Abstract
Uncoupling protein 1 (UCP1) is a crucial protein located in the mitochondrial inner membrane that mediates nonshivering thermogenesis. However, the molecular mechanisms by which enhancer–promoter chromatin interactions control Ucp1 transcriptional regulation in brown adipose tissue (BAT) are unclear. Here, we employed circularized chromosome conformation capture coupled with next-generation sequencing (4C-seq) to generate high-resolution chromatin interaction profiles of Ucp1 in interscapular brown adipose tissue (iBAT) and epididymal white adipose tissue (eWAT) and revealed marked changes in Ucp1 chromatin interaction between iBAT and eWAT. Next, we identified four iBAT-specific active enhancers of Ucp1, and three of them were activated by cold stimulation. Transcriptional repression of the Ucp1-En4 or Ucp1-En6 region significantly downregulated Ucp1 and impaired mitochondrial function in brown adipocytes. Furthermore, depletion of the cohesin subunit RAD21 decreased the interaction intensity between Ucp1-En4 and the Ucp1 promoter and downregulated Ucp1. EBF2 cooperated with the acetyltransferase CBP to regulate Ucp1-En4 activity and increase Ucp1 transcriptional activity. In vivo, lentivirus-mediated repression of Ucp1-En4 was injected into iBAT, resulting in impacted iBAT thermogenic capacity and impaired iBAT mitochondrial function under cold acclimation conditions. Studying the functional enhancers regulating Ucp1 expression in iBAT will provide important insights into the regulatory mechanisms of BAT activity.
Similar content being viewed by others
Introduction
Brown adipose tissue (BAT) is a specialized type of adipose tissue found in mammals that regulates body temperature through nonshivering thermogenesis1,2. The thermogenic program in BAT can be activated by cold exposure or β-adrenergic receptor agonists3,4. The activation of BAT promotes glucose uptake and fatty acid oxidation, leading to increased whole-body energy expenditure5,6. The thermogenic activity of BAT largely depends on the expression of the BAT-specific protein uncoupling protein 1 (UCP1)7. UCP1 serves as a proton transporter localized in the mitochondrial inner membrane, uncoupling oxidative phosphorylation from ATP synthesis to cause mitochondrial proton leakage by dissipating energy as heat8. Ucp1 knockout mice exhibit cold intolerance9 and rely primarily on shivering-induced thermogenesis to maintain body temperature during cold acclimation10. Moreover, Ucp1 knockout mice develop obesity when housed under thermoneutral conditions (30 °C)11. Previous studies have shown the aberrant morphology of mitochondrial cristae and a reduced abundance of the electron transport chain components in the cold-induced BAT of Ucp1 knockout mice12. Understanding the molecular mechanisms that regulate the expression of Ucp1 will provide important insights into driving BAT thermogenesis.
The three-dimensional organization of the genome is essential for regulating gene expression and cellular function by constraining interactions between promoters and regulatory elements13,14,15. Enhancers, as key cis-regulatory elements, drive cell type-specific gene expression programs by serving as transcription factor-binding platforms16,17. Distal enhancers are brought into close proximity with their target gene promoters through chromatin looping to regulate gene expression in a cell type-specific manner18,19,20. Active enhancers typically exhibit features including specific histone modifications such as H3 lysine 27 acetylation (H3K27ac) and H3 lysine 4 monomethylation (H3K4me1) and an open chromatin conformation21,22,23. The cohesin complex (SMC1, SMC3, and RAD21) and the CCCTC binding factor (CTCF) are two important architectural components involved in mediating chromatin looping24,25.
Previous studies have identified a highly conserved enhancer element located 2.5 kb upstream of the Ucp1 transcriptional start site (TSS) in mice26. Cold-induced phosphorylation of JMJD1A alters enhancer–promoter proximity by forming a long-range chromatin loop with the SWI/SNF complex and activating the transcription of the thermogenic gene Adrb1 in brown adipocytes27. A recent study revealed a differentiation-dependent loop between the PPARγ2 promoter and an enhancer sequence located 10 kb upstream, which forms at the onset of PPARγ2 expression in C3H10T1/2 cells28. Although some thermogenesis-related enhancers have been shown to interact with the promoters of the genes they regulate through functional genomics screens, it remains largely unclear whether these enhancers can functionally regulate the expression of thermogenic genes and impact the thermogenic capacity of BAT. UCP1-mediated nonshivering thermogenesis in BAT plays a crucial role in energy homeostasis and body weight regulation in rodents and humans29. UCP1 activated thermogenesis has received significant attention as a potential target for the treatment of obesity and other metabolic diseases. Therefore, identifying functional enhancers regulating Ucp1 expression and elucidating the mechanism by which chromatin loops coordinate the expression of Ucp1 are crucial. Unraveling the molecular mechanisms underlying the transcriptional regulation of Ucp1 may provide novel targets for combating obesity and related metabolic disorders.
In this study, we characterized the dynamic changes in the chromatin interactions of Ucp1 between interscapular brown adipose tissue (iBAT) and epididymal white adipose tissue (eWAT) via circularized chromosome conformation capture coupled with next-generation sequencing (4C-seq). We identified four iBAT-specific active enhancers of Ucp1 and conducted functional studies on the identified iBAT-specific active enhancers Ucp1-En4 and Ucp1-En6. Our results demonstrated that Ucp1-En4 is crucial for Ucp1 expression, the thermogenic capacity of iBAT, and mitochondrial function in iBAT under cold acclimation conditions. A better understanding of the molecular mechanisms underlying Ucp1 expression may provide insights into the treatment of obesity and obesity-related metabolic diseases.
Results
Characterization of Ucp1 chromatin interactomes in iBAT and eWAT
To investigate the genome-wide chromatin interactions of Ucp1 in iBAT and eWAT, we conducted 4C-seq assays using the Ucp1 promoter (−2000 bp to +500 bp from the transcription start site) as the viewpoint (Supplementary Fig. 1a). For 4C-seq libraries, we obtained between 26.31 and 38.24 million aligned reads per dataset, with an average of 32.38 million reads per library. A cis/trans ratio analysis of four 4C datasets revealed that 63.60–80.89% of the mapped reads were distributed on the same chromosome as the viewpoint (Supplementary Data 1, Supplementary Fig. 1b). The 4C datasets conformed to the ‘cis/overall ratio of >40%’ criterion30, indicating high-quality experiments. Ucp1 genome-wide interaction sites were identified via a continuous non-overlapping 2-kb window approach (Supplementary Data 2). Next, we evaluated the reproducibility between replicates by counting the number of cis-interactions in each 1-megabase genomic bin. The Pearson correlation coefficients for chromatin interactions of Ucp1 in iBAT and eWAT were 0.83 and 0.73, respectively (Fig. 1a), reflecting a high consistency between the biological replicates. Principal component analysis revealed that the chromatin interactions of Ucp1 in iBAT and eWAT clustered into distinct groups (Fig. 1b). This result indicates differences in the chromatin interaction profiles of Ucp1 between iBAT and eWAT.
a Scatter plot showing interactions of Ucp1 in interscapular brown adipose tissue (iBAT) and epididymal white adipose tissue (eWAT). The number of interaction sites in each genomic bin (1 Mb cis) is plotted for two replicates. The Pearson correlation coefficient is indicated in each panel. b Principal component analysis of interactions of Ucp1 in iBAT and eWAT. Percentages represent the percent variability explained by each principal component. c Circos plot showing genome-wide interactions of Ucp1 in iBAT and eWAT. Chromosomes are shown in a circular orientation. d Venn diagram showing the number of overlapping and unique interaction sites of Ucp1 between iBAT and eWAT. e Numbers of reliable interaction sites and ratios of cis-/trans-interaction sites of Ucp1 in iBAT and eWAT. f Volcano plot showing differential interaction sites (DISs) of Ucp1 between iBAT and eWAT (left). Ratios of cis-/trans-DISs in upregulated and downregulated DISs (right). g Density plot showing the |Log2(FoldChange)| distribution of up- and downregulated DISs. Vertical red and blue dashed lines indicate the mean values of |Log2(FoldChange)| for upregulated and downregulated DISs, respectively. h Density plot showing the |Log2(FoldChange)| distribution of upregulated DISs. Vertical red and blue dashed lines indicate the mean values of |Log2(FoldChange)| for cis-upregulated and trans-upregulated DISs, respectively.
