Abstract
Endoplasmic reticulum exit sites (ERESs) are ER subdomains where coat protein complex II carriers are assembled for ER-to-Golgi transport. We previously proposed a dynamic capture-and-release model of ERESs by Golgi stacks in plants. However, how ERESs and Golgi stacks maintain a stable interaction in plant cells with vigorous cytoplasmic streaming is unknown. Here, we show that a plant-specific ER transmembrane protein, which we designate as MAG3, plays a crucial role in mediating the capture-and-release of ERESs in Arabidopsis. We isolated a mutant (mag3) defective in protein exit from the ER in seeds. MAG3 localized specifically to the ER-Golgi interface with Golgi-associated ERESs and remained there after ERES release. MAG3 deficiency caused a reduction in the amount of ERESs associated with each Golgi stack. MAG3 interacted with WPP DOMAIN PROTEINs, which are also plant-specific. These results suggest that plants have evolved a unique system to support ER-to-Golgi transport despite intracellular motility.
Similar content being viewed by others
Introduction
Protein transport from the ER to Golgi stacks is mediated by coat protein complex II (COPII) carriers, which are assembled at specific ER subdomains called ER exit sites (ERESs)1. The process begins when the small GTPase SAR1 is activated to associate with the ER membrane2,3. At ERESs, membrane-bound SAR1 recruits the inner-coat-protein complex to facilitate selective cargo loading4,5 and then the outer-coat-protein complex to organize the COPII-coated carriers6,7. Another important factor in this process is SEC16 (plant homologue MAG5), which, although not part of the COPII coat itself, facilitates COPII assembly at ERESs8,9,10,11,12,13. Once fully formed, these COPII carriers detach from the ERESs and fused with Golgi stacks to deliver their cargo proteins1.
The organization of the ERES-Golgi interface exhibits distinct pattern across different organisms. In Saccharomyces cerevisiae (budding yeast), Golgi cisternae exist in an unstacked form and are physically separated from ERESs14. Cargo transfer in yeast occurs when cis-Golgi cisternae or ER-Golgi intermediate compartments (ERGICs) temporarily interact with ERESs14,15. Mammalian cells feature a unique arrangement where multiple Golgi stacks interconnect to form ribbon-like structure near nucleus, spatially distinct from the ERESs distributed throughout the cell periphery16,17. In these cells, ERGICs serve as transport carriers, emerging from ERESs to deliver cargo to the Golgi ribbon15,18. In organisms with discrete Golgi stacks such as Drosophila melanogaster11,19 and plants8,20,21,22, ERESs are typically positioned adjacent to the Golgi stacks. Recent studies have revealed ERGIC-like structures between ERES and Golgi stacks also in these organisms as well23,24.
Plant cells are distinguished by their vigorous cytoplasmic streaming25,26 and ER streaming27, which drives the continuous movement of organelles, including Golgi stacks, throughout the cells. Recent our work has identified a dynamic mechanism where Golgi stacks capture and release ERESs, enabling efficient ER-Golgi transport in the vigorous cytoplasmic streaming20. This process occurs in three distinct steps: (i) free ERESs move around the ER network for cargo loading, (ii) COPII-bound ERESs are associated with a Golgi stack for cargo delivery, and (iii) ERESs are released from Golgi stacks for recycling. However, the molecular mechanisms governing these capture-and-release events remain to be determined.
In this study, we identified MAIGO3 (MAG3), a plant-specific membrane trafficking factor, through forward genetic screening in Arabidopsis thaliana. Our findings demonstrate that MAG3 localizes to the ER-Golgi interface, where it colocalizes with Golgi-associated ERESs, and is essential for the association of ERESs with Golgi stacks. Furthermore, MAG3 interacts with WPP DOMAIN PROTEINs (WPP1 and WPP2)28, which are plant-specific proteins localized to Golgi stacks. We propose that MAG3 plays a critical role in the capture-and-release of ERESs by Golgi stacks at the ER-Golgi interface, thereby promoting efficient protein transport in plant cells with vigorous cytoplasmic streaming.
Results and Discussion
Identification of a plant-specific coiled-coil protein, MAG3, involved in protein transport from the ER
In maturing Arabidopsis seeds, precursors of two major storage proteins (12S globulin and 2S albumin) are actively synthesized on the ER and subsequently transported to protein storage vacuoles (PSVs), where the precursor proteins are converted to mature forms29,30,31,32. We isolated eight Arabidopsis T-DNA insertion mutants that abnormally accumulate storage protein precursors in seeds and named them maigo (mag) mutants (maigo means getting lost in Japanese), because storage proteins are somehow lost on their way to their destination (PSVs) in these mutants33. In this study, we focused on one of the previously uncharacterized mutants, mag3 (Fig. 1a). We found that mag3-1 had a T-DNA insertion in the fourth intron of the AT1G24460 gene (Fig. 1b, upper). We further obtained four publicly available allelic mutants with T-DNA insertions in the AT1G24460 gene from Arabidopsis seed stock centers (Fig. 1b, upper, and Supplementary Fig. 1). Immunoblot analysis of seedlings with specific antibodies against the N-terminal region of MAG3 (Fig. 1b, lower) revealed that mag3-1, mag3-5, and mag3-6 produced truncated MAG3 proteins corresponding to their respective gene sizes (Fig. 1c, arrowheads). Although no MAG3 protein was detected in mag3-3 and mag3-4 (Fig. 1c), transcripts corresponding to the 5′ half of the MAG3 gene were still present (Supplementary Fig. 1a). Immunoblot analysis of seeds with specific antibodies against 12S globulin and 2S albumin showed that all five mag3 mutants with different alleles accumulated precursors of seed storage proteins, whereas the wild types did not (Fig. 1a). These results, together with a complementation assay (see Supplementary Fig. 3c), indicated that the AT1G24460 gene mutation was responsible for the mag3 phenotype. This identified the AT1G24460 gene as MAG3.
