possible score for all histological parameters combined was 1274. Additionally, the loss of alveolar architecture in the lungs of mice was also measured. In this case, this parameter was graded from 0 (normal alveolar architecture) to 4 (severe loss of alveolar architecture).
Statistics and reproducibility
Graphs and statistical analysis were made using GraphPad Prism version 9.3.1 (GraphPad Software, San Diego, California, USA). Conditions were tested in triplicates, and unpaired t-tests were used to compare controls and tested conditions whenever necessary. Some results were analyzed using Kruskal–Wallis non-parametric ANOVA with two or more groups present. Survival curves were analyzed using Log-rank (Mantel–Cox) test. A p value below 0.05 was considered to be significative and the precise p values are indicated in the text or in the figures. Group size was calculated to be four mice in each tested condition the second equation from Dell et al.75, as recommended by the local regulations. The figure legends contain specific information regarding replicates for each experiment. The raw data used for preparing the graphs in this paper are provided in the Supplementary Data set 2.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data used for making the graphs can be found in Supplementary Data set 2. The phages used in this study are not available in biological collections but could be shared after consideration of local regulations and MTA signing with interested parties. Phage genomes are deposited in Genbank under the following accession numbers PQ466444 (VAC1) and PQ466445 (VAC3).
References
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
de Kraker, M. E. A., Stewardson, A. J. & Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLOS Med. 13, e1002184 (2016).
Almeida, G. M., de, F. & Sundberg, L. R. The forgotten tale of Brazilian phage therapy. Lancet Infect. Dis. 20, e90–e101 (2020).
Gordillo Altamirano, F. L. & Barr, J. J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32, e00066–18 (2019).
Fungo, G. B. N. et al. Two Is Better Than One”: The multifactorial nature of phage-antibiotic combinatorial treatments against ESKAPE-induced infections. PHAGE 4, 55–67 (2023).
Ragupathi, N. K. D. et al. Phage-based therapy against biofilm producers in gram-negative ESKAPE pathogens. Microb. Pathog. 178, 106064 (2023).
Chang, R. Y. K. et al. Bacteriophage PEV20 and ciprofloxacin combination treatment enhances removal of pseudomonas aeruginosa biofilm isolated from cystic fibrosis and wound patients. AAPS J. 21, 49 (2019).
Cieślik, M., Bagińska, N., Górski, A. & Jończyk-Matysiak, E. Animal models in the evaluation of the effectiveness of phage therapy for infections caused by gram-negative bacteria from the ESKAPE Group and the reliability of its use in humans. Microorganism 9, 206 (2021).
Papanikolopoulou, A. et al. Ventilator-associated pneumonia, multidrug-resistant bacteremia and infection control interventions in an intensive care unit: analysis of six-year time-series data. Antibiotics 11, 1128 (2022).
Qin, S. et al. Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 7, 199 (2022).
Paprocka, P. et al. Pseudomonas aeruginosa infections in cancer patients. Pathogens 11, 679 (2022).
Akturk, E. et al. Synergistic action of phage and antibiotics: parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics 8, 103 (2019).
Chaudhry, W. N. et al. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS ONE 12, e0168615 (2017).
Jault, P. et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45 (2019).
Pirnay, J. P. et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: a multicentre, multinational, retrospective observational study. Nat. Microbiol. 9, 1434 (2024).
Chanishvili, N. A Literature Review of the Practical Application of Bacteriophage Research. (Nova Biomedical Books, 2012).
Ahmadi, M., Amir Karimi Torshizi, M., Rahimi, S. & Dennehy, J. J. Prophylactic Bacteriophage administration more effective than post-infection administration in reducing Salmonella enterica serovar Enteritidis shedding in quail. Front. Microbiol. 7, 1253 (2016).
Yen, M., Cairns, L. S. & Camilli, A. A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 8, 14187 (2017).
Almeida, G. M. F., Laanto, E., Ashrafi, R. & Sundberg, L. R. Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. MBio 10, e01984–19 (2019).
Wu, Z. et al. Bacteriophage adhering to mucus provide protection against Aeromonas veronii infection in scaleless fish. Aquaculture 586, 740804 (2024).
Wu, J. et al. Bacteriophage defends murine gut from Escherichia coli invasion via mucosal adherence. Nat. Commun. 15, 1–17 (2024).
Chin, W. H. et al. Bacteriophages evolve enhanced persistence to a mucosal surface. Proc. Natl Acad. Sci. USA 119, e2116197119 (2022).
Barr, J. J., Youle, M. & Rohwer, F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage 3, e25857 (2013).
Barr, J. J. et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl Acad. Sci. USA 110, 10771–10776 (2013).
Bakshani, C. R. et al. Evolutionary conservation of the antimicrobial function of mucus: a first defence against infection. npj Biofilms Microbiomes 4, 1–12 (2018).
Almeida, G. et al. Relevance of the bacteriophage adherence to mucus model for Pseudomonas aeruginosa phages. Microbiol. Spectr. 12, e0352023 (2024).
Millard, A. et al. taxmyPHAGE: automated taxonomy of dsDNA phage genomes at the genus and species level. bioRxiv https://doi.org/10.1101/2024.08.09.606593 (2024).