To study the differences in the chromatin interactions of Ucp1 between iBAT and eWAT, we identified 368 and 245 reliable interaction sites of Ucp1 between biological replicates in iBAT and eWAT, respectively (Fig. 1c, Supplementary Data 3). There were 136 shared interaction sites between iBAT and eWAT, including 134 cis-interaction sites and 2 trans-interaction sites (Fig. 1d). A greater number of interaction sites were observed in iBAT than in eWAT (Fig. 1e), suggesting that the specific expression of Ucp1 in iBAT may occur through the activation of tissue-specific regulatory elements. Further, we identified differential interaction sites (DISs) between iBAT and eWAT via DESeq2. A total of 347 DISs were found between iBAT and eWAT, accounting for 72.75% (347/477) of all interaction sites (Fig. 1f, Supplementary Data 4). Among them, 239 DISs were upregulated, 108 DISs were downregulated, and 295 cis-DISs and 52 trans-DISs (Fig. 1f). We then examined the fold change distributions of upregulated DISs and downregulated DISs. There were greater fold changes in upregulated DISs than in downregulated DISs, with mean |Log2(FoldChange)| values of 11.03 and 8.22, respectively (Fig. 1g). This result indicated that the upregulated DISs of Ucp1 in iBAT had a stronger change in the interaction frequency than did the upregulated DISs of Ucp1 in eWAT. In addition, the upregulated cis-DISs of Ucp1 in iBAT presented greater fold changes than did the upregulated trans-DISs of Ucp1 in iBAT, with mean |Log2(FoldChange)| values of 13.68 and 11.43, respectively. This finding suggested that the interaction frequency of the upregulated cis-DISs of Ucp1 in iBAT was changed to a greater degree than that of the upregulated trans-DISs of Ucp1 in iBAT (Fig. 1h). These results indicate that the interaction sites and interaction intensity of Ucp1 markedly changed between iBAT and eWAT.
Identification and characterization of Ucp1 active enhancers
To identify active enhancers of Ucp1 in iBAT and eWAT, we analyzed publicly available datasets of active enhancer-associated ChIP-seq (H3K27ac and H3K4me1) and ATAC-seq data. We identified seven tissue-specific putative active enhancers in iBAT but not in eWAT. Seven putative active enhancer regions were significantly enriched in H3K27ac, H3K4me1, and ATAC-seq (chromatin accessibility) signals (Fig. 2a). To evaluate the enhancer activity of Ucp1 in iBAT, luciferase reporter assays were performed in brown adipocytes. Our results revealed that, compared with the empty vector, Ucp1-En4, Ucp1-En5, Ucp1-En6, and Ucp1-En7 exhibited significantly increased luciferase activity (Fig. 2b). In addition, we performed adrenergic stimulation of brown adipocytes with isoproterenol (ISO) and found that the luciferase transcriptional activity of the Ucp1-En4, Ucp1-En6, and Ucp1-En7 constructs significantly increased after ISO treatment (Fig. 2b). These results indicated that four Ucp1 enhancers were active and that three of these enhancers were activated by cold stimulation. Previous studies have demonstrated that some active enhancers can recruit RNA polymerase II (RNA Pol II)31. Notably, RNA Pol II signals were significantly enriched at these active enhancer regions (Fig. 2a).
a Screenshot from Integrative Genomics Viewer (IGV) showing putative active enhancers of Ucp1. Upper tracks show circularized chromosome conformation capture coupled with next-generation sequencing (4C-seq) chromatin interaction profiles of Ucp1 in iBAT or eWAT. The blue-dashed line represents Ucp1 promoter as a viewpoint. 4C signal is merged across two biological replicates. Lower tracks show ChIP-seq (including H3K27ac, H3K4me1, and RNA Pol II) and ATAC-seq profiles at Ucp1 loci in iBAT and eWAT. Putative active enhancers are indicated by orange rectangles. b Luciferase reporter assay assessing putative active enhancers of Ucp1 in brown adipocytes. On day 8 of differentiation, brown adipocytes were treated with 10 μM isoproterenol (ISO) or vehicle for 4 h before luciferase reporter assay (n = 6). Data are presented as mean ± SEM. Analysis of variance were performed followed by Tukey’s test. c Signal values for chromatin interaction intensity at active enhancer regions in iBAT and eWAT were obtained from bigwig files on IGV. d Signal values for H3K27ac histone enrichment at active enhancer regions in iBAT and eWAT were obtained from bigwig files on IGV.
We then analyzed the differences in chromatin interactions and histone modifications of the four active enhancer regions between iBAT and eWAT. We observed a significantly greater interaction intensity between the Ucp1 promoter and the active enhancer regions in iBAT (Fig. 2c, Supplementary Fig. 2a). Additionally, these active enhancer regions presented significantly greater levels of H3K27ac and H3K4me1 histone modifications in iBAT than in eWAT (Fig. 2d, Supplementary Fig. 2b). These results suggest that chromatin interactions and histone modifications may play crucial roles in the function of these active enhancers, potentially influencing Ucp1 expression in different adipose tissue types.
Ucp1-En4 and Ucp1-En6 regulate Ucp1 expression and affect mitochondrial function
To determine whether the identified active enhancers regulate Ucp1 expression in brown adipocytes, we first examined Ucp1 expression during the adipogenic differentiation of iBAT SVF cells. The results indicated that Ucp1 expression increased on day 6 of differentiation, which persisted at a high level until day 8 (Supplementary Fig. 3a). Next, the empty dCas9-KRAB vector was transiently expressed in SVF cells on day 4 of differentiation to evaluate the expression levels of the dCas9-KRAB epigenetic system in mature brown adipocytes. The qRT-PCR results revealed that the dCas9-KRAB system functioned normally (Supplementary Fig. 3b), with a significant increase in Ucp1 mRNA levels following treatment with ISO, a β-adrenergic receptor agonist, on day 8 of differentiation (Supplementary Fig. 3c).
We subsequently employed dCas9-KRAB to suppress the three enhancers that were activated by cold stimulation, with two sgRNAs designed for each enhancer region (Supplementary Fig. 3d). On day 4 of differentiation, brown adipocytes were transfected with dCas9-KRAB, dCas9-KRAB-En4-sgRNAs, dCas9-KRAB-En6-sgRNAs, or dCas9-KRAB-En7-sgRNAs. On day 8 of differentiation, Ucp1 expression was significantly repressed by 24% and 20% when CRISPRi targeted Ucp1-En4 or Ucp1-En6, respectively, compared to the dCas9-KRAB control (Supplementary Fig. 3e). Notably, inhibition of Ucp1-En4 or Ucp1-En6 resulted in a more significant decrease in Ucp1 expression in brown adipocytes treated with ISO (62% and 46%, respectively) than did expression of the dCas9-KRAB-ISO control (Fig. 3a). Repression of Ucp1-En7 had no significant effect on the expression levels of Ucp1 in brown adipocytes with or without ISO treatment (Fig. 3a, Supplementary Fig. 3e). Immunofluorescence staining revealed that UCP1 fluorescence intensity was significantly reduced when Ucp1-En4 or Ucp1-En6 were repressed with ISO treatment compared to the dCas9-KRAB-ISO control (Fig. 3b). The western blotting results were consistent with the immunofluorescence results (Fig. 3c). These results indicated that repression of Ucp1-En4 and Ucp1-En6 impaired UCP1 expression. Moreover, BODIPY staining revealed that, compared with expression of the dCas9-KRAB-ISO control, the repression of Ucp1-En4 and Ucp1-En6 resulted in larger lipid droplets (Fig. 3d, Supplementary Fig. 3f). Consistently, qRT‒PCR analysis revealed that repression of Ucp1-En4 and Ucp1-En6 significantly increased the expression levels of lipogenesis-related genes (Acly, Acaca, and Fasn) (Fig. 3e). These results indicate that the enlargement of lipid droplets may be related to enhanced lipogenesis in brown adipocytes.
a qRT-PCR analysis of Ucp1 mRNA expression in brown adipocytes on day 8 of differentiation (n = 5). b Immunofluorescence staining of UCP1 (red) in brown adipocytes on day 8 of differentiation. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Quantification of UCP1-positive cells was performed by counting positive cells in five randomly chosen non-overlapping fields. Scale bar = 50 μm. c Western blot analysis of UCP1 in brown adipocytes on day 8 of differentiation (n = 4). Gray values of protein bands were quantified using Image J software, with Vinculin as a normalizer. d BODIPY (green) staining of lipid droplets in brown adipocytes on day 8 of differentiation. The nuclei were stained with DAPI (blue). The lipid sizes were measured as the diameter of individual lipid droplets in five randomly chosen non-overlapping fields. Scale bar = 50 μm. e qRT-PCR analysis of lipogenesis-related genes (Acly, Acaca, and Fasn) mRNA expression in brown adipocytes on day 8 of differentiation (n = 5). f Mitochondrial DNA (mtDNA) content in brown adipocytes on day 8 of differentiation (n = 5). g Western blot analysis of mitochondrial oxidative phosphorylation (OXPHOS) complex subunits in brown adipocytes on day 8 of differentiation (n = 4). Gray values of protein bands were quantified using Image J software, with Vinculin as a normalizer. On day 8 of differentiation, brown adipocytes were treated with 10 μM ISO or vehicle for 4 h before harvest for further experiments. Data are presented as mean ± SEM. P values were calculated by two-tailed unpaired Student’s t-tests.