a Immunoblots of dry seeds of mag3 mutant alleles and their respective wild types with specific antibodies against major storage proteins; anti-12S globulin (anti-12S) and anti-2S albumin (anti-2S). Note that the mag3 seeds accumulated the precursors, pro12S globulin (p12S) and pro2S albumin (p2S), whereas wild-type seeds did not. tno1 is a mag3 mutant allele. b The identified MAG3 gene (AT1G24460) and the mutation sites of mag3 alleles. The MAG3 gene consists of eight exons (gray boxes) and seven introns (solid lines). The T-DNA insertions in six mag3 mutant alleles including tno1 are shown. MAG3 encodes a 198-kD protein composed of multiple coiled-coil regions (green) and a transmembrane domain at the C-terminus (orange). Anti-MAG3 antibodies were raised against the N-terminal region (amino acids 1–497) of the protein. c Immunoblots of 10-day-old seedlings of mag3 mutant alleles and their respective wild types with specific antibodies against MAG3 (anti-MAG3) and tubulin (anti-TUB). Arrowheads indicate truncated MAG3 proteins, while asterisks denote nonspecific bands. d Electron micrographs of the seed cells of the wild type (Ws-2) and mag3-1, showing that mag3 abnormally develop structures with electron-dense cores (circled). PSV, protein storage vacuole; CW, cell wall. See also Supplementary Fig. 1b for mag3-5. e Immunoelectron micrographs of the seed cells of mag3-1 with anti-12S and anti-2S conjugated with 15-nm nanogold particles, showing that the abnormal structures consist of electron-dense cores with 2S albumin and the matrix with 12S globulin. Enlarged images of boxed areas are shown below. Note that the abnormal structures are surrounded by a unit membrane associated with ribosomes. Arrowheads indicate ribosomes associated with the ER membrane.
The AT1G24460 gene product was previously reported as TGN-LOCALIZED SYP41 INTERATING PROTEIN (TNO1)34,35 (see below for its subcellular localization). Similar to other mag3 allele mutants, another T-DNA insertion mutant allele, tno1, accumulated storage protein precursors in seeds (Fig. 1a, b). MAG3/TNO1 encodes a 198-kD protein composed of multiple coiled-coil domains with a transmembrane domain at the C-terminus (Fig. 1b, lower). BLAST searches identified one homologous gene (AT4G31570) in Arabidopsis (Supplementary Fig. 2), but no homologues in fungi or animals. Phylogenetic analysis showed that MAG3 homologues are conserved among land plants (Supplementary Fig. 2). This suggests that MAG3 is a plant-specific membrane trafficking factor.
Transmission electron microscopy showed that wild-type seeds developed PSVs (Fig. 1d, left), whereas mag3-1 seeds developed abnormal structures with electron-dense cores in addition to PSVs (Fig. 1d, right). The structures were surrounded by a unit membrane associated with ribosomes (Fig. 1e, lower), indicating that they are derived from the ER. The ER-derived structures consisted of 2S albumin-positive cores and a 12S globulin-positive matrix (Fig. 1e). Similar abnormal structures were also observed in mag3-5 seeds (Supplementary Fig. 1b). The structures were also observed in three kinds of mutants defective in the ER-Golgi transport: (i) mag2, mip1, and mip2, each of which is defective in a tethering complex on the ER (RINT1–ZW10–NAG/Tip20p–Dsl1p–Sec39p complex)36,37, (ii) mag4, which is defective in a Golgi-localized tethering factor (p115/Uso1p homologue)38, and (iii) mag5, which is defective in a COPII vesicle formation factor (a homologue of Sec16, which is a marker of ERESs)8. These results indicate that MAG3 is involved in protein transport between the ER and Golgi stacks.
MAG3 is an ER membrane protein at the ER-Golgi interface in ER cavities
In a homogenate of wild-type seedlings, MAG3 was present in the microsomal (P100) fraction by differential centrifugation (Fig. 2a, WT, P100). On the other hand, the truncated MAG3, which lacked the C-terminal transmembrane domain in mag3-1, mag3-5, or mag3-6 seedlings (Fig. 1b, c), was not present in the microsomal (P100) fraction, although it was present in the soluble (S100) fraction (Fig. 2a, mag3-5, mag3-6, S100; Supplementary Fig. 3a, mag3-1, S100). Taking into consideration that mag3-6 was also defective in the transport of seed storage proteins (Fig. 1a), this indicates that the transmembrane domain of MAG3 is necessary for its association with microsomal membranes and for protein transport between the ER and Golgi stacks. Sucrose density gradient fractionation of a homogenate of the wild-type seedlings showed that endogenous MAG3 cofractionated with the ER marker BiP, but not with the Golgi marker RGP139 (Fig. 2b). Mg2+ is important for ribosomes to bind to the ER, and hence removal of Mg2+ results in a reduction in the ER membrane density40. The immunopositive signal of MAG3 was shifted in an Mg2+-dependent manner, as was the ER marker (Fig. 2b). These results indicate that MAG3 localizes to the ER membrane.
a mag3-6 has the MAG3 protein lacking a C-terminal transmembrane domain (See Fig. 1b, c). Seedlings of the wild-type (Col-0), mag3-5 and mag3-6 were subjected to differential centrifugation to obtain total homogenate (Total), 1000-g pellet (P1), 100,000-g pellet (P100, microsomes), and 100,000-g supernatant (S100). Each fraction was subjected to immunoblot with anti-MAG3, anti-BRI1 (membrane), and anti-ALEU (soluble) antibodies, showing association of MAG3 with microsomal membrane via its C-terminal transmembrane domain. See also Supplementary Fig. 3a for mag3-1. b Sucrose density gradient fractionation of microsomes prepared from 12-day-old seedlings in the presence of MgCl2 (+Mg2+) or EDTA (−Mg2+). Each fraction was subjected to immunoblots with anti-MAG3, anti-BiP (ER marker), and anti-RGP1 (Golgi marker) antibodies, showing cofractionating MAG3 with the ER. See also Supplementary Fig. 3b for the complemented line (proMAG3:GFP-MAG3 mag3-5). c VAEM images of cotyledon epidermal cells of the transgenic plant expressing GFP-MAG3 driven by native MAG3 promoter together with each of four markers: mCherry-HDEL (ER marker), mCherry-SYP32 (cis-Golgi marker), mRFP-SYP43 (TGN marker), and VHAa1-mRFP (TGN marker). Note that MAG3 is closely associated with ER cavities (arrowheads) and Golgi stacks (arrowheads), but not with TGN (arrows). d Time series of representative VAEM images of GFP-MAG3 and mCherry-SYP32 (cis-Golgi marker) for 7.0 s, showing close association of MAG3 with Golgi stacks during their movements (arrowheads). See also Supplementary Fig. 3e for two replicates and Supplementary Video 1 for the original real-time movie.
