Abstract
Although intense research effort is seeking to address which brain areas fire and connect to each other to produce complex behaviors in a few living primates, little is known about their evolution, and which brain areas or facets of cognition were favored by natural selection. By developing statistical tools to study the evolution of the brain cortex at the fine scale, we found that rapid cortical expansion in the prefrontal region took place early on during the evolution of primates. In anthropoids, fast-expanding cortical areas extended to the posterior parietal cortex. In Homo, further expansion affected the medial temporal lobe and the posteroinferior region of the parietal lobe. Collectively, the fast-expanding cortical areas in anthropoids are known to form a brain network producing mind reading abilities and other higher-order cognitive functions. These results indicate that pursuing complex cognition drove the evolution of Primate brains.

Similar content being viewed by others
Introduction
Mammals are known for possessing relatively large brain volumes compared to their body mass1, and humans, in particular, stand out among all other endothermic vertebrates in this regard. The close link between cognition and brain size made the latter the focus of intense and continuous scientific effort, seeking to describe trends in encephalization and to investigate the link between brain size and cognitive power2,3,4,5. It is clear, though, that absolute (or relative to the body) brain size is not enough to understand cognition in mammals, since brain shape, the relative proportions of different cortical areas and their mutual connections must have had prominent roles in the evolution of mammalian intelligence6,7,8. For instance, Primates are especially large-brained, but they possess more than just a scaled-up version of the average mammalian brain. Primate brains have smaller and more densely packed neurons9. During their evolution, Primates acquired a granular layer in the prefrontal cortex10,11, including a distinct region, the dorsolateral prefrontal cortex, exclusive to the group12,13 and greatly expanded parietal cortex14. Anthropoid primates further stand out for the unusually high neuron count15, and the increased number of cortical areas16. In terms of behavioral attributes, anthropoids (and apes in particular) are especially good at inferring causal relationships, planning future actions, and in the ability to attribute mental states to oneself and to other individuals, forming a ‘theory of mind’ of the intentions and beliefs of their conspecifics17,18,19.
To understand how the primate brain evolved, we tested which regions of the cortex underwent the most profound evolutionary change throughout the evolution of the group, whether these regions’ evolution differs from that of other mammals, and whether the human brain is exceptional in these regards, marking a discontinuity with fellow monkey and apes. Seeking answers to these research questions is usually hampered by limited and hard-to-acquire data, especially for fossil species, and since a wide phylogenetic scope, although crucial to consider11,20 is difficult to come by. To overcome these limitations and to study the evolution of the primate cortex at a fine scale21, we assembled the largest ever, to our knowledge, collection of three-dimensional mammalian brain endocasts completed with phylogenetic information, consisting of 465 individuals referring to 311 different species, of which 34 are extinct. The species on the tree span across mammals from marsupials to hominins and temporally from the Eocene to the present. We registered the position of 208 landmark and semilandmark points (here to fore ‘landmarks’ for simplicity) on the endocast of each specimen by applying geometric morphometrics (Supplementary Fig. 1, Supplementary Table 1). We developed a method we recently devised, rate.map22, which now allows charting the evolutionary rates of shape change directly on the original endocast 3D meshes or even to magnetic resonance image (fMRI) reproductions of the brain, and to associate the rates to individual landmarks5. Since the rates represent phylogenetic ridge regression slopes23 that have a magnitude and a sign, mapping rates allow for the straightforward interpretation of the direction (whether there is an enlargement or a contraction) and the amount of evolutionary shape change across specific areas of the endocast.
Results and discussion
Primates have peculiar brain shapes
We started by applying a relative warp analysis (RWA, the principal component analysis of the landmark coordinates) on the endocast shapes. The distribution of endocast shapes on the Relative Warp 1 (RW1) to RW2 plot (explaining together 65.85% of the total shape variance) indicates that Primates set apart from all mammals in terms of brain shape, along RW1. This axis, which accounts for 56.94% of the total shape variation, describes changes mainly associated to the dorsomedial frontal cortex (Fig. 1), as opposed to changes in the temporal areas and the frontal pole. By repeating RW analysis isolating the non-uniform component to focus directly on small-scale shape variation24,25, Primates were still found to stand apart from other mammals, by virtue of shape changes that still appear concentrated in the medial frontal, as opposed to occipital and frontopolar areas of the cortex (Supplementary Fig. 2).
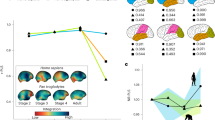
The highest rates of endocast shape evolution pertain to Homo sapiens and Homo neanderthalensis (Fig. 2a). Catarrhini (Old World monkeys) brains evolved at a faster pace compared to other mammals (p value = 0.010). Ferungulata and Marsupialia show an opposite pattern, with significantly low rates (p value = 0.001; Fig. 2b).
a Evolutionary rate values related to the node correspond to the positive and significant shift. b The phylogenetic tree of mammals. The light blue circle indicates the positive shift in Catarrhine. The light red circles indicate the negative shift pertaining to Ferungulata and Marsupialia. The circle size is proportional to the intensity of rate change. c The phylogenetic tree of the Primates (extracted from the tree in b) shows the location of principal primate clades and representative shapes of their respective endocasts. The color gradient represents the map of the evolutionary changes in shape of the endocast from the mammal tree root, in terms of cortical area expansion (blue) and contraction (red). Multivariate rates of brain evolution of individual species are indicated by the colored dots next to the tree tips in both b and c. Brain endocasts of the different species are not to scale. Silhouettes for Homo, Carlito, Lemur, Bradypus, Macropus, Canis, Panthera, and Capreolus are free for reuse under CC0 1.0 Universal Public Domain Dedication license (https://creativecommons.org/publicdomain/zero/1.0/). Silhouettes for Cebus (credits to Sarah Werning) and Ceratotherium (credits to Jan A. Venter, Herbert H. T. Prins, David A. Balfour and Rob Slotow) are free for reuse under the Attribution 3.0 Unported license (https://creativecommons.org/licenses/by/3.0/). Silhouette for Macaca is free for reuse under the Public Domain Mark 1.0 license (https://creativecommons.org/publicdomain/mark/1.0/).
To verify the notion that catarrhine brains evolved faster than in other mammals, we further fitted four different evolutionary models: single rate Brownian Motion model (BM), Ornstein-Uhlenbeck model (OU), and two different multiple rate BM (BMM), one by setting a different rate for anthropoids, and the other by setting a different rate for Catarrhini only. By applying the extended information criterion (EIC)26 we found the latter performs as the best model (EIC = −20865.44) followed by BMM with anthropoids designed to evolve under a different rate (EIC = −20857.24), and then by OU and BM (EIC = −20785.76 and −20789.3, respectively). Clearly, although designing only catarrhine or the entire anthropoid clade as evolving under a different rate regime makes little difference, these nested clades differ from the rate regime experienced by the rest of the mammal tree.
Anthropoids experienced fast cortical expansion across the brain dorsal midline
We charted evolutionary rates on the endocasts to elucidate patterns of cortical expansion and contraction. Primate brain evolution is characterized by enlarging frontal cortex, which is faintly pronounced in stem strepsirrhinae (Adapis) but becomes more evident in anthropoids (Fig. 2c). Towards the human branch of the primate tree, a second area, the posterior parietal cortex (PPC), becomes interested by fast cortical expansion (Fig. 2c). By using the non-uniform component of shape variation, rate shifts coincide in position, direction and magnitude to the analysis of entire shape. Still, the evolutionary enlargement of the posterior parietal area among anthropoids is similarly evident (Supplementary Fig. 3).