BAT-specific UCP1, which is localized in the inner mitochondrial membrane, plays an important role in mitochondrial function12,32. To determine whether repression of Ucp1-En4 and Ucp1-En6 affects mitochondrial function in brown adipocytes, qPCR analysis revealed that the repression of Ucp1-En4 and Ucp1-En6 did not significantly affect the mitochondrial DNA (mtDNA) content in brown adipocytes without ISO treatment (Supplementary Fig. 3g). However, compared with the dCas9-KRAB-ISO control cells, the brown adipocytes with repression of Ucp1-En4 and Ucp1-En6 exhibited significantly decreased mtDNA content after ISO treatment (Fig. 3f). Moreover, we evaluated the mitochondrial mass using MitoTracker Red, a red fluorescent dye that labels the mitochondrial mass. The results demonstrated that cells with repression of Ucp1-En4 and Ucp1-En6 resulted in significantly lower mitochondrial mass than the dCas9-KRAB-ISO control cells (Supplementary Fig. 3h). We subsequently assessed the levels of mitochondrial oxidative phosphorylation (OXPHOS) complex subunits in whole-cell lysates via western blotting. While the expression of the nuclear-encoded subunit of complex V (ATP5A) was not significantly reduced, the expression of the nuclear-encoded subunits of complex I (NDUFB8), complex II (SDHB) and complex III (UQCRC2) and the mtDNA-encoded subunit of complex IV (MT-CO1) was significantly reduced in brown adipocytes with suppressed Ucp1-En4 and Ucp1-En6 following ISO treatment (Fig. 3g), suggesting a reduction in total mitochondrial content. In summary, these results indicate that Ucp1-En4 and Ucp1-En6, which act as functional enhancers, regulate Ucp1 expression and affect mitochondrial function under cold stimulation.
A cohesin-mediated chromatin loop is responsible for Ucp1-En4 and Ucp1 promoter chromatin interaction
Interactions between enhancers and promoters primarily occur within topologically associated domains (TADs)33,34,35, which are delimited by CTCF insulator sites36. TAD organization and CTCF insulator sites are conserved across various cell types and mammalian genomes19,33,37. To determine whether cohesin-mediated chromatin loops are responsible for controlling the interaction between Ucp1-En4 and the Ucp1 promoter, we analyzed publicly available Hi-C and CTCF ChIP-seq datasets. Our analysis revealed that the Ucp1-En4 and Ucp1 promoter are located within an interaction domain whose boundaries are enriched with CTCF binding sites, and the CTCF binding motifs display a convergent orientation (Fig. 4a). This finding suggested that the interaction between Ucp1-En4 and the Ucp1 promoter occurs within a TAD. In addition, MED1, a subunit of the general transcription coactivator complex mediator, facilitates chromatin looping between enhancers and promoters38,39. ChIP-seq analysis also revealed significant MED1 binding at the Ucp1-En4 and Ucp1 promoter regions (Fig. 4a). These results suggest that a cohesin-mediated chromatin loop regulates the interaction between Ucp1-En4 and the Ucp1 promoter.
a Alignment of high-throughput chromosome conformation capture (Hi-C) data from inguinal adipose tissue at Ucp1 locus. Heatmap (upper panel) showing the interaction between Ucp1-En4 and Ucp1 promoter located within the same TAD. Lower panel showing 4C-seq and ChIP-seq (MED1 and CTCF) data. The orange rectangle represents Ucp1-En4, the red rectangle represents Ucp1 promoter, and the gray rectangles represent the boundaries of the interaction domain. CTCF-motif position and orientation are indicated by arrows. b qRT-PCR analysis of Rad21 mRNA expression in differentiated brown adipocytes transfected with control or Rad21 small interfering RNA (siRNA) (n = 5). c Chromatin interactions between Ucp1 promoter and other chromatin sites (−10, −6, and −3 kb) in differentiated brown adipocytes were assessed by quantitative chromosome conformation capture (3C) qPCR assays 48 h after transfection with control or Rad21 siRNA. Chromatin interactions were normalized to the Ercc3 locus. A −6 kb chromatin site was used as a negative control. The triangle represents the 3C-qPCR primer, and the arrow direction represents the 3C-qPCR primer direction (n = 5). d Sanger sequencing of 3C-qPCR product between Ucp1-En4 (−10 kb site) and the Ucp1 promoter. The red base indicates the DpnII digestion site formed. e qRT-PCR analysis of Ucp1 mRNA expression in differentiated brown adipocytes transfected with control or Rad21 siRNA (n = 5). Data are presented as mean ± SEM. P values were calculated by two-tailed unpaired Student’s t-tests.
The cohesin complex plays a critical role in chromatin regulation of gene expression by facilitating enhancer–promoter looping40,41. We chose the RAD21 subunit of the cohesin complex, which facilitates physical contact between genes and enhancers38,42 and is essential for chromatin loop assembly and subsequent TAD formation19,40. To explore the role of cohesin subunits in the chromatin interaction between Ucp1-En4 and the Ucp1 promoter, we transiently depleted RAD21 in brown adipocytes via RAD21-specific siRNA. qRT‒PCR analysis revealed that Rad21 expression significantly decreased by 52% on day 8 of differentiation (Fig. 4b). We used restriction fragments generated by DpnII for chromosome conformation capture (3C) qPCR to assess the impact of RAD21 knockdown on the interaction between active enhancers and the Ucp1 promoter (Fig. 4c). 3C-qPCR analysis revealed that Rad21 knockdown significantly reduced the interaction intensity between the Ucp1 promoter and the −10 kb (Ucp1-En4) region compared to control cells (Fig. 4c). Additionally, the interaction intensity at the −3 kb (Ucp1-En6) site also significantly decreased (Fig. 4c). The 3 C ligation product from the −10 kb site was confirmed by Sanger sequencing (Fig. 4d). These results indicate that depletion of the cohesin subunit RAD21 affected the chromatin interaction between Ucp1-En4 and the Ucp1 promoter. Notably, Ucp1 expression was significantly lower in brown adipocytes with RAD21-knockdown than in control cells (Fig. 4e). In summary, our results indicate that cohesin is essential for the formation of chromatin loop between the Ucp1-En4 and Ucp1 promoter, thereby contributing to the regulation of Ucp1 expression.
EBF2 cooperates with CBP to regulate the activity of enhancers and is conserved between mice and humans
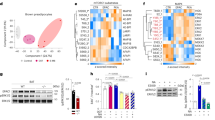
To investigate the molecular mechanisms controlling the activity of Ucp1-En4, a DNA pull-down assay was performed in iBAT nuclear lysates. The proteins that bound to the Ucp1-En4 region were separated via SDS‒polyacrylamide gel electrophoresis (Supplementary Fig. 4), and the entire lane was excised from the gel for identification via mass spectrometry (Supplementary Data 5). We identified 16 TFs that were specific for binding to the Ucp1-En4 region compared to the control group (Fig. 5a). Notably, among these TFs, members of the CCAAT/enhancer-binding protein (C/EBP) family (C/EBPA and C/EBPB), the nuclear factor I (NFI) family (NFIA, NFIB, and NFIX), and EBF2 were found to bind to the Ucp1-En4 region (Fig. 5a). C/EBPs are key regulators of adipocyte cell fate, and their activation drives the BAT-selective gene transcriptional program43,44. NFIA activates cell type-specific enhancers and facilitates the binding of PPARγ to regulate the brown fat gene program45,46. EBF2 binds at enhancers of brown fat-selective genes to activate gene transcription47. We then analyzed publicly available ChIP-seq datasets to visualize TF binding to the Ucp1-En4 region in brown adipocytes or BAT. Our analysis revealed that CEBPA, CEBPB, NFIA, EBF2, and the coactivator CBP bind to the Ucp1-En4 region in brown adipocytes (Fig. 5b). The histone acetyltransferase CBP catalyzes H3K27ac deposition in active enhancer regions48,49. Consistent with our in vitro pull-down findings, ChIP-seq analysis also revealed that EBF2 binds to the Ucp1-En4 region within its endogenous chromatin context (Fig. 5b).
a Venn diagram showing the transcription factors of Ucp1-En4 and control group identified using DNA pulldown assay in two replicates of iBAT. b ChIP-seq profiles showing binding of different transcription factors to Ucp1-En4 region. c ChIP-qPCR analyses showing the binding of EBF2 and CBP to Ucp1-En4 region in control and EBF2-overexpressing brown adipocytes (n = 3). d ChIP-qPCR analyses showing enrichment of H3K27ac on Ucp1-En4 region in control and EBF2-overexpressing brown adipocytes (n = 3). e qRT-PCR analysis of Ucp1 mRNA expression in control and EBF2-overexpressing brown adipocytes (n = 5). f H293T cells were co-transfected with Ucp1-En4 construct with EBF2 or CBP either together or individually (n = 6). g ChIP-seq (including H3K27ac, CBP, and MED1) and DHS-seq profiles defined putative active enhancers of UCP1 in beige adipocytes differentiated from human multipotent adipose-derived stem (hMADS) cells. Putative active enhancer is represented by an orange rectangle. h TF binding site motif analysis on Ucp1-En4 and UCP1-hEn1 sequences using the JASPAR database (https://jaspar.genereg.net/). i H293T cells were co-transfected with UCP1-hEn1 construct with human EBF2 or CBP either together or individually (n = 6). Data are presented as mean ± SEM. P values were calculated by two-tailed unpaired Student’s t-tests.