For a live imaging analysis of MAG3 and a complementation assay, we established the transgenic line (proMAG3:GFP-MAG3 mag3-5) that expressed GFP-MAG3 driven by the native MAG3 promoter in mag3-5. Sucrose density gradient fractionation of this line showed that GFP-MAG3 localized to the ER (Supplementary Fig. 3b), similar to endogenous MAG3 (Fig. 2b). Furthermore, expression of GFP-MAG3 by the native promoter successfully rescued the mag3 phenotype related to transport of seed storage proteins (Supplementary Fig. 3c). These results from the complementation and subcellular localization assays indicate that GFP fusion to MAG3 does not interfere with its function and localization, and hence GFP-MAG3 is functional in vivo.
For colocalization analyses, we coexpressed GFP-MAG3 with each marker [an ER marker, a Golgi marker, and two trans-Golgi network (TGN) markers]. To obtain high temporal resolution of their localizations, we used variable-angle epifluorescence microscopy (VAEM). Cotyledon epidermal cells of the line coexpressing the ER marker (ER-mCherry41) showed that MAG3 localized to beaded-ring structures, each of which was located in an ER cavity (Fig. 2c, first row). Cells of the line coexpressing the cis-Golgi marker (mCherry-SYP3242) showed that each Golgi stack was surrounded by MAG3-labelled beaded-ring structures (Fig. 2c, second row). Quantitative analysis confirmed that MAG3 is located very close to Golgi stacks (Supplementary Fig. 3d). These features are similar to the beaded-ring like signals of ERESs associated with Golgi stacks20. On the contrary, neither of the TGN markers (mRFP-SYP4343 and VHAa1-mRFP44) colocalized with GFP-MAG3 (Fig. 2c, third and fourth rows), indicating that MAG3 does not localize to the TGN. This is different from a previous immunofluorescence result35, which showed that TNO1 (the product of the same gene as MAG3) localized to the TGN. To avoid confusion, we use the name MAG3 instead of TNO1 (TGN-LOCALIZED SYP41 INTERATING PROTEIN) in this study. The VAEM observations indicate that MAG3 localizes exclusively to the ER-Golgi interface in the ER cavities. Time-lapse VAEM imaging of GFP-MAG3 and a cis-Golgi marker (mCherry-SYP32) showed that MAG3 was tightly associated with moving Golgi stacks (Fig. 2d, Supplementary Fig. 3e and Supplementary Video 1). This feature is consistent with the association of ERESs with Golgi stacks in cells with vigorous cytoplasmic streaming20.
MAG3 facilitates the capture-and-release of ERESs by Golgi stacks
In the dynamic capture-and-release model of ERESs by Golgi stacks in plants20, ERESs cycle in three steps (see Fig. 4h): (i) ERESs move along the ER network while loading cargo into COPII carriers, (ii) Golgi stacks capture COPII-bound ERESs within ER cavities for cargo delivery, and (iii) ERESs are released to begin the cycle anew. In this way, ERESs cycle dynamically between the two states: the Golgi-associated and Golgi-free states.
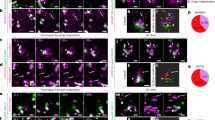
The first question to answer is “in which of these two states is MAG3 involved?” VAEM observations of the line coexpressing GFP-MAG3 and an ERES marker (MAG5-mCherry) showed that the MAG3-positive dots forming a beaded-ring structure clearly colocalized with the Golgi-associated ERESs (Fig. 3a, circled), but not with Golgi-free ERESs (Fig. 3a, arrowheads). Statistical analysis confirmed the specific localization of MAG3 to the Golgi-associated ERESs (Fig. 3b). Real-time VAEM imaging showed the dynamic relationship between MAG3 and a punctate ERES being released by a Golgi stack, in which the ERES in the Golgi-associated state was MAG3-positive and then the ERES released from Golgi stacks was MAG3-negative (Fig. 3c, Supplementary Fig. 4a and Supplementary Video 2). During this process, the MAG3 signal remained colocalized with Golgi-associated ERESs. These results indicate that the ERESs in the Golgi-associated state dissociate from MAG3 when they are released from Golgi stacks.
a Representative VAEM images of cotyledon epidermal cells expressing GFP-MAG3 and MAG5-mCherry (ERES marker), showing MAG3 localizes close to Golgi-associated ERES (circled), but not to Golgi-free ERES (arrowheads). b Colocalization analysis showing Pearson correlation coefficient between MAG3 and ERES marker (MAG5-mCherry). Lines indicate the median and boxes indicate 25% and 75% quartiles. Whiskers indicate the minimum and maximum values within 1.5× the interquartile range. ***P < 0.001 (Two-sided Wilcoxon rank-sum test, P < 2.2 × 10−16). n = 114 Golgi-associated ERESs from 36 cells. n = 114 free ERESs from 36 cells. c Time series of representative VAEM images of cotyledon epidermal cells expressing GFP-MAG3 and MAG5-mCherry (ERES marker) for 1.4 s. Note that an ERES dissociates from MAG3 signals when it is released from a Golgi stack (arrowheads). See also Supplementary Fig. 4a for two replicates and Supplementary Video 2 for the original real-time movie. d Representative VAEM images of cotyledon epidermal cells of mag3-5 and the control line (MAG3), both of which expressed MAG5-GFP (ERES marker) and mCherry-SYP32 (cis-Golgi marker). Circles indicate Golgi-associated ERESs. e Statistical analysis of relative intensity of Golgi-associated ERES in cotyledon epidermal cells of mag3-5 and the control line (MAG3) expressing MAG5-GFP and mCherry-SYP32 (cis-Golgi marker). Relative intensity of Golgi-associated ERES was defined as the ratio of GFP fluorescence intensity of ERESs surrounding a single Golgi stack to the average intensity of a single free ERES. Data are represented as mean ± SD (n = 104 from 21 cells of the control plants and n = 116 from 32 cells of mag3-5). ***P < 0.001 (Two-sided Welch’s t test, P = 1.5 × 10−21). f Time series of representative VAEM images of cotyledon epidermal cells of mag3-5 expressing MAG5-GFP (ERES marker) and mCherry-SYP32 (cis-Golgi marker) for 1.4 s. Arrowheads indicate punctate ERESs being captured by a Golgi stack. See also Supplementary Fig. 4b for two replicates and Supplementary Video 3 for the original real-time movie.