To investigate the patterns of evolutionary cortical expansion at a finer scale, we selected the landmarks in the top quartile distribution of the evolutionary rate values (i.e. the fastest evolving landmarks), that are indicative of cortical expansion in primates. Then, we selected among these landmarks those shared by at least 90% of the primate species. This ‘id90primate’ figure is thus set to recognize the areas of the endocast interested by the largest morphological expansion and shared by most species in the clade. We repeated the procedure for anthropoids (id90anthropoid), for Catarrhini (id90catarrhini), and for the genus Homo (id90Homo). The id90primate is formed by landmarks occurring in the prefrontal area (Fig. 3). This conforms to the strongly held notion that the enlargement of the prefrontal cortex characterized brain evolution in the group5,27,28. In id90anthropoid, the medial PPC adds to the prefrontal, a pattern replicated in catarrhine monkey (id90catarrhini), marking a discontinuity between anthropoids and prosimians. The same discontinuity is evident repeating the analysis using the non-uniform shape component (Supplementary Fig. 4). Finally, in id90Homo, areas of fast cortical expansion further comprise the lateral parietal cortex, including the planum temporale (Fig. 3).
The coloured areas represent the areas of the cortex interested in the greatest evolutionary cortical expansion and the position of the landmarks associated with these areas (from left to right): all primates (id90primate), anthropoids (id90anthropoid), catarrhine (id90catarrhini) and in the genus Homo (id90Homo). The top row indicates the location of the areas in the consensus shape triangle mesh. The bottom row represents the same figure located upon representative species for each group. Silhouettes for Lemur, Gorilla, and Homo are free for reuse under CC0 1.0 Universal Public Domain Dedication license (https://creativecommons.org/publicdomain/zero/1.0/). Silhouette for Alouatta is free for reuse under the Public Domain Mark 1.0 license (https://creativecommons.org/publicdomain/mark/1.0/).
These findings withstand independent verification. We tested whether the areas marked by cortical expansion collectively cover, in primates, a proportionally larger area of the endocast compared to other mammals. We found that id90primate covers 26.2% of the endocast, which is significantly larger than in other mammals (mean percentage for mammals other than Primates: 19.3%, p value of the difference: << 0.001). For id90anthropoid and id90catarrhini the corresponding figures are extremely similar to each other, and both significantly larger than in other mammals (id90anthropoid 36%, p << 0.001; id90catarrhini 35.7%, p << 0.001). Finally, for id90Homo, the inclusion of additional, lateral parietal areas into the fast-expanding zones (Fig. 3) resulted in an overall larger proportion (40.7%), which is significantly larger than in non-human mammals (p << 0.001). In addition to this proof, we extracted the landmark coordinates defining id90anthropoid for all primate species and reprojected the coordinates to a common ordination space, that allows replicating PCA on Primates only. In this new PCA space anthropoids are set apart from non-anthropoids along PC1 (Fig. 4). The difference in shape along this axis concentrates in the frontal and PPC, confirming that the patterns we found with rate.map are genuine (Fig. 4). The same applies when restricting the shape analysis to the non-uniform component (Supplementary Fig. 5).
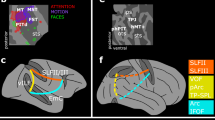
To identify where the greatest evolutionary expansion occurs over the brain cortex of Homo sapiens, we devised a procedure to associate the top quartile landmarks interested by cortical expansion in our species to the digital MRI reproduction of a human individual’s brain. These landmarks coincide with the prefrontal and, especially, the dorsomedial prefrontal cortex (dmPFC), with the lateral prefrontal cortex (lPFC), including the inferior frontal gyrus (IFG), with the PPC (in coincidence to the superior parietal lobule), with the angular gyri within the inferior parietal lobules, and eventually with the medial temporal lobe (MTL, Fig. 5). By inspecting the non-uniform component of shape variation, dmPFC and PPC at least still appears the main loci for cortical expansion in Homo sapiens (Supplementary Fig. 6).
Selection for higher-order cognition?
Early mammals had relatively small neocortex3 containing a single layer of pyramidal neurons. The neocortex later extended to six layers in present-day mammals29. Several derived features of the primate PFC are known to confer them complex cognitive capacities, playing crucial roles in the representation of the self, working memory, decision-making, and social interactions12,30,31,32,33. Therefore, most investigations of brain evolution in Primates focused on the relative size of PFC and tend to indicate this large area of the brain greatly expanded towards the human branch of the tree5,28,30,34. Here, we found evidence supporting this same pattern (Figs. 2 and 3, Supplementary Figs. 3 and 4). Yet, we were also able to highlight the regions of the endocast, and the clades, interested by the highest rate of cortical expansion. This trend starts with dmPFC (Fig. 2c), a brain region involved in managing emotions and understanding others in both humans and other primates31,35,36,37,38. In anthropoids, expanding cortical regions extend to the superior parietal lobule in the PPC. Finally, in Homo sapiens, enlarging cortical areas further extend to the angular gyri and the MTL (Fig. 5). dmPFC and PPC are strongly connected to each other and participate in brain networks involved in social cognition19,32,39,40,41. These overlapping networks include the default mode network (DMN)42,43,44, the mirror neuron system (MNS)45, and the theory of mind network (ToM)19. In humans, DMN, MNS and ToM extend to IFG, MTL and the angular gyri19,39,42. ToM is only present in humans and to a lower level in greater apes, whereas MNS does not include IFG in non-human primates and includes the inferior- but not the superior parietal lobules45. In contrast, the DMN is present and well-developed in both humans and other primates and extends to further, non-social higher-order cognitive tasks such as mind-wandering, long-term memory consolidation and mental time travel32,42,44,46,47. Resting state networks functionally equivalent to the human DMN were identified in a variety of mammals besides primates, including rats, ferrets, and dogs48,49,50,51. Yet, our results suggest that cortical areas expansion in DMN-related regions of the brain played a special role in primate evolution, indicating the behavioral outputs produced by the network were a primary target of selection during the evolution of anthropoids in particular. This interpretation further agrees with earlier findings suggesting that the precuneus52, medial PFC47, PFC in general11,30 and PPC20,53,54 are all larger in anthropoids as compared to prosimians. However, there is momentous variation even within the group. The IFG is important to integrate facial and vocal information in catarrhines but is not nearly as specialized in platyrrhines55,56. Albeit humans and chimpanzees may have similar DMNs49, in non-hominoid Primates (macaques, marmosets, and mouse lemurs) the connection between dmPFC and PPC is less strongly developed57,58 suggesting that although a shared pattern for evolution towards DMN-related areas occurs in anthropoids, it develops further in Homo (Figs. 3 and 5).
We propose that the special features of anthropoid cognition, including their standout ability for mind-wandering, communication, logical inference, and mindread others, affected brain evolution in the group. The reasons for selection in favor of DMN-related brain activities are readily apparent, given the obvious fitness advantages provided by higher-order cognition, including creativity, complex sociality, and Machiavellian intelligence, which all link to DMN31,59,60. This interpretation derives from the projection of the landmarks associated with the highest rate of cortical expansion on the human brain and, thence, the cortical areas they intersect and their functions. As such, there are potential caveats to consider which may undermine its validity. First, we measured cortical expansion in terms of an increased proportion of the cortex surface occupied by a specific portion of the endocast. Cortical areas, though, develop in three dimensions, which means our metric is a proxy for cortical area expansion at best. Secondly, although we strove to place landmarks according to anatomical homology (Supplementary Fig. 1, Supplementary Table 1), and slid semilandmarks on the endocasts keeping the frontal, cerebellar and neocortex (exclusive of the frontal portion) regions separate in order to maintain topological consistency (Methods), we implicitly assumed that the positioning of major cortical territories is the same across mammals. There is good evidence that this is true at the very least for medial PFC across mammals3,11 and for both medial PFC and PPC across primates54,61 meaning the evolutionary trend for cortical expansion in these areas is genuine.