Chromatin-modifying enzymes are recruited to specific genomic regions through interactions with TFs to regulate gene expression50. We hypothesized that EBF2 and CBP synergistically regulate the activity of Ucp1-En4 to increase the transcriptional activity of Ucp1. Chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis confirmed that overexpression of EBF2 significantly increased the binding of both EBF2 and CBP to the Ucp1-En4 region in brown adipocytes (Fig. 5c). We then assessed the impact of EBF2 overexpression on H3K27ac levels in the Ucp1-En4 region. The ChIP-qPCR results revealed a significant increase in H3K27ac enrichment at the Ucp1-En4 region of brown adipocytes (Fig. 5d). In addition, overexpressed EBF2 significantly increased the expression of Ucp1 in brown adipocytes (Fig. 5e). To further investigate the functional significance of the EBF2 and CBP interaction in the activation of Ucp1-En4, we conducted luciferase reporter assays using the Ucp1-En4 construct. EBF2 and CBP expression constructs were cotransfected with the Ucp1-En4 construct into H293T cells. Compared with transfection of the empty vector, transfection of the EBF2 expression construct alone increased Ucp1-En4 luciferase activity 3.7-fold, whereas transfection of the CBP expression construct alone caused a 1.6-fold increase (Fig. 5f). Notably, cotransfection with the EBF2 and CBP expression constructs significantly increased Ucp1-En4 luciferase activity, leading to a robust 4.8-fold increase (Fig. 5f). These results suggest that CBP is recruited to Ucp1-En4 via cooperation with EBF2 to increase the transcriptional activity of Ucp1 via H3K27 acetylation at the Ucp1-En4 region.
Surprisingly, ChIP-seq data of beige adipocytes differentiated from human multipotent adipose-derived stem cells (hMADS) cells revealed a putative active enhancer element (UCP1-hEn1) located 6.4 kb upstream of the TSS of UCP1, which was significantly enriched with H3K27ac and DHS-seq (chromatin accessibility) signals (Fig. 5g). We performed TF binding site motif analysis on the Ucp1-En4 and UCP1-hEn1 sequences via the JASPAR database with the default settings. Consistent with the above results, both the Ucp1-En4 and UCP1-hEn1 enhancer sequences contain putative DNA-binding sites for EBF2, CEBPA, CEBPB, and NFIA (Fig. 5h). ChIP-seq analysis of hMADS beige adipocytes also revealed significant binding of the acetyltransferases CBP and mediator complex subunit 1 (MED1) to the UCP1-hEn1 region (Fig. 5g). Next, we used a luciferase reporter assay to examine whether EBF2 cooperates with CBP to regulate the activity of UCP1-hEn1. Compared with the transfection of the empty vector control, transfection of the EBF2 expression construct alone significantly increased UCP1-hEn1 luciferase activity by 2.0-fold, whereas transfection of the CBP expression construct alone did not significantly affect luciferase activity (Fig. 5i). However, a robust 3.2-fold increase in UCP1-hEn1 luciferase activity was observed when cotransfected with the EBF2 and CBP constructs (Fig. 5i). Together, these results suggest that EBF2 cooperates with CBP to regulate the activity of enhancers is conserved between mice and humans. Moreover, we evaluated the conservation of the mouse Ucp1-En4 enhancer sequence using the UCSC Genome Browser (http://genome-asia.ucsc.edu/) (Supplementary Fig. 5a). The analysis revealed that the mouse Ucp1-En4 enhancer sequence was not conserved with the human UCP1-hEn1 enhancer sequence, but was relatively conserved with the sequence located 40 kb upstream of the TSS of human UCP1 (Supplementary Fig. 5b).
Repression of Ucp1-En4 affects thermogenic capacity and mitochondrial function in iBAT
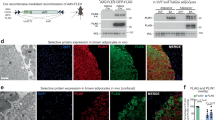
To investigate the effect of Ucp1-En4 on BAT function in vivo, we injected the dCas9-KRAB-En4 lentiviral system directly into iBAT. The mice were housed at different ambient temperatures (30 °C or 4 °C), and the lentiviral injection strategy is illustrated in Fig. 6a. Two weeks postinjection, immunofluorescence microscopy analysis showed successful infection and expression of the lentiviral GFP vector in iBAT expressing UCP1 (Supplementary Fig. 6a), confirming successful lentiviral infection. The Ucp1 expression levels in iBAT from iBAT-En4 and control mice were determined. The results demonstrated that Ucp1 expression was moderately reduced at 30 °C but significantly decreased at 4 °C (Supplementary Fig. 6b, Fig. 6b). In addition, the expression levels of lipogenesis-related genes (Acly, Acaca, and Fasn) were significantly increased in iBAT-En4 mice at both 30 °C and 4 °C (Supplementary Fig. 6c, Fig. 6c). Consistent with the changes in Ucp1 expression levels, the western blotting results revealed a more pronounced decrease in UCP1 expression in iBAT-En4 mice at 4 °C (Fig. 6d, Supplementary Fig. 6d). These results suggested that repression of Ucp1-En4 reduced UCP1 expression in vivo.
a Schematic of experimental design of lentiviral injection experiments. b, c qRT-PCR analysis of Ucp1 and lipogenesis-related genes (Acly, Acaca, and Fasn) mRNA expression in iBAT from control and iBAT-En4 mice at 4 °C (n = 5). d Western blot analysis of UCP1 in iBAT from control and iBAT-En4 mice at 4 °C (n = 4). Gray values of protein bands were quantified using Image J software, with Vinculin as a normalizer. e, f Oxygen consumption (VO2) and heat production of control and iBAT-En4 mice in metabolic cages at 30 °C (n = 5). g, i Representative infrared images (left) and quantification of average dorsal skin temperature (right) of control and iBAT-En4 mice at 30 °C or 4 °C (n = 5). h, j Rectal temperature of control and iBAT-En4 mice at 30 °C or 4 °C (n = 5). k, l Representative photographs of iBAT from control and iBAT-En4 mice at 30 °C or 4 °C (left). Representative hematoxylin and eosin staining of iBAT from control and iBAT-En4 mice at 30 °C or 4 °C (right). Scale bar = 50 μm. m, n Immunofluorescence staining of iBAT from control and iBAT-En4 mice at 30 °C or 4 °C for UCP1 (red) and nuclei (blue). Scale bar = 50 μm. o Transmission electron microscopy images of control and iBAT-En4 mice at 4 °C. Scale bar = 1 μm. p Assessment of mitochondrial DNA content by qPCR for iBAT from control and iBAT-En4 mice at 4 °C (n = 5). q Western blot analysis of mitochondrial OXPHOS complex subunits in iBAT from control and iBAT-En4 mice at 4 °C (n = 4). Gray values of protein bands were quantified using Image J software, with Vinculin as a normalizer. Data are presented as mean ± SEM. P values were calculated by two-tailed unpaired Student’s t-tests.
There were no significant differences in oxygen consumption, heat production, or body weight between iBAT-En4 and control mice at either 30 °C or 4 °C (Fig. 6e, f, Supplementary Fig. 6e–g). However, the dorsal skin temperature in the interscapular area and the rectal temperature were significantly lower in iBAT-En4 mice than in control mice at both 30 °C and 4 °C, with a more significant decrease observed at 4 °C (Fig. 6g–j). Additionally, we observed that the iBAT of iBAT-En4 mice appeared whitish in color at both 30 °C and 4 °C compared with that of control mice (Fig. 6k, l). Further, hematoxylin and eosin (H&E) staining revealed an increased diameter of lipid droplets with more unilocular, white-like adipocytes in iBAT from iBAT-En4 mice than in that from control mice at both 30 °C and 4 °C (Fig. 6k, l). Immunofluorescence analysis showed a moderate reduction in UCP1 expression in iBAT from iBAT-En4 mice at 30 °C, with the greatest decrease in expression at 4 °C compared to control mice (Fig. 6m, n). These results suggest that the inhibition of iBAT-En4 impacts the thermogenic capacity of iBAT, particularly under cold acclimation conditions, although it did not affect whole-body energy expenditure.
To determine whether the repression of Ucp1-En4 affects mitochondrial morphology and function, transmission electron microscopy was performed and revealed swollen mitochondria with irregular cristae in iBAT from iBAT-En4 mice at 4 °C, but not at 30 °C, compared with normal mitochondria in control mice (Fig. 6o, Supplementary Fig. 6h). Consistently, quantification of the mtDNA content and protein levels of complexes I–V revealed significantly reduced mtDNA content and levels of mitochondrial complex I (NDUFB8) and complex II (SDHB) in iBAT from iBAT-En4 mice at 4 °C but not at 30 °C (Fig. 6p, q, Supplementary Fig. 6i, j). These results suggest that the repression of iBAT-En4 impairs mitochondrial function in iBAT under cold acclimation conditions. Overall, our results demonstrate that iBAT-En4 regulates Ucp1 expression and is important for the thermogenic capacity and mitochondrial function of iBAT.
Discussion
UCP1-mediated nonshivering thermogenesis in BAT is crucial for maintaining energy homeostasis and regulating adiposity51. The activation of UCP1-mediated thermogenesis increases whole-body energy expenditure, resulting in reduced body fat in both rodents and humans52,53. Understanding the molecular mechanisms by which functional enhancers regulate Ucp1 expression may reveal new targets for the treatment of obesity and metabolic disorders. In this study, we characterized the Ucp1 chromatin interactomes in iBAT and eWAT. We identified four iBAT-specific active enhancers of Ucp1. Further functional analysis revealed that Ucp1-En4 affects thermogenesis and mitochondrial function in brown adipocytes and iBAT.