Shariati, A. et al. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. N. Microbes N. Infect. 21, 75–80 (2017).
Blanchard, A. C. & Waters, V. J. Opportunistic pathogens in cystic fibrosis: epidemiology and pathogenesis of lung infection. J. Pediatr. Infect. Dis. Soc. 11, S3–S12 (2022).
Orenti, A., Mei-Zahav, M., Boracchi, P., Lindblad, A. & Shteinberg, M. Prevalence, trends and outcomes of long-term inhaled antibiotic treatment in people with cystic fibrosis without chronic Pseudomonas aeruginosa infection—ā European cystic fibrosis patient registry data analysis. J. Cyst. Fibros. 22, 103–111 (2023).
Kunz Coyne, A. J., El Ghali, A., Holger, D., Rebold, N. & Rybak, M. J. Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 11, 661–682 (2022).
Horcajada, J. P. et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 32, e00031–19 (2019).
Ahiwale, S. et al. In vitro management of hospital Pseudomonas aeruginosa biofilm using indigenous T7-like lytic phage. Curr. Microbiol. 62, 335–340 (2011).
Kim, S., Rahman, M., Seol, S. Y., Yoon, S. S. & Kim, J. Pseudomonas aeruginosa bacteriophage PA1Ø requires type IV Pili for infection and shows broad bactericidal and biofilm removal activities. Appl. Environ. Microbiol. 78, 6380 (2012).
Mapes, A. C., Trautner, B. W., Liao, K. S. & Ramig, R. F. Development of expanded host range phage active on biofilms of multi-drug resistant Pseudomonas aeruginosa. Bacteriophage 6, e1096995 (2016).
Latz, S. et al. Differential effect of newly isolated phages belonging to PB1-Like, phiKZ-Like and LUZ24-like viruses against multi-drug resistant Pseudomonas aeruginosa under varying growth conditions. Viruses 9, 315 (2017).
Adnan, M. et al. Isolation and characterization of bacteriophage to control multidrug-resistant Pseudomonas aeruginosa planktonic cells and biofilm. Biologicals 63, 89–96 (2020).
Waters, E. M. et al. Phage therapy is highly effective against chronic lung infections with Pseudomonas aeruginosa. Thorax 72, 666–667 (2017).
Jeon, J. & Yong, D. Two novel bacteriophages improve survival in Galleria mellonella infection and mouse acute pneumonia models infected with extensively drug-resistant Pseudomonas aeruginosa. Appl. Environ. Microbiol. 85, e02900–e02918 (2019).
Ferry, T. et al. Personalized bacteriophage therapy to treat pandrug-resistant spinal Pseudomonas aeruginosa infection. Nat. Commun. 13, 4239 (2022).
Racenis, K. et al. Use of phage cocktail BFC 1.10 in combination with ceftazidime-avibactam in the treatment of multidrug-resistant Pseudomonas aeruginosa femur osteomyelitis—a case report. Front. Med. 9, 851310 (2022).
Camens, S. et al. Preclinical development of a bacteriophage cocktail for treating multidrug resistant Pseudomonas aeruginosa infections. Microorganisms 9, 2001 (2021).
Sundberg, L. R., Rantanen, N. & De Freitas Almeida, G. M. Mucosal environment induces phage susceptibility in Streptococcus mutans. PHAGE Ther. Appl. Res. 3, 128 (2022).
Shan, J. et al. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci. Rep. 8, 1–8 (2018).
Green, S. I. et al. Targeting of mammalian glycans enhances phage predation in the gastrointestinal tract. MBio 12, 1–18 (2021).
Fakih, D. et al. Normal murine respiratory tract has its mucus concentrated in clouds based on the Muc5b mucin. Am. J. Physiol. Lung Cell. Mol. Physiol. 318, L1270–L1279 (2020).
Carbone, A., Vitullo, P., Di Gioia, S. & Conese, M. Lung inflammatory genes in cystic fibrosis and their relevance to cystic fibrosis transmembrane conductance regulator modulator therapies. Genes 14, 1966 (2023).
Testa, I., Crescenzi, O. & Esposito, S. Gut dysbiosis in children with cystic fibrosis: development, features and the role of gut-lung axis on disease progression. Microorganisms 11, 9 (2022).
Dupont, C. et al. Highly diverse dynamics of Pseudomonas aeruginosa colonization from initial detection in cystic fibrosis patients: a 7-year longitudinal genetic diversity study. Infect. Genet. Evol. 115, 105513 (2023).
Perikleous, E. P. et al. Antibiotic resistance in patients with cystic fibrosis: past, present, and future. Antibiotics 12, 217 (2023).
DEL REY, M. Armata Pharmaceuticals Announces Positive Topline Data from Phase 1b/2a SWARM-P.a. Clinical Trial of Inhaled AP-PA02 in Patients with Cystic Fibrosis. (Armata Pharmaceuticals, Inc., 2023) https://www.prnewswire.com/news-releases/armata-pharmaceuticals-announces-positive-topline-data-from-phase-1b2a-swarm-pa-clinical-trial-of-inhaled-ap-pa02-in-patients-with-cystic-fibrosis-301762961.html.