It is worth noticing that we avoided referring to specific areas of the cortex, such as Brodmann areas, and their functions in interpreting the results because neither topological nor functional homology is maintained across species at this small level of investigation62,63. Still, there are valid statistical reasons not to interpret landmark variation one landmark at a time64. This implies that we can infer function only at a coarse scale of investigation. Moreover, our insights only hold if the expanding areas held similar functions over the evolutionary time. Consistently with this assumption, dmPFC is involved in decision-making, self-representation and social behavior in all mammals and plays a crucial role in processing social information and the representation of the self in Primates31,41,65. The PPC is involved in decision making, working memory, and motor planning66 across mammals. For what concerns the DMN, it is feasible to assume that monkeys engage in cognition detached from goal-oriented behavior, mostly like humans do46. Their DMN recruits PFC and PPC as ours, although the overlap between brain areas active during resting states between humans and model monkey species is not perfect46,58. A study on captive macaques found that the medial frontal cortex progressively activates more and more as the test individual is exposed to an increasing number of conspecifics during resting state experiments67. This further points to the preeminent role of dmPFC in particular31,60,65 and medial PFC in general31,35, to primate social lives. Still, on the functional grounds, the paracingulate sulcus within the dmPFC provides the ability to mentalize (that is to endow the individual’s mind with metarepresentations of the world), which is a highest-order cognitive function, in Hominoidea (rather than just in humans as previously thought). To a rudimentary level, this skill can be traced to Old World monkey (Catarrhini)47. The role of the DMN in mentalization and social cognition seems especially linked to dmPFC and PPC19,60,65.
Most studies point to the precuneus and posteromedial cortex (rather than just PPC) as the main areas involved in DMN in primates40,42,46,58. We cannot study the brain inner structures by looking at the endocasts, let alone the understanding of how the connections between different cortical areas evolved. Yet, in keeping with our results, it was demonstrated that humans have greatly expanded PPC68 and the expansion of the precuneus52 within the posteriomedial cortex can only have expanded the posteriomedial cortex outer surface.
Despite the comforting evidence bolstering our insight that dmPFC and PPC expansion in anthropoids were under selection, it is worth considering that primates evolved different new cortical areas and novel connections between them, that may have produced new or improved functions and behavioral outputs regardless of the relative cortical surface extension. For instance, in anthropoid primates, the rostral portion of the PPC is expanded compared to strepsirrhine (galagoes)16. Our results are consistent with this finding, and with the notion that strong connections occurring between dorsolateral PFC and the parietal and temporal cortices are typical of Primates12. Yet, these connections are embodied in the dense corticotectal projections originating from the frontal, parietal and temporal areas12 and reach the dorsal pulvinar and superior collicus, something that our data cannot reveal. Interestingly, Kaas16 highlighted that expanding PPC in the human lineage may have been involved the use of tools and gestures for communication.
One additional caveat regards the MTL. This region is associated with DMN in humans42,43. Yet, the relationship between MTL and DMN is complex42, and MTL shows reduced centrality within DMN compared to both dmPFC and PPC43. In the chimpanzee’s and monkeys’ DMNs, the greatest overlap with human’s occurs across the dmPFC and PPC46,49, rather than MTL. Our results indicate that the recruitment of MTL within the DMN could be confined to Homo sapiens (Fig. 5).
The findings we report here point to the emphasis on DMN-related brain activity during anthropoid evolution. They further indicate that the human DMN does not need, and probably is not the same as in the anthropoid brain. Overall, the monkey and ape DMNs are especially developed along the cortical midline10,46, which is entirely consistent with our findings, but also suggests that there are features of the human DMN not shared with other species. In keeping with this, the PFC in Homo sapiens follows a different allometry than in other Primates, is more lateralized27, and the PPC has a richer array of connections compared to other primates, most notably mapping to tool use and speech centers (in the IFG)40. Still, the human brain has overall stronger patterns of covariation among the brain cortical areas than any other primate alive69 and the dmPFC-to-PPC connection is better developed in our species58. These non-trivial differences suggest that in Homo sapiens the DMN activity is maximized and most probably includes novel functions that are not present in other species. Yet, the evolutionary drive towards DMN-related behavioral outputs is shared among anthropoids, suggesting a difference in grade, rather than a sheer innovation, subtended the evolution of the human mind.
Methods
Reproducing the mammalian endocasts
We collected 93 mammalian endocasts either from websites or previous studies (Supplementary Data 1). Three-hundred and seventy-two additional specimens refer to three-dimensional skulls originally obtained via CT scanning. For these specimens, we applied the function endomaker from the package Arothron70 to retrieve the endocast shape. endomaker is a fast endocast extraction tool which is as precise as manual segmentation in terms of endocast reproduction and endocast volume calculation71,72. Most of the specimens were completed with gender information, so that we could get a fair representation of each sex per species. The specimens refer to 311 different species, 34 of them (unsexed) refer to fossil taxa. For the latter, since the fossilization process comes with natural deformation, we limited the sampling collection to symmetric and undeformed specimens only (Supplementary Data 1).
Landmarks and semilandmarks positioning
The landmarking procedure begun by sampling 18 landmark points on each specimen, focusing on both anatomical and geometrical similarities (Supplementary Table 1). The study sample includes specimens belonging to Marsupialia, Carnivora, Artiodactyla, Perissodactyla, Notoungulata, Xenarthra, and Primates. Although most of the landmarks were placed following geometric criteria (i.e., frontal pole, occipital pole, see Supplementary Table 1, Supplementary Fig. 1), we conducted an in-depth revision of the relevant literature pertaining to the brain anatomy of each group to identify homologies aiding the landmarks placement. The whole procedure was repeated three times testing whether the inter-operator error was significant. We found the average error around each landmark to be 2.77% (1.1–4.3%).
In eutherian mammals, one of the largest frontal areas is the motor cortex, which can be identified by robust criteria in most cases73. Hence, it is possible to partition the brain into three main regions, the frontal portion, the rest of the cortex and the cerebellum (which is morphologically quite distinctive and separated by the telencephalon by the sinus transversus). In Primates, the inferior convolution of the frontal lobe is delimited by the lateral sulcus (LS; i.e. the Sylvian scissure), while the posterior limit is demarcated by the central sulcus (CS). The CS separates the motor cortex (anteriorly to CS) from the somatosensory cortex (posteriorly). However, there are some notable exceptions. As noted in ref. 74 tarsiers and galagoes do not have a CS, and owl monkeys have only a shallow dimple in the cortex where the CS would occur. We, therefore, decided to mark the boundary of the frontal portion as medially appearing above the anterior margin of LS in these species and used the CS otherwise14,75,76,77,78,79,80,81.
The cruciate sulcus is a distinctive carnivoran feature82, known to coincide with the premotor and motor areas83,84 and, hence, to separate the frontal lobe from the rest of the cortex. There are exceptions among carnivores as well. Some viverrid species (i.e., Genetta) lack the cruciate sulcus but may have an incipient ansate sulcus medially to the coronal sulci to guide the placement of the landmark identifying the rearmost limit of the frontal lobe85,86. To recognize the correct positioning of the cruciate sulcus we referred to detailed reports illustrating carnivoran brain anatomy in both living and fossil carnivore species87,88,89,90,91,92.
Among Artiodactyla, one of the best-studied species for neurological research is Ovis aries. Its brain is claimed to be analogous to the human brain in many regards and can be similarly divided into four lobes (frontal, parietal, temporal, and occipital)93,94. The sheep brain is further very well-known because of extensive MRI analyses and detailed atlases95. As stated by John and co-authors94, the frontal lobe lies anterior to the ansate (cruciate) sulcus in Ovis. Similarly, the pig brain matches human brain’s gross anatomy96, and the limit of the motor cortex can be located between the cruciate and the ansate sulci. It is to be noticed that the nomenclature of the cruciate and ansate sulcus is not universally accepted and can differs among institutions97, and even for the same species, as pointed out by Graïc and co-author in describing the giraffe brain98. The pattern of cerebral sulci differs among Artiodactyla (figure 13.3 in ref. 99). Yet, the major sulci can always be recognized100,101,102,103,104.
Among Perissodactyla, the neuroanatomy of horses and donkeys is well-studied105,106, and the main delimitations between brain lobes were readily identified. The position of the motor cortex can be safely recognized by the presence of the cruciate sulcus107. For the identification of cerebral sulci on the rhinoceros brain, we followed ref. 108 which provides a description of its gross anatomy. As stated in ref. 109, living Xenarthra show many cortical architectural traits in common with the stem eutherian mammal, including the motor cortex. We followed the sulcal pattern in Xenarthra described in ref. 110.