Previous studies have shown that an increase in the number of promoter interactions is positively correlated with gene expression54,55. Similarly, our results revealed an increase in the number of chromatin interactions in iBAT, suggesting that Ucp1 expression in iBAT may occur through the activation of tissue-specific regulatory elements. Chromatin interactions are spatial structures that can bring distant regulatory elements into close proximity to target gene promoters to regulate gene expression56,57. Our results indicated that four Ucp1 active enhancers exhibited higher chromatin interaction frequencies with the Ucp1 promoter in iBAT, implying that chromatin interaction is involved in the regulation of Ucp1 expression. Histone modification of chromatin influences gene expression by modulating chromatin accessibility58,59. Chromatin regions with active histone modifications (H3K27ac and H3K4me1) are associated with an open chromatin structure and active transcription. The four Ucp1 active enhancer regions displayed higher levels of active histone modifications in iBAT, suggesting that chromatin opening in these enhancer regions may increase the binding capacity of TFs, thereby leading to increased Ucp1 expression. Our findings suggest that active enhancers exhibit a high frequency of local chromatin interactions and high levels of local active histone modifications, which are necessary for driving Ucp1 expression. Upon cold exposure, sympathetic system-derived norepinephrine activates β-adrenergic receptors on brown adipocytes, triggering the release of free fatty acids and increasing UCP1 expression60. We further explored the effect of cold on enhancer activity. Treatment of brown adipocytes with ISO revealed that the activity of three of the four Ucp1 active enhancers was induced by cold stimulation. This result suggested that the increase in Ucp1 expression under cold stimulation may be due to the increased activity of these enhancers.
We investigated the function of the three enhancers activated by cold stimulation via the CRISPR-dCas9-KRAB epigenetic system. Repression of Ucp1-En4 and Ucp1-En6 significantly reduced Ucp1 expression, indicating their importance as functional enhancers of Ucp1. Enhancer redundancy, a common feature of mammalian genomes, implies that the loss of a single enhancer does not necessarily impact gene expression61,62. Repression of Ucp1-En7 did not significantly affect Ucp1 expression, implying that Ucp1-En7 is a functionally redundant enhancer in the regulation of Ucp1 transcription. Recently, an increasing number of studies have focused on super-enhancers, which are clusters of enhancers occupied by master TFs that facilitate the recruitment of RNA Pol II or writers of activation marks to target genes63,64. Notably, there appears to be a Ucp1 super-enhancer locus that spans from the distal −27 kb region to −2.2 kb upstream of the Ucp1 promoter65. We investigated the effects of a single functional enhancer located within this super-enhancer region on the expression of Ucp1 and the thermogenic capacity of BAT, providing a reference for future research on the potential functional role of the Ucp1 super-enhancer. Studying the molecular mechanisms of a single Ucp1 enhancer and super-enhancer will contribute to understanding the synergistic regulation of Ucp1 expression by enhancers in brown fat and their comprehensive impact on brown fat thermogenesis.
The loop extrusion model of chromatin loop formation suggests that the cohesin protein complex slides along chromatin to generate a DNA loop until it is paused by two CTCF molecules bound in a convergent orientation66,67. Previous studies have demonstrated that chromatin loops mediate enhancer–promoter interactions to regulate gene transcription19,68. Different types of chromatin loops are involved in these interactions, which encompass CTCF-mediated chromatin loops69, cohesin-mediated chromatin loops70, and TF-mediated chromatin loops71. In our study, analysis of published Hi-C data revealed that the interaction between Ucp1-En4 and the Ucp1 promoter occurs within a TAD. Further, siRNA and 3C-qPCR assays revealed that depletion of the cohesin subunit RAD21 decreased the interaction intensity between iBAT-En4 and the Ucp1 promoter and reduced Ucp1 expression. These findings indicate that a cohesin-mediated chromatin loop regulates the functional interaction between iBAT-En4 and the Ucp1 promoter. Although our results demonstrated that Ucp1-En4 and Ucp1-En6 play key roles in regulating Ucp1 expression and influencing mitochondrial function, this study aimed to screen and identify the enhancer with the most pronounced impact on Ucp1 expression and mitochondrial function. Consequently, we conducted further mechanistic studies on Ucp1-En4. We also recognize the importance of Ucp1-En6, and future studies will examine Ucp1-En6 in greater detail.
EBF2, an important regulator of brown fat cell commitment, is essential for maintaining the thermogenic function of BAT under standard conditions47,72. The absence of the EBF repressor ZFP423 or the overexpression of EBF2 in mouse WAT can stimulate browning and suppress the development of obesity73,74. Previous studies have shown that the −6 kb Ucp1 enhancer element contains putative DNA-binding sites for EBF2 and ERR and that EBF2 cooperates with ERRα to increase enhancer activity72. Our study revealed that EBF2 cooperates with CBP to regulate the activity of Ucp1-En4, thereby increasing Ucp1 transcriptional activity. Activating thermogenic gene programs in brown and beige fat enhances systemic energy expenditure and can help alleviate or prevent obesity-related metabolic disorders75. Increasing the activity of thermogenic adipocytes through regulating the expression of thermogenic genes has the potential to be a promising strategy for preventing and treating obesity and associated metabolic disorders. Our results demonstrated that EBF2 cooperates with CBP to regulate the activity of iBAT Ucp1-En4 and that hMADS beige adipocyte UCP1-hEn1 is conserved between mice and humans. Studying enhancers of thermogenic genes in mouse brown fat will provide important insights into the regulatory mechanisms of human BAT activity.
UCP1 is activated in BAT during cold stress and dissipates the mitochondrial proton motive force generated by the electron transport chain to produce heat76. Previous studies have shown that the protein abundance of mitochondrial respiratory chain complexes in BAT from UCP1-KO mice is significantly lower than that in BAT from wild-type mice12. Notably, more pronounced mitochondrial alterations were observed in BAT from UCP1-KO mice exposed to low environmental temperatures than in that from UCP1-KO mice at thermoneutrality12,77,78. Consistently, our transmission electron microscopy results revealed swollen mitochondria with irregular cristae in iBAT from iBAT-En4 mice at 4 °C. Compared with those of control mice, the mtDNA content and protein levels of respiratory chain complexes in iBAT from iBAT-En4 mice at 4 °C were significantly lower, and the levels of mitochondrial complex I (NDUFB8) and complex II (SDHB) were also significantly lower. These results suggest that the repression of iBAT-En4 impairs mitochondrial function in iBAT under cold acclimation conditions. Only CI and CII protein levels were significantly reduced in iBAT from iBAT-En4 mice where Ucp1 expression was significantly downregulated, which may indicate that specific mitochondrial OXPHOS complex subunits are selectively affected in response to the altered metabolic demands of iBAT. Future studies could further investigate these complex-specific responses to clarify how Ucp1-En4 impacts the regulation of mitochondrial complexes and iBAT thermogenesis. We believe that elucidating the transcriptional regulatory mechanisms underlying the repression of Ucp1 expression under conditions such as obesity, aging, and other pathologies in mice is critical. However, there is an absence of publicly available epigenetic data on brown fat from mice with these conditions. More investigations are needed to further clarify these potential mechanisms.
Methods
Ethics statement
All animal experiments were conducted according to the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in March 2017) and approved by the Animal Ethical and Welfare Committee (AEWC) of Sichuan Agricultural University under permit No. DKY-B3022102011.
4C-seq assay
4C-seq was performed following previously described30,79 with some modifications. Briefly, 1 g of interscapular brown adipose tissue (iBAT) and epididymal white adipose tissue (eWAT) were obtained from 8-week-old C57BL/6J male mice (n = 2 biological replicates) at room temperature (RT) under ambient environment. The samples were crushed using a pestle and mortar in liquid nitrogen. The powder was re‐suspended into 2% formaldehyde solution in PBS and cells cross‐linked at RT for 30 min, then cross-linking was quenched with glycine buffer at a final concentration of 0.13 M. The cells were lysed for 10 min on ice in 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, and protease inhibitor. Nuclei were isolated by 10 min centrifugation at 4 °C at 500 × g. Chromatin was digested using 500 U of primary restriction enzyme DpnII (New England Biolabs) at 37 °C overnight. Ligation was performed using 400 U of T4 DNA ligase (New England Biolabs) at 16 °C overnight. After the reversal of cross-linking, DNA was purified by phenol-chloroform extraction followed by ethanol precipitation. Purified DNA was digested using 500 U of secondary restriction enzyme Csp6I (Thermo Fisher Scientific) at 37 °C overnight and religated by T4 DNA ligase at 16 °C overnight. DNA was extracted using phenol-chloroform and further purified using a QIAquick PCR Purification Kit (QIAGEN). 4C PCR was performed using a total input of 3.2 μg, divided into 16 reactions. The PCR products were separated on a 2% agarose gel, and fragments between 200 to 800 bp were excised and purified. The 4C-seq libraries were sequenced on an Illumina Novaseq 6000 platform with 150 bp paired-end reads. The primers used for the 4C-seq library are listed in Supplementary Data 6.