A second question is “Is MAG3 involved in the capture and release of ERESs by Golgi stacks?” To answer this question, we introduced both the ERES marker (MAG5-GFP8) and the cis-Golgi marker (mCherry-SYP32) in mag3-5. Multiple ERESs are associated with each Golgi stack to form a beaded-ring structure20. VAEM observations showed a drastic change in Golgi-associated ERESs in mag3-5 (Fig. 3d, compare between mag3 and MAG3). For quantitative analysis, we calculated the relative signal intensity of Golgi-associated ERESs by dividing the total signal intensity of ERESs surrounding a single Golgi stack by the average signal intensity of a single free ERES. The quantitative analysis demonstrated that the relative intensity of ERESs surrounding a Golgi stack was significantly lower in mag3-5 than in the control (Fig. 3e), indicating that MAG3 deficiency caused a reduction in the amount of Golgi-associated ERES. Real-time VAEM imaging revealed that ERES capture by the Golgi stack still occurred in mag3-5, indicating that ERESs can be captured by Golgi stacks even in the absence of MAG3 (Fig. 3f, Supplementary Fig. 4b and Supplementary Video 3). Taken together, these results suggest that MAG3 regulates the capture-and-release process of ERESs by Golgi stacks at the ER-Golgi interface (see Fig. 4h).
a Immunoblots (left) and silver staining (right) of pulled-down products with anti-GFP antibody from seedlings of the transgenic plant expressing either GFP or GFP-MAG3. Arrowheads indicate full-length GFP-MAG3. b Mass spectrometry-based proteomics of the pulled-down products with anti-GFP antibody from different organs of the transgenic plants identified two WPP domain proteins (WPP1 and WPP2) with high scores as well as MAG3. Scores were calculated using the Mascot program (Matrix Science). n.d. not detected. c Yeast two-hybrid analysis of the strain expressing MAG3 lacking the transmembrane domain (MAG3ΔTMD) fused with the GAL4 DNA binding domain (BD) and each of WPPs (WPP1, WPP2, and WPP3) fused with the GAL4 activation domain (AD). Three independent transformants were incubated on SD/–Leu/–Trp/–His/–Ade/+Aureobasidin A/ + X-α-Gal medium (–LWHA +AbA +X-α-Gal) or SD/–Leu/–Trp medium (–LW). Negative controls (empty vector) were also examined. d Representative VAEM images of cotyledon epidermal cells of two transgenic plants (proMAG3:GFP-MAG3 proWPP1:WPP1-mCherry mag3-5 and proMAG3:GFP-MAG3 proWPP2:WPP2-mCherry mag3-5), showing close localization of MAG3 with WPP1 or WPP2 (arrowheads). Arrows show WPP1 and WPP2 detected on plasma membrane (see Supplementary Fig. 5 and Supplementary Video 4). e Representative VAEM images of cotyledon epidermal cells of wpp1-1 wpp2-1 and the respective control line (WPP1 WPP2) expressing GFP-MAG3 and mCherry-SYP32 (cis-Golgi marker), showing no effects of deficiency of both WPP1 and WPP2 on MAG3 signal patterns (arrowheads). f The MAG3 protein levels in seedlings (arrowhead) of wpp1-1, wpp2-1, and wpp1-1 wpp2-1 were similar to that of the wild-type (Col-0). g No accumulation of storage protein precursors (pro12S globulin and pro2S albumin) in seeds of wpp1-1, wpp2-1, and wpp1-1 wpp2-1, as in the wild type (Col-0). h A hypothetical model of the MAG3 function in the dynamic capture-and-release of ERESs by Golgi stacks at the ER-Golgi interface. COPII-bound ERESs (green) are captured by a Golgi stack in an ER cavity, where COPII vesicles bud from ERESs and then fuse with cis-Golgi cisternae. After cargo transport, ERESs are released by Golgi stacks for recycling. The coiled-coil ER membrane protein MAG3 (blue), in cooperation with WPPs (orange), facilitates the cargo transport from the ER to Golgi stacks. Adapted from Takagi et al. 20.
MAG3 interacts with the plant-specific proteins WPPs
To identify proteins that interact with MAG3, we prepared immunoprecipitates with anti-GFP antibodies from three different organs of the transgenic plant (seedlings, shoots, and roots) and subjected them to mass spectrometry. A seedling homogenate of the line (proMAG3:GFP-MAG3 mag3-5) gave a positive band of full-length of GFP-MAG3 on an immunoblot with anti-GFP antibodies (Fig. 4a, left) and the respective band on an SDS gel with silver staining (Fig. 4a, right). From these pull-down products, two WPP DOMAIN PROTEINs (WPP1 and WPP2) and MAG3 were identified (Fig. 4b), while neither of them were identified in the three organs of the control plant that expressed GFP only. This result was supported by yeast two-hybrid assays (Fig. 4c), indicating that MAG3 interacts with WPP1 and WPP2. Although the Arabidopsis genome has three WPP proteins (WPP1-3), WPP3, which has less similarity to WPP1 and WPP228, was not detected in the pull-down products.
Previous confocal microscopic analyses observed WPP1 and WPP2 at the dot-like structures, plasma membrane, and nuclear envelope28,45. Our VAEM observations showed the colocalization of both WPP1-mCherry and WPP2-mCherry with the cis-Golgi marker (mGFP-SYP3220) (Supplementary Fig 5a, arrowheads). VAEM also detected smaller punctate WPP signals that did not colocalize with the cis-Golgi marker (Supplementary Fig. 5a, arrows). These signals were immobile (Supplementary Fig. 5b and Supplementary Video 4) and showed no colocalization with mRFP-SYP43 (TGN), VHAa1-mRFP (TGN), or MAG5-mCherry (ERES) (Supplementary Fig. 5c), suggesting that they localize to the plasma membrane rather than to particulate organelles that move as a result of cytoplasmic streaming.
To examine the colocalization of MAG3 and WPP proteins, we individually expressed WPP1-mCherry and WPP2-mCherry in the line (proMAG3:GFP-MAG3 mag3-5). VAEM observations showed that MAG3 colocalized with WPP1 and WPP2 at Golgi stacks (Fig. 4d, arrowheads), but not with punctate WPP signals that were possibly on the plasma membrane (Fig. 4d, arrows), which supports the close relationship between MAG3 and the WPPs on Golgi stacks (Supplementary Fig 5a, arrowheads). A deficiency of MAG3 did not affect the distribution pattern of the WPPs (Supplementary Fig. 6a).