Unlike placental mammals, marsupials do not have a well-distinguished motor cortex. Yet, the somatosensory cortex (S1) is recognizable73,111. Kaas112 proposed that a so-called sensory-motor amalgam represents the primitive form of the mammalian neocortex, which is segregated into separate sensory and motor areas in placentals. Karlen and Krubitzer113 illustrated that starting with early studies of the motor cortex in mammals the amalgam62,114 is topographically organized and located at the rostral pole of the neocortex115,116. They also illustrated two different hypotheses for the evolution of sensorimotor cortex in mammal, which show that, unlike what is known for monotremes, in marsupials S1 and the motor cortex have the same orientation, and there is a complete overlap in less derived groups. Karlen and Krubitzer113 eventually proposed that the somatosensory rostral field of marsupial brain may be homologous to area 3a of primates and flying foxes. With this knowledge at hand, and thanks to the Comparative Brain Anatomy atlas by Prof. Kenneth Ashwell and Dr. Yamila Gurovich (https://www.comparativebrainanatomy.org/), we were able to identify sulcal patterns on the marsupial endocasts.
Sliding procedure
The endocast morphology shows high variability among mammals. The sliding procedure for semilandmark placing can be affected by this variability. On the other hand, maintaining the homology between configurations is crucial. To provide the best fit for semilandmarks, we elaborated a new sliding protocol which works as follows. We performed a preliminary alignment of the landmarks configurations to identify the specimen closest to the consensus shape, and its 3D surface was chosen as reference, in this case Eulemur mongoz USNM35260. Then, we cut the 3D surface of the neocortex into three different patches by using Geomagic Studio software (version 2014.3.0; © 2014 Geomagic): 1. frontal portion, 2. non-frontal and 3. cerebellum. The frontal portion was separated from the rest of the neocortex along the coronal plane considering landmarks number 16–18 as boundary. The cerebellum was separated along the transverse sinuses.
Semilandmarks were automatically sampled on each patch separately through the k-means clustering methods (kmeans function, ‘stats’ R package) as follow:
-
1.
the points of the patch surface are clustered and partitioned into k groups following the Voronoi tessellation (which minimizes the sum of squares distance from each 3D mesh triangle vertex to the cluster it belongs);
-
2.
all cluster centers are at the mean of their Voronoi sets (the set of data points which are nearest to the cluster center), and the center of each group returned.
At the end of this process, we produced three different bilateral semilandmark patches (Supplementary Fig. 1): one for the frontal region (60 semilandmarks), one for the non-frontal neocortex (100 semilandmarks), and one for the cerebellum (30 semilandmarks).
Based on these templates, the semilandmarks were then transferred to all other Primates specimens by following the sliding procedure described in ref. 117:
-
1.
create a template with semilandmarks placed outside the 3D surface (see above)
-
2.
create an atlas which can work as a guideline in which points that are allowed to slid are specified (createAtlas function, Morpho R package)
-
3.
transfer the semilandmarks to all other specimens (placePatch function, Morpho R package)
-
4.
slide semilandmarks along surfaces (slider3d function, Morpho R package)
In dealing with Primates, the atlas was the original template generated on Eulemur mongoz surface. In any other case, the template was warped on the closest-to-mean specimen per group via thin-plate spline (tps3d, Morpho R package). With this step, it was possible to maintain the number and the identity of semilandmarks.
The iterations for the sliding function can be set with the argument iterations. We set the argument equal to 0, which means iteration until convergence, except for marsupials for which the empirically derived value of 10 was used.
Once that all the separate sliding sessions were performed, we put all sets together and we performed a final and global sliding to minimize the noise which could have been generated from the group-wise and cortical region-wise procedures, setting the iteration parameter to 10.
Then, we performed a global sliding to minimize the noise which could have been generated from the group-wise and cortical region-wise procedures. Finally, all the configurations were symmetrized. The symmetrization was performed via symmetrize, a function embedded in the Morpho R package based on the reflection and relabeling procedure.
Phylogenetic tree and comparative methods
We built an informal supertree including all the species in our sample. The tree was constructed by means of the tree.merger tool118 available within the RRphylo R package (v. 2.8.0). Our main references for species phylogenetic positions and extinction ages are ref. 119 for primates and ref. 120 for all other living species. Locations and extinction ages for missing species were individually found from different sources (Supplementary Data 1). The final tree includes 311 species (277 extant, 34 extinct). The tree root is set at 141.8 Ma120.
Relative warp analyses
The standard principal component analysis (PCA) is an effective way to reduce variable dimensionality. In geometric morphometrics (GM), the variables are represented by the coordinates of the superimposed landmarks (and semilandmarks), or warp scores, calculated through mathematical decomposition of the deformations computed via Thin-Plate Spline (TPS)121. The TPS is a mathematical algorithm used to predict the difference in shape between a reference and another shape and can address affine and non-affine deformations separately.
We analyzed the shape variation via RWA (a PCA of warp scores) with the function relWarp included in the Morpho R package122. Procrustes superimposition applied to remove the size, orientation, and translation effect is part of the function.
As we are interested in analyzing the entire shape variation (affine and non-affine deformation taken together), we set the alpha parameter equal to 0. The alpha parameter is a means to emphasize the large-scale/small-scale variation when set equal to 1/−1. When the parameter is set to 0, a standard PCA is performed.
Besides using ordinary RWA with alpha set to zero, we implemented a second shape analysis to look for shape variation occurring at the local scale. Following Grunstra and colleagues25, we replicated the analyses separating the non-affine from the affine shape variation, as non-uniform deformation has different effects over different regions of the form. Grunstra and colleagues developed the function prWarp in R to compute principal warps, partial warp scores, and the non-affine component of shape variation for two-dimensional data25,123. We implemented their algorithm create.pw.be to work with three-dimensional data.
The partial warps are obtained through the decomposition of the bending energy matrix, which is a function of the distance between landmarks in the reference shape. Usually, the chosen reference is the consensus shape retrieved from the Procrustes superimposition. In this study, since we were interested in evolutionary changes, we decided to use as reference the shape of mammalian ancestor reconstructed by applying the RRphylo function23, available within RRphylo R package toolkit (v. 2.8.0). RRphylo applies phylogenetic ridge regression to the tree and data (see below) to derive branch-wise rates of phenotypic evolution and estimate ancestral phenotypes at internal nodes, while minimizing within-clades and maximizing between-clades phenotypic differences. Here the phenotypes are the RW scores. RRphylo provides ancestral character estimation at nodes, from which we retrieved the vector of RW scores at the root of the tree124. To compute the root estimate, we first performed a preliminary PCA and supplied the PC scores to RRphylo, to retrieve the PC scores at the tree root. Then, we use restoreShapes, an algorithm embedded in the package Morpho, to restore the shape coordinates at the root from the PC scores estimates. The coordinates estimated at the root were used to obtain the coordinates of the non-uniform shape, and to perform a RWA (from now on RWA, which is a PCA weighted by the bending energy125) by using custom functions from prWarp package126 .
Eventually, we extracted the relative warp scores (RW scores) and averaged the values per species, which were used for further analyses as representing the endocast ‘phenotype’. We applied this procedure both to the full-shape and to the non-uniform component datasets.
Evolutionary shape analysis
We used RRphylo to locate instances of rate shift in endocast shape evolution, by applying the function search.shift23.
To map the intensity and direction of evolutionary change directly on endocast models, we used the rate.map function embedded in the brand-new R package RRmorph5. For a node-species pair, rate.map identifies the RW axes linked to the highest (lowest) evolutionary rate values for the species and reconstructs its endocast morphology weighted on such axes. Then, it selects the RW axes associated with the highest magnitudes in evolutionary rate by using the extreme distance estimator approach127 and uses the RW scores estimated by RRphylo at the focal node to reconstruct the ancestor’s 3D surface mesh. Eventually, the algorithm calculates the area differences between corresponding triangles of the species’ and the ancestor’s reconstructed surfaces. Finally, the values of differences associated to each triangle of the 3D surface are transferred to the vertices via an interpolation procedure (interpolMesh function from RRmorph). Since the species reconstructed surface is based on RWs with extremely positive and negative rate values, the magnitude of the area differences is directly related to the magnitude of the rates, so that the latter are conveniently translated into shape changes.