4C-seq data analysis
Demultiplexing, trimming, and alignment were performed using the pipe4C pipeline79. 4C-seq data analysis was performed using the r3Cseq package80. Bowtie2 (v2.2.5) was used to map the trimmed reads to the mm10 mouse genome. The reads mapped to sequences containing the DpnII restriction site GATC and the Csp6I restriction site GTAC were termed 4C fragment-ends. SAM files were converted to BAM files, sorted, and indexed using Samtools (v1.11). Statistical analyses were performed using the r3Cseq by counting and normalization mapped reads in 2 kb windows to identify interaction regions. Reliable interaction sites were defined by biological reproducibility and sufficient read counts (nReads > 10). Significant interaction sites (SISs) were defined as those with a q-value < 0.05. DESeq2 (v1.36.0) with the ‘ashr’ algorithm was used to analyze the differential interaction sites81,82. Differential interaction sites (DISs) were defined with the threshold |Log2(FoldChange)| > 1 and q-value < 0.05.
ChIP-seq, ATAC-seq, and DHS-seq
Publicly available ChIP-seq, ATAC-seq, and DHS-seq data used in this study were downloaded from the NCBI GEO and SRA databases. ChIP-seq, ATAC-seq, and DHS-seq reads were aligned to mouse (mm10) or human (hg38) genome using Bowtie2 (v2.4.2) with the default parameters83. Peak calling was performed using MACS2 (v2.2.7.1) with the default parameters84. BedGraph files were converted to BigWig files using bedGraphToBigWig (v4), and then BigWig was visualized by Integrative Genomics Viewer (IGV). Detailed information about the public datasets used in this study can be found in Supplementary Data 7.
Primary cell culture and differentiation
Stromal vascular fraction (SVF) cells were isolated from iBAT of 5-week-old C57BL/6J male mice, cultured, and differentiated into brown adipocytes according to published methods85 with slight modifications. Briefly, iBAT was minced and digested with 0.1% collagenase type I (Gibco) in HBSS buffer containing 2% penicillin-streptomycin for 60 min at 37 °C. An equal volume of DMEM/F-12 medium containing 2% pen/strep and 15% fetal bovine serum (FBS) was added to neutralize collagenase. The digested tissues were filtered through a 100 μm cell strainer and centrifuged at 500 × g for 5 min. The cell pellets were resuspended in DMEM/F-12 medium containing 1% pen/strep and 15% FBS, filtered through a 40 μm cell strainer and plated on cell culture flasks. For induction of brown adipogenesis, confluent cultures were exposed to induction DMEM/F-12 medium containing 0.5 mM 3-isobutyl-1-methylxanthine, 5 μM dexamethasone, 20 nM insulin, 1 nM T3, 1 μM rosiglitazone, and 10% FBS for two days. The cells were then maintained in a differentiation medium supplemented with 20 nM insulin, 1 nM T3, and 1 μM rosiglitazone for another 6 days.
Identification of active enhancers of Ucp1
Putative active enhancer regions were defined as genomic regions with significant chromatin interactions with the Ucp1 promoter (measured by 4C-seq) and overlapping with H3K27ac, H3K4me1, and ATAC-seq peaks, located at least 2 kb away from annotated TSSs. To evaluate the activity of the putative enhancers, we employed a luciferase reporter assay. Putative active enhancer fragments were PCR amplified from C57BL/6J mouse genomic DNA. These fragments were then cloned into the Firefly luciferase reporter vector pGL3-Promoter (Promega). On the 6th day after the induction of differentiation, differentiated brown adipocytes were cotransfected using Lipofectamine 3000 (Invitrogen) with pGL3-Promoter-enhancer constructs and a Renilla luciferase expression vector (as an internal control). 48 h post-transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (Vazyme, Nanjing, China) on a Glomax 96 Microplate Luminometer. The PCR primers used to amplify putative active enhancers are shown in Supplementary Data 8.
The sgRNA design and plasmid construction
CRISPR single guide RNAs (sgRNA) were designed using CHOPCHOP86 and CRISPOR87. The sgRNAs were evaluated for their potential off-target effects using the Cas-OFFinder (http://www.rgenome.net/cas-offinder/)88 and CRISPR Finder (https://wge.stemcell.sanger.ac.uk/)89 Single sgRNAs with predicted low off-target activity and high on-target activity were chosen. If the guide sequence does not begin with a ‘G’, an added ‘G’ is added to the end of the 5′ universal flanking sequence.
For the dCas9-KRAB expressing construct, we used KRAB-dCas9 (Addgene #112195)90,91 for transient expression. To generate sgRNA expression vectors, oligonucleotides were annealed and inserted into the BsaI site of the pGL3-U6-sgRNA-PGK-puromycin vector (Addgene #51133). On the 4th day after the induction of differentiation, differentiated brown adipocytes were cotransfected using Lipofectamine 3000 (Invitrogen) with KRAB-dCas9 vector and pGL3-U6-sgRNA-PGK-puromycin vector. On day 8 of differentiation, brown adipocytes were treated with 10 μM isoproterenol (ISO) or vehicle for 4 h before harvest for further experiments. We used pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP (Addgene #71237) for lentivirus stable expression. The sgRNA oligonucleotides were synthesized, annealed, and inserted into the pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP vector via the BsmBI-v2 restriction site. The sequences of all sgRNAs are provided in Supplementary Data 9.
RNA isolation and qRT-PCR
Total RNA was isolated from brown adipocytes or iBAT using Trizol reagent (Invitrogen). The RNA was reverse transcribed into cDNA using the HiScript III RT SuperMix (Vazyme, Nanjing, China). qRT-PCR was performed using the ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on the CFX Connect Real-Time System (Bio-Rad). Relative gene expression levels were calculated using the 2–ΔΔCt method92, with the Rplp0 gene used as an internal control for data normalisation. Primer sequences used for qRT-PCR are shown in Supplementary Data 10.
Quantification of the mtDNA copy number
Total DNA was isolated from brown adipocytes or iBAT using FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme, Nanjing, China). The relative mitochondrial DNA (mtDNA) copy number was determined by the ratio of mtDNA-encoded genes (ND1 and 16S) to a nuclear DNA-encoded gene (HK2) via qPCR. Primer sequences for the qPCR are provided in Supplementary Data 10.
Immunocytochemistry, BODIPY 493/503 staining, and MitoTracker staining
Differentiated brown adipocytes were fixed in 4% paraformaldehyde for 15 min at RT, and permeabilized with 0.25% Triton X-100 for 10 min. Cells were then blocked with PBS containing 10% normal goat serum for 30 min at 37 °C. Cells were incubated with primary UCP1 antibody (ABclonal, A5857, 1:100) at 4 °C overnight. The secondary antibody Cy3-conjugated Goat anti-Rabbit IgG (ABclonal, AS007, 1:100) was applied for 60 min at 37 °C. Lipid droplets were stained with 5 μM BODIPY 493/503 (Invitrogen) for 20 min at RT. To stain the mitochondria, 200 nM MitoTracker Red CMXRos (Solarbio, Beijing, China) was applied for 30 min at 37 °C. Nuclei were counterstained with 4′-6-diamidino-2-phenylindole (DAPI) (Beyotime, Shanghai, China) for 5 min at RT.
Western blot
Brown adipocytes or iBAT were homogenized and lysed in RIPA lysis buffer (Solarbio, Beijing, China) for 30 min on ice, and the lysates were centrifuged at 12,000 × g for 10 min at 4 °C. Protein concentrations were determined using a BCA protein assay kit (Beyotime, Shanghai, China). Protein was separated by 10% SDS‒polyacrylamide gel electrophoresis and transferred to PVDF membrane (Millipore). Membranes were then blocked with 5% non-fat milk in TBST buffer for 1 h at RT and incubated with primary antibody overnight at 4 °C. The following primary antibodies were used: UCP1 (Cell Signaling Technology, 72298, 1:1000), total OXPHOS cocktail (Abcam, ab110413, 1:1000) and Vinculin (Cell Signaling Technology, 13901, 1:1000). Subsequently, the membranes were incubated with HRP-conjugated anti-mouse or -rabbit secondary antibodies at 37 °C for 1 h. Protein bands were visualized using the ECL detection kit (Beyotime, Shanghai, China) and analyzed using ImageJ software.
Small interfering RNA transfection
Rad21 small interfering RNA (siRNA) duplexes were synthesized by Sangon Biotechnology (Shanghai, China). Differentiated brown adipocytes were transfected with Rad21-siRNA (sense, 5′-GGAAUAACCGGCUACUGAATT-3′, antisense, 5′-UUCAGUAGCCGGUUAUUCCTT-3′) and negative control siRNA (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) using Lipofectamine 3000 (Invitrogen) at a final concentration of 50 nM. Cells were harvested 48 h after transfection.