We established single and double mutants lacking WPP1 and/or WPP2 (Supplementary Fig. 6b). VAEM of the double mutant wpp1-1 wpp2-1 expressing both GFP-MAG3 and the cis-Golgi marker (mCherry-SYP32) showed that a deficiency of both WPP1 and WPP2 did not affect MAG3 localization to the ER-Golgi interface (Fig. 4e). In the single and double mutants, the MAG3 protein levels were similar to the level in the wild type (Fig. 4f). Seeds of these mutants accumulated no precursors of major storage proteins (Fig. 4g). These results suggest that WPPs have some supportive effects on MAG3. WPPs have been shown to act as cytosolic chaperones/receptors toward tail-anchored coiled-coil proteins46. This implies that WPP1 and WPP2 have chaperone-like activities toward the tail-anchored coiled-coil protein MAG3.
A plant-specific MAG3 system supports the dynamic capture-and-release of ERESs by Golgi stacks in the presence of ER streaming and cytoplasmic streaming
In this study, we provide evidence for the existence of a MAG3 system to support the dynamic capture-and-release model of ERESs by Golgi stacks20 (Fig. 4h). Non-plant organisms also have ERESs that are closely associated with a Golgi stack or ERGICs11,17,47,48, although they involve proteins that differ from MAG3 in their domain structures. In animals, the coiled-coil transmembrane proteins TANGO1 and cTAGE5 localize to ring-shaped ERESs adjacent to Golgi stacks23,49,50,51,52, where they mediate ERES formation by recruiting COPII machineries53. The proline-rich domains (PRDs) of TANGO1 and cTAGE5 and the intrinsically disordered region of SEC16 are involved in this ERES formation by protein phase separation54.
The coiled-coil transmembrane protein MAG3 has no PRDs, although it has a function similar to that of TANGO1 or cTAGE5. Instead, MAG3 interacts with WPPs, which contain several prolines in their N- and C-terminal predicted IDRs (Supplementary Fig. 6c) and have the ability to form a large complex45. Thus, it is possible that MAG3 modulates the organization of ERESs at the ER-Golgi interface in plants, in cooperation with WPP1 and WPP2. Like MAG3, WPPs are plant-specific proteins28. Therefore, plants may have evolved a unique system for the dynamic capture-and-release of ERESs by Golgi stacks in cells exposed to vigorous cytoplasmic streaming25,26, thereby facilitating protein transport from the ER. Our findings also provide a valuable insight into inter-organelle communications.
Methods
Plant materials and growth conditions
A. thaliana, accessions Columbia-0 (Col-0), Landsberg erecta (Ler), and Wassilewskija-2 (Ws-2) were used as the wild-type plant. Transfer DNA (T-DNA) mutants were obtained from the Arabidopsis Biological Resource Center at Ohio State University (https://abrc.osu.edu/) or The Nottingham Arabidopsis Stock Centre (https://arabidopsis.info/): GT_3_4205 (mag3-2), SALK_021815 (mag3-3), WiscDsLox457-460P14 (mag3-4), SALK_148610 (mag3-5), SAIL_1179_H08 (mag3-6), SALK_151417 (wpp1-1), and SALK_206215 (wpp2-1). mag3-1 was isolated from the K. Feldmann’s T-DNA insertion lines (CS6501). SALK_112503 [tno135], mRFP-SYP4343, and VHAa1-mRFP44 were provided by Dr. D. Bassham, Dr. T. Uemura, and Dr. K. Schumacher, respectively. proMAG5:MAG5-GFP proUBQ10:mCherry-SYP32 mag5-120 and GFP-BE55 were described previously. The primer sets for genotyping by PCR are shown in Supplementary Table 1. Seeds were surface sterilized with 70% ethanol and then sown onto 1× MS medium that contained 0.5% [w/v] Gellan Gum, 1% [w/v] sucrose, 1× Murashige-and-Skoog medium (Wako), and 2.5 mM MES-KOH, pH 5.7. The seeds were incubated at 4 °C for 2 days to break seed dormancy, followed by growth at 22 °C under continuous light or under a 16 h:8 h light:dark photoperiod.
Molecular cloning
sGFP56, mGFP57 and mCherry58 were amplified by PCR. The genomic sequence of MAG5 including 2.1 kb of upstream sequence and 0.7 kb of downstream sequence, the genomic sequence of WPP1 including 2.7 kb of upstream sequence and 0.5 kb of downstream sequence, and the genomic sequence of WPP2 including 2.2 kb of upstream sequence and 0.5 kb of downstream sequence were PCR-amplified using genomic DNA of wild-type plants. The amplified fragments were inserted into pENTER 1 A dual selection (Invitrogen) using an In-Fusion HD cloning kit (Clontech) to produce proMAG5:MAG5-mCherry, proWPP1:WPP1-mGFP, proWPP1:WPP1-mCherry, proWPP2:WPP2-mGFP and proWPP2:WPP2-mCherry. proMAG5:MAG5-mCherry, proWPP1:WPP1-mGFP, proWPP1:WPP1-mCherry, proWPP2:WPP2-mGFP and proWPP2:WPP2-mCherry were transferred into the destination vector pHGW or pBGW59 using LR clonase II (Invitrogen). The genomic sequence of MAG3 including 1.7 kb of upstream sequence and 0.5 kb of downstream sequence was PCR-amplified using genomic DNA of wild-type plants. The amplified sequence was fused with sGFP using overlap extension PCR and then inserted into pENTR/D-TOPO (Invitrogen). First 0.8 kb of MAG3 upstream sequence was deleted using site-directed mutagenesis to produce proMAG3:GFP-MAG3. proMAG3:GFP-MAG3 was transferred into the destination vector pHGW59 using LR clonase II (Invitrogen). proUBQ10:mCherry-SYP32 was PCR-amplified using the WAVE22R plasmid42 and inserted into pENTR/D-TOPO (Invitrogen). proUBQ10:mCherry-SYP32 was transferred into the destination vector pGWB60160 using