Testing for different evolutionary models
We tested the fitting of four different evolutionary models: single rate BM, Ornstein-Uhlenbeck model (OU), multiple rate BM (BMM) by setting a different rate for anthropoids, and BMM by setting a different rate for Catarrhini. The models were fitted by the function mvgls embedded in the R package mvMORPH128.
Models’ comparison was performed via EIC26 using the function EIC (mvMORPH R package).
Selection of landmarks occurring in high-rate areas
To find which landmarks (and semilandmarks) occur in areas of the endocast experiencing significant relative expansion in a particular clade, we collated the area difference between the mesh triangles (the reconstructed mesh minus the common reference mesh triangle area) from the largest to the smallest. Then, we calculated the 75th percentile (90th percentile when the non-affine variation is considered) of the area differences and selected all landmarks and semilandmarks that fall in the selected triangles in at least 90% of the species of the focal clade. This procedure serves the aim of selecting those landmarks and semilandmarks coinciding with the areas of maximum expansion for most of the species in the clade.
Testing expansion of landmarks occurring in high-rate areas with aligned coordinates
We tested the enlargement of areas involved in the DMN resulting from multivariate statistics and phylogenetic ridge regression by measuring from raw data (shape variables and 3D endocranial meshes) their relative surface areas.
We used the three-dimensional coordinate of the consensus shape to build a 3D surface with Geomagic Studio software (v. 2014.3.0). We used this template together with the averaged coordinates to reconstruct the surface of each species. Subsequently, we isolated the triangles defined by selected vertices occurring in high-rate areas, and their area was calculated. We compared the relative isolated areas selecting in hierarchical manner all the species belonging to Primates, anthropoids and Catarrhini.
Analyzing selected areas via GM
We decided to delimitate the shape variation analysis among anthropoids to the areas subjected to high rates of cortical expansion. The areas were treated as separated modules, and their morphological information was combined into a single dataset under a GM context with combinland, a method written in R language (Arothron R package), which builds a common ordination space considering the entire shape information encoded in the starting configurations129.
Transferring cortical expansion/contraction patterns on the fMRI digital reproduction of Homo sapiens brain
To visualize the cortical areas affected by high rate of evolution, we used a digital reproduction of fMRI image of the same individual of Homo sapiens showed in Fig. 3 whose endocast was included in the analyses. To this aim, we sampled the same 18 landmarks (Supplementary Table 1) directly on the fMRI brain surface. Then, we transferred the semilandmarks patch of the corresponding H. sapiens endocast specimen on the brain surface by using the thin-plate spline algorithm (tps3d function in Morpho R package). Eventually, we used Morpho’s slider3d on both endocast and fMRI semilandmarks sets to minimize the differences between them. The cortical expansion/contraction patterns on the fMRI were eventually interpolated for the endocasts by using the interpolMesh algorithm in RRmorph5.
Statistics and reproducibility
Statistical analyses were performed by using R Studio Software version 2023.06.0 based on R 4.4.0. The R packages used to run the analyses were RRphylo23, RRmorph5, Morpho, and Rvcg122. Digital models of the skulls were either downloaded from public repositories, repositories available upon request, or provided by colleagues. The full list of specimen provenances is available as Supplementary Data 1. The complete list of the species with their status (living/extinct), extinction age, reference for phylogenetic position, and reference for extinction age are provided in Supplementary Data 2. R script associated with this study is available as a Supplementary Data 3.zip file. The files include annotated R scripts that describe the entire procedure used to reproduce the analysis with both alpha = 0 and by using the non-affine shape. Data to reproduce the analysis and results that take too long time are also provided as .rda files. All the materials to reproduce the analyses (along with information about specimen provenance and references) are available via Zenodo130.
Endocasts were extracted via the R software endomaker71 available within the R package Arothron70. CT and laser scans were processed in Amira131 (version 5.4.5) and landmarked with this same software. Rate analysis was performed with the function RRphylo23. We reproduced and analyzed 465 specimens overall, referring to 311 different species, inclusive of 34 extinct taxa. Figures and plots were prepared using the R package ggplot2132 and the Photoshop suite.
Data availability
All data necessary to reproduce the findings presented in the main manuscript and in the supplementary material are available at Zenodo, https://doi.org/10.5281/zenodo.14614902.
Code availability
Input data to reproduce the analyses in R and the supporting R code are available at Zenodo, https://doi.org/10.5281/zenodo.14614902.
References
Tsuboi, M. et al. Breakdown of brain–body allometry and the encephalization of birds and mammals. Nat. Ecol. Evol. 2, 1492–1500 (2018).
Smaers, J. B. et al. The evolution of mammalian brain size. Sci. Adv. 7, 1–12 (2021).
Kaas, J. H. The evolution of brains from early mammals to humans. Wiley Interdiscip. Rev. Cogn. Sci. 4, 33–45 (2013).
Venditti, C., Baker, J. & Barton, R. A. Co-evolutionary dynamics of mammalian brain and body size. Nat. Ecol. Evol. 1–9 https://doi.org/10.1038/s41559-024-02451-3 (2024).
Melchionna, M. et al. RRmorph—a new R package to map phenotypic evolutionary rates and patterns on 3D meshes. Commun. Biol. 7, 1009 (2024).
Smaers, J. B. & Soligo, C. Brain reorganization, not relative brain size, primarily characterizes anthropoid brain evolution. Proc. R. Soc. B: Biol. Sci. 280, 20130269 (2013).
Schwartz, E. et al. Evolution of cortical geometry and its link to function, behaviour and ecology. Nat. Commun. 14, 2252 (2023).
Pang, J. C. et al. Geometric constraints on human brain function. Nature https://doi.org/10.1038/s41586-023-06098-1 (2023).
Herculano-Houzel, S., Collins, C. E., Wong, P. & Kaas, J. H. Cellular scaling rules for primate brains. Proc. Natl Acad. Sci. 104, 3562–3567 (2007).
Hutchison, R. M. & Everling, S. Monkey in the middle: why non-human primates are needed to bridge the gap in resting-state investigations. Front. Neuroanat. 6, 29 (2012).
Preuss, T. M. & Wise, S. P. Evolution of prefrontal cortex. Neuropsychopharmacology 47, 3–19 (2022).
Preuss, T. M. Evolutionary specializations of primate brain systems. In: Primate origins: adaptations and evolution 625–675 (Springer). https://doi.org/10.1007/978-0-387-33507-0_18 (2007).
Ma, S. et al. Molecular and cellular evolution of the primate dorsolateral prefrontal cortex. Science (1979) 377, (2022).
Kaas, J. H., Qi, H.-X. & Stepniewska, I. The evolution of parietal cortex in primates. In: Handbook of Clinical Neurology, vol. 151 31–52 (Elsevier B.V., 2018).
Kverková, K. et al. The evolution of brain neuron numbers in amniotes. Proc. Natl. Acad. Sci. 119, e2121624119 (2022).
Kaas, J. H. The evolution of neocortex in primates. In: Progress in Brain Research. Vol. 195, 91–102 (Elsevier B.V., 2012).
Hayashi, T. et al. Macaques exhibit implicit gaze bias anticipating others’ false-belief-driven actions via medial prefrontal cortex. Cell Rep. 30, 4433–4444.e5 (2020).
Watanabe, S., Hofman, M. A. & Shimizu, T. Evolution of the brain, cognition, and emotion in vertebrates. (Springer Japan, Tokyo). https://doi.org/10.1007/978-4-431-56559-8., 2017
Wang, Y. et al. A large-scale structural and functional connectome of social mentalizing. Neuroimage 236, 118115 (2021).
Garin, C. M., Garin, M., Silenzi, L., Jaffe, R. & Constantinidis, C. Multilevel atlas comparisons reveal divergent evolution of the primate brain. Proc. Natl. Acad. Sci. 119, e2202491119 (2022).
de Sousa, A. A. et al. From fossils to mind. Commun. Biol. 6, 636 (2023).