3C assay
Quantitative chromosome conformation capture (3C) qPCR assays were performed as described93,94. Briefly, 1 × 107 brown adipocytes were crosslinked with 2% formaldehyde for 10 min at RT, and the reaction was quenched with 0.125 M glycine for 5 min. Cells were lysed on ice using the lysis buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 1% NP-40, and protease inhibitor) for 10 min. After centrifuging at 400 × g for 5 min, nuclei were resuspended in 500 μL of 1.2 × NEBuffer2.1 with 0.3% SDS and incubated at 37 °C for 1 h, followed by 2% Triton X-100 for another 1 h. DNA was then digested with 800 U DpnII (New England Biolabs) at 37 °C overnight. Digested DNA was diluted with 6.125 mL of 1.15 × T4 DNA Ligase Reaction Buffer and ligated at 16 °C overnight using 1600 U T4 DNA ligase. Cross-links were reversed by incubation with proteinase K overnight at 65 °C. The qPCR was performed using purified 3C samples, with 250 ng of DNA templates in a 10 μL reaction volume. After normalization to a cross-linking control at the Ercc3 locus95,96, the relative cross-linking frequency at the same site across different samples was calculated. The primers used for 3C-qPCR are listed in Supplementary Data 11.
DNA pull-down assay
DNA pull-down assay was performed with a DNA pull-down kit (BersinBio, Guangdong, China). Briefly, the Ucp1-En4 enhancer region was amplified by PCR using 5′-desthio biotin labeled or non-labeled primers. The primer sequences were as follows: forward primer, GGGACTAACGAAGCCTGCAT; reverse primer, AGGCTCTCCACTCAGTTTCTG. Two biological replicates were performed using nuclear lysates of iBAT from 8-week-old C57BL/6J male mice (four mice were pooled per biological replicate) maintained at RT. The control group was mixed with streptavidin magnetic beads and non-labeled DNA, and the experimental group was mixed with streptavidin magnetic beads and 5′-desthio biotin labeled DNA. The desthio biotin labeled or non-labeled DNA was incubated with streptavidin magnetic beads at RT for 30 min with agitation. Nuclear protein and DNA beads then were incubated for 1 h at 4 °C. After washing four times with 800 μL binding buffer, the pulled-down complex was eluted using 70 μL of protein elution buffer for 2 h at 37 °C. The silver stain was carried out using a Fast Silver Stain Kit (Beyotime, Shanghai, China). The digested peptides were analyzed by an UltiMate 3000 RSLCnano System (Thermo Fisher Scientific) coupled to a Q Exactive HF mass spectrometer (Thermo Fisher Scientific).
ChIP assay
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP Assay Kit (Beyotime, Shanghai, China). Briefly, brown adipocytes on day 6 of differentiation were transfected with the pEGFP-N1-EBF2 vector. On day 8 of differentiation, the cells were crosslinked with 2% formaldehyde for 10 min, followed by quenching with 0.125 M glycine for 5 min at RT. Brown adipocytes were harvested and lysed in cell lysis buffer supplemented with protease inhibitor (Roche) and 2 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich). Subsequently, chromatin was sheared to a size of 200–1000 bp using a Q800R3 Sonicator (QSonica). The indicated antibodies (2 μg) were added to the chromatin solution and immunoprecipitated at 4 °C overnight. Antibody-bound chromatins were washed, eluted, and reverse crosslinked. Quantification of immunoprecipitated DNA was achieved by qPCR on a CFX Connect Real-Time System (Bio-Rad). The percentage of immunoprecipitate signals was calculated over the input signals. The antibodies used in ChIP assays were EBF2 (R&D Systems, AF7006), CBP (Cell Signaling Technology, 7389), and H3K27ac (Abcam, ab4729). Primer sequences used for qPCR are shown in Supplementary Data 10.
Construction of overexpression vectors and luciferase reporter assay
The coding sequences of EBF2 and CBP were PCR amplified from brown adipocytes cDNA or H293T cells cDNA, respectively. All the PCR products were cloned into the pEGFP-N1 vector using KpnI (New England Biolabs) restriction sites. H293T cells were cultured in 96-well plates and were cotransfected with a mixture of 50 ng of PGL3-Promoter-Ucp1-En3/-UCP1-hEn1 luciferase reporter construct, 5 ng of Renilla vector, and 50 ng of pEGFP-N1-EBF2 and/or CBP expression vector using the Lipofectamine 3000 (Invitrogen). 48 h post-transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System. Primer sequences used for vector construction are shown in Supplementary Data 8.
Lentivirus production and mice infection
To produce lentivirus, H293T cells were cultured in the T175 flask. H293T cells grown to approximately 70% confluence were cotransfected with the mixture (95 μg) of transfer plasmid pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP (Addgene #71237), packaging plasmid psPAX2 (Addgene #12260), and envelope plasmid pVSV-G (Addgene #138479) at a weight ratio of 4:3:1 using a Calcium Phosphate Transfection Kit (Beyotime, Shanghai, China). The culture medium was changed 8 h post transfection and viral supernatant was harvested at 24, 48, and 72 h post transfection. The viruses were concentrated using Amicon Ultra-15 100 kDa centrifugal filter units (Millipore). The viral titre was determined using the colloidal gold kit (Biodragon, Beijing, China).
The 5-week-old male C57BL/6J mice (n = 20) were acclimated to thermoneutrality (30 °C) or cold (4 °C) for 1 week before starting experiments. During the study, mice were kept on a 12 h light/dark cycle and had free access to standard chow and water. Mice under each experimental condition were randomly divided into two groups (treatment group: injection of dCas9-KRAB-En4 lentivirus; control group: injection of dCas9-KRAB-GFP lentivirus; n = 5 mice per group). The 6-week-old C57BL/6J male mice were anaesthetized with 1% sodium pentobarbital via intramuscular injection. Then, 100 μL of lentiviral particles (6 × 107 lentiviral transducing particles (TU)/mL, 200 μL per mouse) were administered by sight into the iBAT by 4 point injections on each side according to previous methods97. Mice were injected with lentivirus once every 3 days for a total of 4 injections. A total of 800 μL of lentivirus was injected into each animal raised at 30 °C or 4 °C. For cold stimulation, mice recovered for 12 h at 23 °C after each injection of lentivirus and then exposed to 4 °C for 60 h. After the fourth injection of lentivirus, mice were acclimated to thermoneutrality (30 °C) or cold (4 °C) for 1 week later for subsequent experimental analyses. Animal fluorescence imaging was performed using an IVScope 8500 small animal imaging system (CLiNX Science Instruments, Shanghai, China).
Metabolic study and infrared imaging
Whole-body energy metabolism was measured using an Animal Metabolism Monitoring System (EM-8M-WA, Tow-Int Tech, Shanghai, China) in a temperature- and humidity-controlled room, set at either 4 °C or 30 °C, 50% relative humidity and a 12 h light/dark cycle. Mice were acclimated for 24 h in metabolic cages before monitoring. Oxygen consumption (VO2) and heat production were measured after acclimation. Mice were shaved at the interscapular regions and allowed to recover for 2 days. Thermal images were obtained by an infrared thermal imaging camera (Fotric 348A, Shanghai, China) and analyzed by Analyz IR software.
Morphological and histological study
After fixation in 4% paraformaldehyde, iBAT samples were embedded in paraffin, and sections were prepared. Samples were embedded in paraffin, and sectioned, followed by hematoxylin and eosin staining. For immunofluorescence, slides were fixed with 4% paraformaldehyde, and permeabilized with ice-cold methanol. Heat-mediated antigen retrieval with 0.01 M citric acid (pH 6.0) was performed for 5 min in a microwave. After blocking with 3% BSA for 30 min, sections were incubated with anti-UCP1 primary antibody (Servicebio, GB112174, 1:200) overnight at 4 °C. Stained using CY3-goat anti-rabbit secondary antibody (Servicebio, GB21303, 1:300) for 50 min at RT. The nuclei were counterstained with DAPI (Beyotime, Shanghai, China). The adipocyte diameter and relative UCP1 intensity were measured using ImageJ software.
Transmission electron microscopy
Samples of iBAT (1 mm × 1 mm × 1 mm) were rapidly harvested from the mice, washed in fresh 0.1 M phosphate buffer (pH 7.4), and fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at RT for 4 h. After three washes in fresh 0.1 M phosphate buffer (pH 7.4), the tissues were fixed in 1% osmium tetroxide at RT for 2 h and then washed three times in 0.1 M phosphate buffer (pH 7.4). The tissues were then embedded in Epon using standard techniques following dehydration with ethanol and propylene oxide. Ultrathin sections (60 nm) were cut from the blocks, collected, and stained with uranyl acetate followed by lead citrate. Images were acquired using a JEM 1400 Transmission Electron Microscope (JEOL Ltd., Tokyo, Japan).
Statistical analysis
All data are presented as mean ± SEM. Error bars are SEM. Two-tailed unpaired Student’s t-tests were performed for the comparison of two groups. For multiple comparisons, analysis of variance were performed followed by Tukey’s post hoc comparison. For all experiments, p < 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA017780) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa. The processed datasets generated in the present study are deposited in the OMIX (China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences), under accession number (OMIX007817, https://ngdc.cncb.ac.cn/omix). The numerical source data behind the graphs and the corresponding statistical analysis are added as Supplementary Data 12. The original and unprocessed pictures of the western blots are added as Supplementary Fig. 7.
References
Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiological Rev. 84, 277–359 (2004).
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Cypess, A. M. et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab. 21, 33–38 (2015).
Bartelt, A. et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 17, 200–205 (2011).