LR clonase II (Invitrogen). pBGW/proSYP32:mGFP-SYP3220 and pFGC19/pro35S:mCherry-HDEL41 were described previously. These binary vectors were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. The binary vectors were transformed into the wild-type (Col-0), mag5-3, mag3-5 or proMAG3:GFP-MAG3 mag3-5 plants with Agrobacterium using the floral dip method61 to generate proMAG5:MAG5-mCherry mag5-3, proMAG3:GFP-MAG3 mag3-5, proMAG3:GFP-MAG3 pro35S:mCherry-HDEL mag3-5, proMAG3:GFP-MAG3 proUBQ10:mCherry-SYP32 mag3-5, proWPP1:WPP1-mGFP, proWPP2:WPP2-mGFP, proMAG3:GFP-MAG3 proWPP1:WPP1-mCherry mag3-5, proMAG3:GFP-MAG3 proWPP2:WPP2-mCherry mag3-5, proSYP32:mGFP-SYP32 proWPP1:WPP1-mCherry, and proSYP32:mGFP-SYP32 proWPP2:WPP2-mCherry. The following transgenic lines used in this study were generated by crossing: proMAG3:GFP-MAG3 proSYP43:mRFP-SYP43, proMAG3:GFP-MAG3 proVHAa1:VHAa1-mRFP, wpp1-1 wpp2-1, proMAG3:GFP-MAG3 proUBQ10:mCherry-SYP32 mag3-5 wpp1-1 wpp2-1, proSYP32:mGFP-SYP32 proWPP2:WPP2-mCherry mag3-5, proWPP1:WPP1-mGFP proSYP43:mRFP-SYP43, proWPP1:WPP1-mGFP proVHAa1:VHAa1-mRFP, proWPP1:WPP1-mGFP proMAG5:MAG5-mCherry, proWPP2:WPP2-mGFP proSYP43:mRFP-SYP43, proWPP2:WPP2-mGFP proVHAa1:VHAa1-mRFP, proWPP2:WPP2-mGFP proMAG5:MAG5-mCherry, proMAG5:MAG5-GFP proUBQ10:mCherry-SYP32 mag3-5 mag5-1, and proMAG3:GFP-MAG3 proMAG5:MAG5-mCherry. The primer sets for molecular cloning are shown in Supplementary Table 1.
Isolation of mag3 mutant and TAIL-PCR
Arabidopsis T-DNA–tagged lines were screened according to the previously established method33 as follows. Dry seeds (0.1 mg) from a mixture of 20 lines were subjected to SDS-PAGE and then to immunoblot analysis with anti-12S globulin and anti-2S albumin antibodies to detect storage protein precursors. The mag3-1 line described in this article was derived from a pool of T-DNA–tagged lines (CS6501) that was prepared with the 3850:1003 Ti plasmid62. The T-DNA insertion site in the MAG3 gene was determined through TAIL-PCR33. The flanking sequence of the T-DNA was amplified from genomic DNA of the mag3-1 mutant using primers specific for the T-DNA border, pGVAK-RB1 and pGVAK-RB2, and a degenerate primer, TAIL-1. The pGVAK-RB1 and pGVAK-RB2, were used for the first and second amplification, respectively. The pGVAK-RB2 primer was used for the nucleotide sequencing of the resulting PCR products. The primer sets for TAIL-PCR are shown in Supplementary Table 1.
SDS-PAGE and immunoblotting
Protein samples were analyzed by SDS-PAGE and immunoblot63. Blots were probed with anti-12S globulin α-subunit63 (anti-12S, diluted 10,000-fold), anti-2S albumin8 (anti-2S3M, diluted 5000-fold), anti-MAG3 (diluted 200-fold), anti-α-tubulin (diluted 2000-fold; Sigma-Aldrich, T5168), anti-BRI1 (diluted 1000-fold; Agrisera), anti-ALEU64 (diluted 5000-fold), anti-BiP65 (diluted 5000-fold), anti-RGP139 (diluted 5000-fold), or anti-GFP (diluted 5000-fold; Clontech, 632380), followed by incubation with secondary antibodies, HRP-linked anti-rabbit IgG (Cell Signaling Technology, 7074), HRP-linked anti-rabbit IgG (Invitrogen, 31461), or HRP-linked anti-mouse IgG (Cytiva, NA931). Unprocessed original images of immunoblots are shown in Supplementary Fig. 7.
Antibodies
Anti-MAG3 antibodies were raised against an N-terminal region (amino acids 1–497) of MAG3 according to the previously used method32 as follows. N-terminal region of MAG3 was amplified by PCR. The PCR product was subcloned into pDONR221 (Invitrogen) and then transferred into the GATEWAY vector pET32GW, in which the recombination site is located at the EcoRV site33. The recombinant protein composed of the N-terminal region of MAG3 and His-tag was expressed in Escherichia coli BL21(DE3) cells. The recombinant protein was purified with a HiTrap chelating column (Cytiva) and then used for the production of specific antibodies against MAG3. The primer sets for molecular cloning are shown in Supplementary Table 1.
Electron microscopy
Ultrastructural analysis was performed using chemical fixation method8, or conducted by the Tokai Electron Microscopy service. Dry seeds were fixed, dehydrated, and embedded in LR white resin (London Resin). Ultrathin sections were examined with a transmission electron microscope (model JEM-1015B; JEOL) at 100 kV. Immunocytochemistry was performed with the following antibody dilutions: anti-12S, 1:2000; anti-2S, 1:1000; and AuroProbe EM anti-rabbit IgG (H + T), 1:30 (15-nm gold; GE Healthcare).
Subcellular fractionation
Membrane proteins were purified with the subcellular fractionation according to the previously used method66 as follows. Seedlings of 12-day-old or 14-day-old wild-type (Col-0), wild-type (Ws-2), mag3-1, mag3-5 or mag3-6 were homogenized in a mortar on ice in triple volume of buffer (100 mM HEPES-KOH, pH 7.5, 0.3 M sucrose, 5 mM MgCl2, 5 mM EGTA and protease inhibitor [cOmplete EDTA-free Protease Inhibitor Cocktail; Roche]). The homogenate was filtered through double layered Miracloth (CalBiochem). An aliquot of the filtrate was used as a total fraction. The filtrate was centrifuged at 1000 × g and 4 °C for 20 min. The supernatant was ultracentrifuged at 100,000 × g and 4 °C for 1 h to obtain supernatant (S100) and pellet (P100) fraction. Each of total fractions, P1, P100, and S100, was subjected to SDS-PAGE and immunoblot analyses.