Castiglione, S. et al. Human face‐off: a new method for mapping evolutionary rates on three‐dimensional digital models. Palaeontology 65, 1–10 (2022).
Castiglione, S. et al. A new method for testing evolutionary rate variation and shifts in phenotypic evolution. Methods Ecol. Evol. 9, 974–983 (2018).
Mitteroecker, P. & Schaefer, K. Thirty years of geometric morphometrics: achievements, challenges, and the ongoing quest for biological meaningfulness. Am. J. Biol. Anthropol. 178, 181–210 (2022).
Grunstra, N. D. S., Bartsch, S. J., Le Maître, A. & Mitteroecker, P. Detecting phylogenetic signal and adaptation in papionin cranial shape by decomposing variation at different spatial scales. Syst. Biol. 70, 694–706 (2021).
Ishiguro, M., Sakamoto, Y. & Kitagawa, G. Bootstrapping log likelihood and EIC, an extension of AIC. Ann. Inst. Stat. Math. 49, 411–434 (1997).
Smaers, J. B. et al. Primate prefrontal cortex evolution: human brains are the extreme of a lateralized ape trend. Brain Behav. Evol. 77, 67–78 (2011).
Donahue, C. J., Glasser, M. F., Preuss, T. M., Rilling, J. K. & Van Essen, D. C. Quantitative assessment of prefrontal cortex in humans relative to nonhuman primates. Proc. Natl. Acad. Sci. 115, E5183–E5192 (2018).
Hofman, M. A. Size and shape of the cerebral cortex in mammals. Brain Behav. Evol. 32, 17–26 (1988).
Smaers, J. B., Gómez-Robles, A., Parks, A. N. & Sherwood, C. C. Exceptional evolutionary expansion of prefrontal cortex in great apes and humans. Curr. Biol. 27, 714–720 (2017).
Isoda, M. The role of the medial prefrontal cortex in moderating neural representations of self and other in primates. Annu. Rev. Neurosci. 44, 295–313 (2021).
Yokoyama, C. et al. Comparative connectomics of the primate social brain. Neuroimage 245, 118693 (2021).
Baetens, K. L. M. R., Ma, N. & Van Overwalle, F. The dorsal medial prefrontal cortex is recruited by high construal of non-social stimuli. Front. Behav. Neurosci. 11, 44 (2017).
Levy, R. The prefrontal cortex: from monkey to man. Brain 147, 794–815 (2024).
Cirillo, R., Ferrucci, L., Marcos, E., Ferraina, S. & Genovesio, A. Coding of self and other’s future choices in dorsal premotor cortex during social interaction. Cell Rep. 24, 1679–1686 (2018).
Gangopadhyay, P., Chawla, M., Dal Monte, O. & Chang, S. W. C. Prefrontal–amygdala circuits in social decision-making. Nat. Neurosci. 24, 5–18 (2021).
Le Petit, M. et al. Functional connectivity of the medial prefrontal cortex related to mindreading abilities. Cereb. Cortex Commun. 3, 1–13 (2022).
Ferrucci, L. et al. Dedicated representation of others in the macaque frontal cortex: from action monitoring and prediction to outcome evaluation. Cereb. Cortex 32, 891–907 (2022).
Alcalá-López, D. et al. Computing the social brain connectome across systems and states. Cereb. Cortex 28, 2207–2232 (2018).
Dadario, N. B. & Sughrue, M. E. The functional role of the precuneus. Brain https://doi.org/10.1093/brain/awad181 (2023).
Báez-Mendoza, R., Mastrobattista, E. P., Wang, A. J. & Williams, Z. M. Social agent identity cells in the prefrontal cortex of interacting groups of primates. Science 374, eabb4149 (2021).
Smallwood, J. et al. The default mode network in cognition: a topographical perspective. Nat. Rev. Neurosci. 22, 503–513 (2021).
Alves, P. N. et al. An improved neuroanatomical model of the default-mode network reconciles previous neuroimaging and neuropathological findings. Commun. Biol. 2, 370 (2019).
Menon, V. 20 years of the default mode network: a review and synthesis. Neuron 111, 2469–2487 (2023).
Bonini, L., Rotunno, C., Arcuri, E. & Gallese, V. Mirror neurons 30 years later: implications and applications. Trends Cogn. Sci. 26, 767–781 (2022).
Mantini, D. et al. Default mode of brain function in monkeys. J. Neurosci. 31, 12954–12962 (2011).
Amiez, C. et al. Sulcal organization in the medial frontal cortex provides insights into primate brain evolution. Nat. Commun. 10, 3437 (2019).
Thomas, J., Sharma, D., Mohanta, S. & Jain, N. Resting-State functional networks of different topographic representations in the somatosensory cortex of macaque monkeys and humans. Neuroimage 228, 117694 (2021).
Barks, S. K., Parr, L. A. & Rilling, J. K. The default mode network in Chimpanzees (Pan troglodytes) is similar to that of humans. Cereb. Cortex 25, 538–544 (2015).
Garin, C. M. et al. Resting state functional atlas and cerebral networks in mouse lemur primates at 11.7 Tesla. Neuroimage 226, 117589 (2021).
Robinson, J. L. et al. Characterization of structural connectivity of the default mode network in dogs using diffusion tensor imaging. Sci. Rep. 6, 36851 (2016).
Bruner, E., Preuss, T. M., Chen, X. & Rilling, J. K. Evidence for expansion of the precuneus in human evolution. Brain Struct. Funct. 222, 1053–1060 (2017).
Goldring, A. B. & Krubitzer, L. A. Evolution of parietal cortex in mammals: from manipulation to tool use. In: Evolution of nervous systems: second edition. vols 3–4, 259–286 (Elsevier Inc., 2016).
Kaas, J. H. & Stepniewska, I. Evolution of posterior parietal cortex and parietal-frontal networks for specific actions in primates. J. Comp. Neurol. 524, 595–608 (2016).
Romanski, L. M. Integration of faces and vocalizations in ventral prefrontal cortex: implications for the evolution of audiovisual speech. Proc. Natl. Acad. Sci. USA 109, 10717–10724 (2012).
Wang, J., Yang, Y., Zhao, X., Zuo, Z. & Tan, L.-H. Evolutional and developmental anatomical architecture of the left inferior frontal gyrus. Neuroimage 222, 117268 (2020).
Xu, T. et al. Cross-species functional alignment reveals evolutionary hierarchy within the connectome. Neuroimage 223, 117346 (2020).
Garin, C. M. et al. An evolutionary gap in primate default mode network organization. Cell Rep. 39, 110669 (2022).
Raichle, M. E. Creativity and the brain’s default mode network. In: Secrets of creativity 107–123 (Oxford University Press). https://doi.org/10.1093/oso/9780190462321.003.0006, (2019).
Li, W., Mai, X. & Liu, C. The default mode network and social understanding of others: what do brain connectivity studies tell us. Front. Hum. Neurosci. 8, 52017 (2014).
Cruz-Rizzolo, R. J., De Lima, M. A. X., Ervolino, E., de Oliveira, J. A. & Casatti, C. A. Cyto-, myelo- and chemoarchitecture of the prefrontal cortex of the Cebus monkey. BMC Neurosci. 12, 6 (2011).
Fischl, B. et al. Cortical folding patterns and predicting cytoarchitecture. Cereb. Cortex 18, 1973–1980 (2008).
Sherwood, C. C., Broadfield, D. C., Holloway, R. L., Gannon, P. J., & Hof, P. R. Variability of Broca’s area homologue in African great apes: Implications for language evolution. The Anatomical Record Part A: Discoveries in Molecular, Cellular, and Evolutionary Biology 271A, 276–285 (2003).
Cardini, A. & Marco, V. A. Procrustes shape cannot be analyzed, interpreted or visualized one landmark at a time. Evol. Biol. 49, 239–254 (2022).
Putnam, P. T. & Chang, S. W. C. Social processing by the primate medial frontal cortex. Int. Rev. Neurobiol. 158, 213–248 (2021).