Yu, X. X., Lewin, D. A., Forrest, W. & Adams, S. H. Cold elicits the simultaneous induction of fatty acid synthesis and β‐oxidation in murine brown adipose tissue: prediction from differential gene expression and confirmation in vivo. FASEB J. 16, 155–168 (2002).
Wang, W. & Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. cell Biol. 17, 691–702 (2016).
Fedorenko, A., Lishko, P. V. & Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151, 400–413 (2012).
Enerbäck, S. et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997).
Golozoubova, V. et al. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 15, 2048–2050 (2001).
Feldmann, H. M., Golozoubova, V., Cannon, B. & Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009).
Kazak, L. et al. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proc. Natl Acad. Sci. 114, 7981–7986 (2017).
Bonev, B. & Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 17, 661–678 (2016).
Schoenfelder, S. & Fraser, P. Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet. 20, 437–455 (2019).
Zheng, H. & Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. cell Biol. 20, 535–550 (2019).
Spitz, F. & Furlong, E. E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613–626 (2012).
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011).
Zhang, Y. et al. Chromatin connectivity maps reveal dynamic promoter–enhancer long-range associations. Nature 504, 306–310 (2013).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Ong, C.-T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011).
Visel, A. et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 (2009).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. 107, 21931–21936 (2010).
Stępniak, K. et al. Mapping chromatin accessibility and active regulatory elements reveals pathological mechanisms in human gliomas. Nat. Commun. 12, 3621 (2021).
Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013).
Li, Y. et al. The structural basis for cohesin–CTCF-anchored loops. Nature 578, 472–476 (2020).
Shore, A., Karamitri, A., Kemp, P., Speakman, J. & Lomax, M. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia 53, 1164–1173 (2010).
Abe, Y. et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat. Commun. 6, 7052 (2015).
LeBlanc, S. E., Wu, Q., Lamba, P., Sif, S. & Imbalzano, A. N. Promoter–enhancer looping at the PPARγ2 locus during adipogenic differentiation requires the Prmt5 methyltransferase. Nucleic acids Res. 44, 5133–5147 (2016).
Chouchani, E. T., Kazak, L. & Spiegelman, B. M. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 29, 27–37 (2019).
Van de Werken H. J., et al. 4C technology: protocols and data analysis. In: Methods in enzymology). Elsevier (2012).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. cell Biol. 16, 144–154 (2015).
Jastroch, M. Uncoupling protein 1 controls reactive oxygen species in brown adipose tissue. Proc. Natl Acad. Sci. 114, 7744–7746 (2017).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Ong, C.-T. & Corces, V. G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 (2014).
Merkenschlager, M. & Odom, D. T. CTCF and cohesin: linking gene regulatory elements with their targets. Cell 152, 1285–1297 (2013).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Ramasamy, S. et al. The Mediator complex regulates enhancer-promoter interactions. Nat. Struct. Mol. Biol. 30, 991–1000 (2023).
Rao, S. S. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e324 (2017).
Chien, R. et al. Cohesin mediates chromatin interactions that regulate mammalian β-globin expression. J. Biol. Chem. 286, 17870–17878 (2011).
Heidari, N. et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 24, 1905–1917 (2014).
Kajimura, S., Seale, P. & Spiegelman, B. M. Transcriptional control of brown fat development. Cell Metab. 11, 257–262 (2010).
Inagaki, T., Sakai, J. & Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. cell Biol. 17, 480–495 (2016).
Hiraike, Y. et al. NFIA co-localizes with PPARγ and transcriptionally controls the brown fat gene program. Nat. Cell Biol. 19, 1081–1092 (2017).
Hiraike, Y. et al. NFIA differentially controls adipogenic and myogenic gene program through distinct pathways to ensure brown and beige adipocyte differentiation. PLoS Genet. 16, e1009044 (2020).
Rajakumari, S. et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 17, 562–574 (2013).
Jin, Q. et al. Distinct roles of GCN5/PCAF‐mediated H3K9ac and CBP/p300‐mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 (2011).
Tie, F. et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 (2009).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Kozak, L., Koza, R. & Anunciado-Koza, R. Brown fat thermogenesis and body weight regulation in mice: relevance to humans. Int. J. Obes. 34, S23–S27 (2010).
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009).
Inokuma, K.-i et al. Indispensable role of mitochondrial UCP1 for antiobesity effect of β3-adrenergic stimulation. Am. J. Physiol.-Endocrinol. Metab. 290, E1014–E1021 (2006).
Siersbæk, R. et al. Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol. cell 66, 420–435.e425 (2017).
Rubin, A. J. et al. Lineage-specific dynamic and pre-established enhancer–promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017).
Choy, M.-K. et al. Promoter interactome of human embryonic stem cell-derived cardiomyocytes connects GWAS regions to cardiac gene networks. Nat. Commun. 9, 2526 (2018).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41–45 (2000).
Li, K., Han, J. & Wang, Z. Histone modifications centric-regulation in osteogenic differentiation. Cell Death Discov. 7, 91 (2021).
Kajimura, S., Spiegelman, B. M. & Seale, P. Brown and beige fat: physiological roles beyond heat generation. Cell Metab. 22, 546–559 (2015).
Osterwalder, M. et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243 (2018).
Frankel, N. et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493 (2010).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lai, B. et al. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic acids Res. 45, 6388–6403 (2017).
Emmett, M. J. et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature 546, 544–548 (2017).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. 112, E6456–E6465 (2015).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Zhou, Q. et al. ZNF143 mediates CTCF-bound promoter–enhancer loops required for murine hematopoietic stem and progenitor cell function. Nat. Commun. 12, 43 (2021).
Guo, Y. et al. CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc. Natl Acad. Sci. 109, 21081–21086 (2012).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer-promoter loops. Cell 171, 1573–1588.e1528 (2017).
Angueira, A. R. et al. Early B cell factor activity controls developmental and adaptive thermogenic gene programming in adipocytes. Cell Rep. 30, 2869–2878.e2864 (2020).
Stine, R. R. et al. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol. Metab. 5, 57–65 (2016).
Shao, M. et al. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab. 23, 1167–1184 (2016).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Lowell, B. B. & Spiegelman, B. M. Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 (2000).
Bal, N. C. et al. Both brown adipose tissue and skeletal muscle thermogenesis processes are activated during mild to severe cold adaptation in mice. J. Biol. Chem. 292, 16616–16625 (2017).
Keipert, S. et al. Long-term cold adaptation does not require FGF21 or UCP1. Cell Metab. 26, 437–446.e435 (2017).
Krijger, P. H., Geeven, G., Bianchi, V., Hilvering, C. R. & de Laat, W. 4C-seq from beginning to end: A detailed protocol for sample preparation and data analysis. Methods 170, 17–32 (2020).
Thongjuea, S., Stadhouders, R., Grosveld, F. G., Soler, E. & Lenhard, B. r3Cseq: an R/Bioconductor package for the discovery of long-range genomic interactions from chromosome conformation capture and next-generation sequencing data. Nucleic acids Res. 41, e132–e132 (2013).
Love M., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 Genome Biology 15 (12), 550.) (2014).
Stephens, M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, 1 (2008).
Oeckl, J., Bast-Habersbrunner, A., Fromme, T., Klingenspor, M. & Li, Y. Isolation, Culture, and Functional Analysis of Murine Thermogenic Adipocytes. STAR Protoc. 1, 100118 (2020).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic acids Res. 47, W171–W174 (2019).
Haeussler, M. et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 1–12 (2016).
Bae, S., Park, J. & Kim, J.-S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475 (2014).
Hodgkins, A. et al. WGE: a CRISPR database for genome engineering. Bioinformatics 31, 3078–3080 (2015).
O’Geen, H. et al. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics chromatin 12, 1–20 (2019).
O’Geen, H., Tomkova, M., Combs, J. A., Tilley, E. K. & Segal, D. J. Determinants of heritable gene silencing for KRAB-dCas9+ DNMT3 and Ezh2-dCas9+ DNMT3 hit-and-run epigenome editing. Nucleic acids Res. 50, 3239–3253 (2022).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 25, 402–408 (2001).
Hagege, H. et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 (2007).
Naumova, N., Smith, E. M., Zhan, Y. & Dekker, J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods 58, 192–203 (2012).
Deng, W. et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 149, 1233–1244 (2012).
Montavon, T. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011).
Balkow, A. et al. Direct lentivirus injection for fast and efficient gene transfer into brown and beige adipose tissue. J. Biol. Methods 3, e48 (2016).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2023YFD1300802) and the National Natural Science Foundation of China (32472852).
Author information
Authors and Affiliations
Contributions
L.W. and D.S. designed and supervised the experiments. D.S., T.J., Y.S. and D.L. conducted the experiments and analyzed data. S.Z., T.Z. and J.G. helped to conduct animal experiments. L.W. was responsible for funding acquisition and project administration. D.S., L.L. and H.Z. wrote and edited the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Aaron Brown, Lorena Aguilar-Arnal and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Christopher Hine and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, D., Jiang, T., Song, Y. et al. Identification of a distal enhancer of Ucp1 essential for thermogenesis and mitochondrial function in brown fat. Commun Biol 8, 31 (2025). https://doi.org/10.1038/s42003-025-07468-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07468-3