Sucrose density gradient assay
Subcellular localization was analyzed using the sucrose density gradient assay27. Twelve-day-old wild-type (Col-0) or 13-day-old proMAG3:GFP-MAG3 seedlings were homogenized in a mortar on ice in quadruple volume of +Mg2+ or −Mg2+ homogenize buffer (100 mM HEPES-KOH, pH 7.5, 0.4 M sucrose, 1 mM DTT, 5 mM EGTA, 5 mM MgCl2 or 5 mM EDTA, and protease inhibitor [cOmplete EDTA-free Protease Inhibitor Cocktail; Roche]). The homogenate was filtered through a 70 μm Cell Strainer by centrifugation at 50 × g and 4 °C. An aliquot of the filtrate was used as the total fraction. The microsomal fractions were prepared by sequential centrifugations of the filtrate at 1000 × g and 4 °C for 20 min and 100,000 × g and 4 °C for 1 h. The microsomal fractions were resuspended in 600 μL of the homogenize buffer without protease inhibitor and layered directly on top of a 16-mL linear sucrose density gradient (14–50%, w/w). After centrifugation at 100,000 × g and 4 °C for 18 h (L-100XP, SW32.1Ti, Beckman), 700-μL fractions were collected with a piston gradient fractionator (TOWA LABO). Each fraction was concentrated with acetone and subjected to SDS-PAGE and immunoblot analyses.
Immunoprecipitation
Immunoprecipitation was performed using µMACS GFP isolation kits (Miltenyi Biotec). Whole seedlings, shoots or roots of 22-day-old or 21-day-old transgenic plants expressing GFP or GFP-MAG3 were homogenized in a mortar on ice in triple volume of extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor [cOmplete EDTA-free Protease Inhibitor Cocktail; Roche]). After centrifugation at 10,000 × g and 4 °C for 10 min, the supernatants were mixed with anti-GFP microbeads (Miltenyi Biotec) and then incubated on ice for 30 min. Immunoaffinity complexes were purified with µColumns (Miltenyi Biotec) in a magnetic field according to the manufacturer’s protocol.
Mass spectrometry analysis
Mass spectrometry analysis was performed according to the previously used method8 as follows. The immunoprecipitates were subjected to SDS-PAGE for in-gel digestion. The SDS gel of each lane was separated into five fractions based on molecular mass: <30 kDa, 30–50 kDa, 50–60 kDa, 60–100 kDa, and >100 kDa. Each gel fraction was dehydrated using 100% (v/v) acetonitrile for 15 min, and subsequently dried using a vacuum concentrator. The gels were treated with 10 mM DTT in 50 mM NH4HCO3 at 56 °C for 45 min, followed by incubation with 55 mM 2-iodoacetamide in 50 mM NH4HCO3 for 30 min in the dark. After washing with 25 mM NH4HCO3, the gels were again dehydrated with 50% (v/v) acetonitrile in 50 mM NH4HCO3, and then dried. The dried gels were incubated with 0.01 mg/mL of trypsin in 50 mM NH4HCO3 at 37 °C for 16 h. Digested peptides were recovered using two serial extractions: 50 μL of 5% (v/v) formic acid in 50% (v/v) acetonitrile followed by 50 μL of 5% (v/v) formic acid in 70% (v/v) acetonitrile. The combined extracts were concentrated to 10 μL using a vacuum concentrator and then filtered through the Ultrafree-MC Centrifugal Filters (PVDF 0.45 µm; Millipore).
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses were performed using the LTQ-Orbitrap XL-HTC-PAL system. Trypsin digests were loaded onto a 100-µm internal diameter, 15-cm L-Column (CERI) using the Paradigm MS4 HPLC pump (Michrom BioResources) and HTC-PAL Autosampler (CTC Analytics), and eluted with 5–45% (v/v) acetonitrile gradient in 0.1% (v/v) formic acid for 26 min. Eluted peptides were directly introduced into an LTQ-Orbitrap XL mass spectrometer (Thermo) at 500 nL/min with 2.0 kV spray voltage. The mass spectrometry scan was conducted in the mass-to-charge ratio range of 450–1500, and the top three peaks were selected for MS/MS analysis. MS/MS spectra were analyzed by the Mascot server (version 2.3) in house67 (http://www.matrixscience.com/) and searched against the Arabidopsis protein database TAIR10 (The Arabidopsis Information Resource). The Mascot search parameters were set as follows: threshold of the ion score cutoff, 0.05; peptide tolerance, 10 ppm; MS/MS tolerance, ±0.5 Da; and peptide charge, 2+ or 3 + ; allowing one missed cleavage by trypsin; carbamidomethylation on Cys residues as a fixed modification; oxidation on Met residues as a variable modification.
Yeast two-hybrid assay
The coding regions of WPP1, WPP2, and WPP3 were PCR-amplified with cDNA prepared from wild-type seedlings. The amplified fragments were inserted into pENTER/D-TOPO (Invitrogen) to produce the entry clones. The coding regions of MAG3 lacking a C-terminal transmembrane domain was PCR-amplified with cDNA prepared from wild-type seedlings. The amplified fragments were inserted into pENTER 1 A dual selection (Invitrogen) to produce the entry clones. These fragments were transferred into the destination vector either pDEST-GBKT7 (BD-) or pDEST-GADT7 (AD-)68. These destination vectors were transformed into S. cerevisiae Y2H Gold strain (Clontech) using Frozen-EZ Yeast Transformation II kit (Zymo Research). Transformed yeasts were selected on SD/-Leu/-Trp plates and the interactions were examined on SD/-Leu/-Trp/-His/-Ade/+Aureobasidin A/ + X-α-Gal plates according to the manufacturer’s protocol. pGBKT7 and pGADT7 vectors (Clontech) were used for negative control. The primer sets for molecular cloning are shown in Supplementary Table 1.
Live cell imaging
Live cell imaging was performed using the VAEM system20. Fluorescence images from cotyledon epidermal cells of 3-d-old seedlings were captured using an inverted fluorescence microscope (IX83; Olympus) equipped with a total internal reflection fluorescence microscopy system (IX2-RFAEVA-2; Olympus), solid-state lasers at 488 nm (Sapphire 488 LP; Coherent) and 561 nm (Sapphire 561 LP; Coherent), 100× oil immersion objectives (UApoN 100× OTIRF, numerical aperture (NA) = 1.49; UPlanApo 100× OHR, NA = 1.50; Olympus), Dual View 2 emission splitting system (Photometrics) and a EM-CCD camera (iXon3 897; Andor). The pixel size corresponds to 80 nm. For real-time imaging, time-sequential images were captured at 100 ms intervals for 10–20 s. Images were processed using MetaMorph (Molecular Devices) and Fiji 2.14.0/1.54f69.