Whitlock, J. R. Posterior parietal cortex. Curr. Biol. 27, R681–R701 (2017).
Mars, R. B. et al. On the relationship between the ‘default mode network’ and the ‘social brain’. Front. Hum. Neurosci. 6, 1–9 (2012).
Hill, J. et al. Similar patterns of cortical expansion during human development and evolution. Proc. Natl Acad. Sci. 107, 13135–13140 (2010).
Sansalone, G. et al. Homo sapiens and Neanderthals share high cerebral cortex integration into adulthood. Nat. Ecol. Evol. 7, 42–50 (2023).
Profico, A. et al. Arothron: an R package for geometric morphometric methods and virtual anthropology applications. Am. J. Phys. Anthropol. 176, 144–151 (2021).
Profico, A., Buzi, C., Melchionna, M., Veneziano, A. & Raia, P. Endomaker, a new algorithm for fully automatic extraction of cranial endocasts and the calculation of their volumes. Am. J. Phys. Anthropol. 172, 511–515 (2020).
Profico, A. et al. Reproducing the internal and external anatomy of fossil bones: two new automatic digital tools. Am. J. Phys. Anthropol. 166, 979–986 (2018).
Ebbesen, C. L. & Brecht, M. Motor cortex — to act or not to act? Nat. Rev. Neurosci. 18, 694–705 (2017).
Wong, P., Collins, C. E. & Kaas, J. H. Overview of sensory systems of Tarsius. Int J. Primatol. 31, 1002–1031 (2010).
Gonzales, L. A., Benefit, B. R., McCrossin, M. L. & Spoor, F. Cerebral complexity preceded enlarged brain size and reduced olfactory bulbs in old world monkeys. Nat. Commun. 6, 7580 (2015).
Harrington, A. R., Silcox, M. T., Yapuncich, G. S., Boyer, D. M. & Bloch, J. I. First virtual endocasts of adapiform primates. J. Hum. Evol. 99, 52–78 (2016).
Roumazeilles, L. et al. Cortical morphology and white matter tractography of three phylogenetically distant primates: evidence for a simian elaboration. Cereb. Cortex 32, 1608–1624 (2022).
Pereira‐Pedro, A. S., Beaudet, A. & Bruner, E. Parietal lobe variation in cercopithecid endocasts. Am. J. Primatol. 81, e23025 (2019).
Ho, C. L. A., Fichtel, C. & Huber, D. The gray mouse lemur (Microcebus murinus) as a model for early primate brain evolution. Curr. Opin. Neurobiol. 71, 92–99 (2021).
Ni, X., Flynn, J. J., Wyss, A. R. & Zhang, C. Cranial endocast of a stem platyrrhine primate and ancestral brain conditions in anthropoids. Sci. Adv. 5, eaav7913 (2019).
Kirk, E. C., Daghighi, P., Macrini, T. E., Bhullar, B.-A. S. & Rowe, T. B. Cranial anatomy of the Duchesnean primate Rooneyia viejaensis: New insights from high resolution computed tomography. J. Hum. Evol. 74, 82–95 (2014).
Dozo, M. T., Paulina-Carabajal, A., Macrini, T. E. & Walsh, S. Paleoneurology of amniotes. paleoneurology of amniotes: new directions in the study of fossil endocasts (Springer International Publishing, Cham). https://doi.org/10.1007/978-3-031-13983-3, (2023).
Arsznov, B. M. & Sakai, S. T. Pride diaries: sex, brain size and sociality in the African Lion (Panthera leo) and Cougar (Puma concolor). Brain Behav. Evol. 79, 275–289 (2012).
Sakai, S. T., Arsznov, B. M., Hristova, A. E., Yoon, E. J. & Lundrigan, B. L. Big cat coalitions: a comparative analysis of regional brain volumes in felidae. Front. Neuroanat. 10, 99 (2016).
Radinsky, L. Viverrid neuroanatomy: phylogenetic and behavioral implications. J. Mammal. 56, 130–150 (1975).
Lyras, G. A., van der Geer, A. A. E. & Werdelin, L. Paleoneurology of carnivora. In: Paleoneurology of Amniotes 681–710 (Springer International Publishing, Cham). https://doi.org/10.1007/978-3-031-13983-3_17, (2023).
Flink, T. & Werdelin, L. Digital endocasts from two late Eocene carnivores shed light on the evolution of the brain at the origin of Carnivora. Pap. Palaeontol. 8, e1422 (2022).
Sienkiewicz, T. et al. The brain anatomy of the brown bear (Carnivora, Ursus arctos L., 1758) compared to that of other carnivorans: a cross-sectional study using MRI. Front. Neuroanat. 13, 439655 (2019).
Ahrens, H. E. Morphometric study of phylogenetic and ecologic signals in Procyonid (Mammalia: Carnivora) endocasts. Anat. Rec. 297, 2318–2330 (2014).
Dong, W. Virtual cranial endocast of the oldest giant panda (Ailuropoda microta) reveals great similarity to that of its extant relative. Naturwissenschaften 95, 1079–1083 (2008).
de Bonis, L., Grohé, C., Surault, J. & Gardin, A. Description of the first cranium and endocranial structures of Stenoplesictis minor (Mammalia, Carnivora), an early aeluroid from the Oligocene of the Quercy Phosphorites (southwestern France). Hist. Biol. 34, 1672–1684 (2022).
Czeibert, K., Sommese, A., Petneházy, Ö., Csörgő, T. & Kubinyi, E. Digital endocasting in comparative canine brain morphology. Front Vet. Sci. 7, 565315 (2020).
Kendrick, K. M., da Costa, A. P., Leigh, A. E., Hinton, M. R. & Peirce, J. W. Sheep don’t forget a face. Nature 414, 165–166 (2001).
John, S. E. et al. The ovine motor cortex: a review of functional mapping and cytoarchitecture. Neurosci. Biobehav Rev. 80, 306–315 (2017).
Ella, A., Delgadillo, J. A., Chemineau, P. & Keller, M. Computation of a high-resolution MRI 3D stereotaxic atlas of the sheep brain. J. Comp. Neurol. 525, 676–692 (2017).
Sauleau, P., Lapouble, E., Val-Laillet, D. & Malbert, C. H. The pig model in brain imaging and neurosurgery. Animal 3, 1138–1151 (2009).
Grisham, W. Resources for teaching Mammalian neuroanatomy using sheep brains: a review. J. Undergrad. Neurosci. Educ. 5, R1–R6 (2006).
Graïc, J.-M., Peruffo, A., Ballarin, C. & Cozzi, B. The brain of the giraffe (Giraffa Camelopardalis): surface configuration, encephalization quotient, and analysis of the existing literature. Anat. Rec. 300, 1502–1511 (2017).
Orliac, M. J., Maugoust, J., Balcarcel, A. & Gilissen, E. Paleoneurology of Artiodactyla, an overview of the evolution of the artiodactyl brain. In: Paleoneurology of Amniotes 507–555 (Springer International Publishing, Cham). https://doi.org/10.1007/978-3-031-13983-3_13, 2023
Butti, C. et al. The cerebral cortex of the Pygmy Hippopotamus, Hexaprotodon liberiensis (Cetartiodactyla, Hippopotamidae): MRI, cytoarchitecture, and neuronal morphology. Anat. Rec. 297, 670–700 (2014).
Thiery, G. & Ducrocq, S. Endocasts and brain evolution in Anthracotheriidae (Artiodactyla, Hippopotamoidea). J. Anat. 227, 277–285 (2015).
Fontoura, E., Ferreira, J. D., Bubadué, J., Ribeiro, A. M. & Kerber, L. Virtual brain endocast of Antifer (Mammalia: Cervidae), an extinct large cervid from South America. J. Morphol. 281, 1223–1240 (2020).
Dozo, M. T. & Martínez, G. First digital cranial endocasts of late oligocene Notohippidae (Notoungulata): implications for endemic south american ungulates brain evolution. J. Mamm. Evol. 23, 1–16 (2016).