Sanz, P. & Moss, B. Identification of a transcription factor, encoded by two vaccinia virus early genes, that regulates the intermediate stage of viral gene expression. Proc. Natl Acad. Sci. USA 96, 2692–2697 (1999).
Maddocks, S. E. & Oyston, P. C. F. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623 (2008).
Schneider, H., Fsihi, H., Kottwitz, B., Mygind, B. & Bremer, E. Identification of a segment of the Escherichia coli Tsx protein that functions as a bacteriophage receptor area. J. Bacteriol. 175, 2809–2817 (1993).
Nieweg, A. & Bremer, E. The nucleoside-specific Tsx channel from the outer membrane of Salmonella typhimurium, Klebsiella pneumoniae and Enterobacter aerogenes: functional characterization and DNA sequence analysis of the tsx genes. Microbiology 143, 603–615 (1997). (Pt 2).
Kortright, K. E., Doss-Gollin, S., Chan, B. K. & Turner, P. E. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front. Microbiol. 12, 658374 (2021).
Li, P. et al. Screening of polyvalent phage-resistant Escherichia coli strains based on phage receptor analysis. Front. Microbiol. 10, 439623 (2019).
Bertani, G. STUDIES ON LYSOGENESIS I: the mode of phage liberation by lysogenic Escherichia coli1. J. Bacteriol. 62, 293 (1951).
Santos, M. A. An improved method for the small scale preparation of bacteriophage DNA based on phage precipitation by zinc chloride. Nucleic Acids Res. 19, 5442 (1991).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. https://home.liebertpub.com/cmb (2012).
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A. & Korobeynikov, A. Using SPAdes de novo assembler. Curr. Protoc. Bioinforma. 70, e102 (2020).
Wang, R. H. et al. PhageScope: a well-annotated bacteriophage database with automatic analyses and visualizations. Nucleic Acids Res. 52, D756–D761 (2024).
Hyatt, D. et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 11, 1–11 (2010).
Söding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33, W244–W248 (2005).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344–D354 (2021).
Quevillon, E. et al. InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120 (2005).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 3, 361–362 (2009).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., Von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296 (2021).
Schenck, L. P. et al. Nasal tissue extraction is essential for characterization of the murine upper respiratory tract microbiota. mSphere 5, e00562–20 (2020).
Cigana, C. et al. Efficacy of the novel antibiotic POL7001 in preclinical models of Pseudomonas aeruginosa pneumonia. Antimicrob. Agents Chemother. 60, 4991 (2016).
Yamanel, L. et al. Ozone therapy and hyperbaric oxygen treatment in lung injury in septic rats. Int. J. Med. Sci. 8, 48–55 (2011).
Dell, R. B., Holleran, S. & Ramakrishnan, R. Sample size determination. ILAR J. 43, 207–213 (2002).
Acknowledgements
This work was funded by the Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG – APQ-05218-23; APQ-00544-23 and APQ-04845-23); The Coordenação de Aperfeicoamento de Pessoal de Nivel Superior, Brazil (CAPES) (Finance Code 001); the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq – Grants Number 408147/2022-9 and 308468/2021-0) and the Centre for New Antibacterial Strategies (CANS) of the Arctic University of Norway (project ID #2520855).
Funding
Open access funding provided by UiT The Arctic University of Norway (incl University Hospital of North Norway).
Author information
Authors and Affiliations
Contributions
L.F.L.C., L.C.C.M., and G.M.F.A. designed the experimental setup, analyzed the data, and drafted the paper. M.S.T., S.P.L.N., R.P.A., A.L.C.S., W.P.G., B.A.C., G.A.C., M.F.R.D., L.S., G.S.M., and M.L.C.G. were responsible for in vitro and in vivo phage experiments. C.E.O. was responsible for histopathology analysis. E.L.H.C., G.S.A., and J.P.A.J. sequenced phage genomes. V.F.Q. was responsible for bioinformatic analysis. All authors contributed to the writing.
Corresponding author
Ethics declarations
Competing interests
We declare that G.M.F.A. is one of the owners of a patent titled “Improved methods and culture media for production, quantification and isolation of bacteriophages” (FI20185086, PCT/FI2019/050073). All the other authors have no competing interests as defined by Nature Portfolio, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Ethics statement
The Institutional Animal Care and Use Committee approved all animal experimental protocols (0041/2021) of the Federal University of Alfenas (Minas Gerais, Brazil). We have complied with all relevant ethical regulations for animal use. All eight-week-old female Mus musculus C57BL/6 mice used in this study were maintained with free access to food and water according to the local animal welfare regulations (protocol 0041/2021).
Peer review
Peer review information
Communications Biology thanks Deepto Mozumdar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Si Ming Man and Tobias Goris.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Coelho, L.F.L., de Souza Terceti, M., Neto, S.P.L. et al. Mucosal-adapted bacteriophages as a preventive strategy for a lethal Pseudomonas aeruginosa challenge in mice. Commun Biol 8, 13 (2025). https://doi.org/10.1038/s42003-024-07269-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-07269-0