Balcarcel, A. M., Bastiaans, D. & Orliac, M. J. Endocranial casts of camelops hesternus and palaeolama sp.: new insights into the recent history of the camelid brain. Brain Behav. Evol. 98, 107 (2023).
Johnson, P. J. et al. Equine stereotaxtic population average brain atlas with neuroanatomic correlation. Front. Neuroanat. 13, 459102 (2019).
Abdel Maksoud, M. K. M., Halfaya, F. M., Mahmoud, H. H. & Ibrahim, A. A. H. Morphological characteristics of the forebrain in the donkey (Equus asinus): a compared atlas of magnetic resonance imaging and cross-sectional anatomy. Anat. Histol. Embryol. 50, 974–984 (2021).
Cozzi, B. et al. The laminar organization of the motor cortex in monodactylous mammals: a comparative assessment based on horse, chimpanzee, and macaque. Brain Struct. Funct. 222, 2743–2757 (2017).
Bhagwandin, A., Haagensen, M. & Manger, P. R. The brain of the black (Diceros bicornis) and white (Ceratotherium simum) African Rhinoceroses: morphology and volumetrics from magnetic resonance imaging. Front. Neuroanat. 11, 285850 (2017).
Sherwood, C. C. et al. Neocortical neuron types in Xenarthra and Afrotheria: implications for brain evolution in mammals. Brain Struct. Funct. 213, 301–328 (2009).
Boscaini, A. et al. Digital cranial endocasts of the extinct sloth glossotherium robustum (Xenarthra, Mylodontidae) from the late pleistocene of argentina: description and comparison with the extant sloths. J. Mamm. Evol. 27, 55–71 (2020).
Dooley, J. C., Franca, J. G., Seelke, A. M. H., Cooke, D. F. & Krubitzer, L. A. Evolution of mammalian sensorimotor cortex: thalamic projections to parietal cortical areas in Monodelphis domestica. Front. Neuroanat. 8, 125293 (2015).
Kaas, J. H. Evolution of somatosensory and motor cortex in primates. Anat. Rec. 281A, 1148–1156 (2004).
Karlen, S. J. & Krubitzer, L. The functional and anatomical organization of marsupial neocortex: evidence for parallel evolution across mammals. Prog. Neurobiol. 82, 122–141 (2007).
Sherwood, C. C., Broadfield, D. C., Holloway, R. L., Gannon, P. J. & Hof, P. R. Variability of Broca’s area homologue in African great apes: Implications for language evolution. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 271A, 276–285 (2003).
Goldby, F. An experimental investigation of the motor cortex and its connexions in the phalanger, Trichosurus vulpecula. J. Anat. 74, 12–33 (1939).
Abbie, A. A. The excitable cortex in Perameles, Sarcophilus, Dasyurus, Trichosurus and Wallabia (Macropus). J. Comp. Neurol. 72, 469–487 (1940).
Schlager, S. Morpho and Rvcg – shape analysis in R. In: Statistical Shape and Deformation Analysis 217–256 (Elsevier). https://doi.org/10.1016/B978-0-12-810493-4.00011-0 (2017).
Castiglione, S., Serio, C., Mondanaro, A., Melchionna, M. & Raia, P. Fast production of large, time‐calibrated, informal supertrees with tree.merger. Palaeontology 65, e12588 (2022).
Wisniewski, A. L., Lloyd, G. T. & Slater, G. J. Extant species fail to estimate ancestral geographical ranges at older nodes in primate phylogeny. Proc. R. Soc. B: Biol. Sci. 289, 20212535 (2022).
Álvarez-Carretero, S. et al. A species-level timeline of mammal evolution integrating phylogenomic data. Nature https://doi.org/10.1038/s41586-021-04341-1 (2021).
Webster, M. & Sheets, D. A practical introduction to landmark-based geometric morphometrics. In: Quantitative Methods in PaleobIology (eds. Alroy, J. & Hunt, G.) vol. 16 163–188 (2010).
Schlager, S. Morpho & Rvcg - shape analysis in R: R-packages for geometric morphometrics, shape analysis and surface manipulations. In: Statistical Shape and Deformation Analysis: Methods, Implementation and Applications 217–256. https://doi.org/10.1016/B978-0-12-810493-4.00011-0 (2017).
Mitteroecker, P. et al. Morphometric variation at different spatial scales: coordination and compensation in the emergence of organismal form. Syst. Biol. 69, 913–926 (2020).
Castiglione, S. et al. Ancestral state estimation with phylogenetic ridge regression. Evol. Biol. 47, 220–232 (2020).
Bookstein, F. L. Morphometric tools for landmark data: geometry and biology. (Cambridge University Press). https://doi.org/10.1017/CBO9780511573064 (1991).
Le Maitre, A., Bartsch, S., Grunstra, N. & Mitteroecker, P. prWarp: warping landmark configurations. R package. https://CRAN.R-project.org/package=prWarp (2024).
Christopoulos, D. T. On the efficient identification of an inflection point. Int. J. Math. Sci. Comput. 6, 13–20 (2016).
Clavel, J., Escarguel, G. & Merceron, G. mvmorph: an R package for fitting multivariate evolutionary models to morphometric data. Methods Ecol. Evol. 6, 1311–1319 (2015).
Profico, A. et al. Seeing the wood through the trees. Combining shape information from different landmark configurations. Hystrix 30, 1–43 (2019).
Raia, P., Melchionna, M. & Castiglione, S. Cortical areas associated to higher cognition drove primate brain evolution [Data set and R Codes]. Zenodo https://doi.org/10.5281/zenodo.14614902 (2024).
Stalling, D., Westerhoff, M. & Hege, H. C. Amira: a highly interactive system for visual data analysis. Vis. Handb. 1, 749–767 (2005).
Wickham, H. et al. ggplot2: create elegant data visualisations using the grammar of graphics. R package. https://ggplot2.tidyverse.org (2023).
Acknowledgements
PR was supported by project OMEGA (P2022RZ4PL, CUP:E53D23014440001), financed by the European Union NextGenerationEU (Mission 4, Component 1) fund. We are grateful to multiple institutions and museum curators and clinics for providing access to the mammal cranial material held under their care, and in particular to Stephen Hall, Giuseppe Bianco and Maria Mostadius from Lund University (Sweden), Alexandre Mille, Yves Laurent and Maud Dahlem from the Museum of Toulouse (France), Agustin Ruella, Sergio Vizcaino (MLP, Argentina), Alejandro Kramarz (MACN, Argentina) and Bernardino Mamani Quispe (MNHN Bol, Bolivia), to Guillaume Billet and Helder Gomes Rodrigues (MNHN, France) and the AST-RX facilities for access to MF-M to holotypes. We thank Peter Rask Møller (Associate Professor, Staff Curator) and Daniel Klingberg Johansson (Collection Manager for Mammalogy and Herpetology at Natural History Museum of Denmark) for collaborating in the project of Alejandro Pérez-Ramos, for access to study material. We also thank Henrik Lauridsen (Institut for Klinisk Medicin - Comparative Medicine Lab. Palle Juul-Jensens) for participating in the project by generating the CT scans of the study specimens. The Homo sapiens scan and fMRI labeled as NMDID was taken from the Free Access Decedent Database funded by the National Institute of Justice grant number 2016-DN-BX-0144.
Author information
Authors and Affiliations
Contributions
P.R., M.M., and S.C. designed the study. M.M., S.C., A.P., and G.S. conducted the analyses and made the figures with significant inputs by G.G., C.S., A.M., M.D.F., and J.D. Digital endocasts were prepared and landmarked by M.M.C., A.D.C., L.M., G.G., A.E., and C.S. under the supervision of M.M. N.C., M.F.M., L.P., and A.P.R. scanned and provided several digital endocasts used in this study. P.R. conducted the writing with significant output by all other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Katherine Bryant and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Jasmine Pan.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Melchionna, M., Castiglione, S., Girardi, G. et al. Cortical areas associated to higher cognition drove primate brain evolution. Commun Biol 8, 80 (2025). https://doi.org/10.1038/s42003-025-07505-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07505-1
This article is cited by
-
Human dexterity and brains evolved hand in hand
Communications Biology (2025)