RT-PCR
Total RNA was isolated from wild-type (Col-0), wild-type (Ws-2), mag3-1, mag3-3, mag3-4, mag3-5, mag3-6, tno1, wpp1-1, wpp2-1, and wpp1-1 wpp2-1 plants using TRIzol Reagent (Invitrogen). RNA (500 ng) was treated with RQ1 RNase-Free DNase (Promega) to reduce genomic DNA contamination. Reverse transcription was performed using ReverTra Ace (Toyobo) with an oligo(dT)15 primer (Promega). Quantitative Real-Time PCR (qRT-PCR) was performed using AriaMx Real-Time PCR System (Agilent Technologies) with TB Green premix EX Taq II kit (TaKaRa) and gene-specific primer sets (Supplementary Table 1). For RT-PCR, ACTIN2 was used as a control. Unprocessed original images of gels are shown in Supplemental Fig. 7. The primers used for RT-PCR are shown in Supplementary Table 1.
Phylogenetic analysis
Amino acid sequences of MAG3-like proteins in each plant were retrieved from public databeses TAIR10 (https://www.arabidopsis.org/) and Phytozome v13 (https://phytozome-next.jgi.doe.gov/) with MAG3 and At4g31570 as queries. These amino acid sequences were aligned using MAFFT v.6.86470 (https://www.genome.jp/tools-bin/mafft) with the default parameters, followed by trimming with ClipKIT v.1.4.171. Phylogenetic tree constructions were performed using the maximum-likelihood algorithm on RAxML-NG v.1.0.072 with the JTT + G + I + F substitution model selected by ModelTest-NG v.0.1.673. To assess statistical support for the topology, we performed bootstrap analyses with 1000 replicates for each analysis. The graphic representation of the tree was generated using the FigTree v.1.4.4 software (http://tree.bio.ed.ac.uk/software/figtree).
Protein domain search
Coiled-coil domain search was performed with Multicoil274 (https://cb.csail.mit.edu/cb/multicoil2/cgi-bin/multicoil2.cgi), and transmembrane domains were predicted with DeepTMHMM75 (https://dtu.biolib.com/DeepTMHMM). IDRs were predicted with flDPnn76 (http://biomine.cs.vcu.edu/servers/flDPnn/). Protein structures were predicted using AlphaFold277 within ColabFold v1.5.578 with default parameters.
Statistics and reproducibility
For colocalization analysis, circular regions of interest (ROIs) of 20- and 10- pixels in diameter were selected for Golgi-associated ERES clusters (n = 114 from 36 cells) and free ERESs (n = 114 from 36 cells), respectively. Pearson’s R-value of each ROIs was calculated with the Coloc2 plugin of Fiji 2.14.0/1.54f69.
To quantify the relative ERES intensity associated with a single Golgi stack in control and mag3 plants, Fiji 2.14.0/1.54f69 was used. Circular ROIs of 20-pixels in diameter were selected for single isolated Golgi stack (n = 104 from 21 cells of control plants, n = 116 from 32 cells of mag3-5). After subtraction of background, total Golgi-associated ERES signal of each ROIs were divided by average total signal of free ERES selected using circular ROIs of 10-pixels in diameter (3 ~ 8 free ERESs from each cell) to calculate the amount of ERES surrounding a Golgi stack.
To quantify the distance between MAG3 signals and cis-Golgi, object-based distance analysis was performed using the imageJ plugin DiAna79. MAG3 punctate structures (n = 1417 MAG3 signals from 48 cells) were extracted using the spots segmentation method of DiAna-segmentation, while cis-Golgi regions were extracted using global thresholding of DiAna-segmentation after denoising with a Gaussian blur filter. Following the segmentation process, the edge-to-edge distance between each MAG3 punctate structure and its nearest cis-Golgi compartment was calculated using DiAna-analysis. Due to the limitations of fluorescence imaging, the size of each structure might be overestimated, consequently leading to an underestimation of the actual distance between MAG3 punctate structures and Golgi stacks. The pixel size corresponds to 80 nm, and the bin width was set to 80 nm.
Statistical analyses were performed using R version 4.2.3. Two-sided Wilcoxon rank-sum test and two-sided Welch’s t test were used for the colocalization analysis and the comparison of the relative intensity of Golgi-associated ERES, respectively.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Sequence data from this article can be found in The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org) with the accession numbers as follows: MAG3, AT1G24460; WPP1, AT5G43070; WPP2, AT1G47200; WPP3, AT5G27940; MAG5, AT5G47480; SYP32, AT3G24350; SYP43, AT3G05710; VHA-a1, AT2G28520. Source data underlying graphs can be obtained in Supplementary Data 1–4. Proteome data were deposited in jPOSTrepo (JPST003034) and ProteomeXchange (PXD051419). The data that supports the findings of this study are available from the corresponding author on request.
References
Brandizzi, F. & Barlowe, C. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392 (2013).
Barlowe, C. & Schekman, R. SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365, 347–349 (1993).
Nakano, A. & Muramatsu, M. A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 109, 2677–2691 (1989).
Miller, E. A. et al. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 (2003).
Bi, X., Corpina, R. A. & Goldberg, J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419, 271–277 (2002).
Stagg, S. M. et al. Structural basis for cargo regulation of COPII coat assembly. Cell 134, 474–484 (2008).
Bi, X., Mancias, J. D. & Goldberg, J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev. Cell 13, 635–645 (2007).
Takagi, J. et al. MAIGO5 functions in protein export from Golgi-associated endoplasmic reticulum exit sites in Arabidopsis. Plant Cell 25, 4658–4675 (2013).
Kung, L. F. et al. Sec24p and Sec16p cooperate to regulate the GTP cycle of the COPII coat. EMBO J. 31, 1014–1027 (2012).
Yorimitsu, T. & Sato, K. Insights into structural and regulatory roles of Sec16 in COPII vesicle formation at ER exit sites. Mol. Biol. Cell 23, 2930–2942 (2012).
Ivan, V. et al. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell 19, 4352–4365 (2008).
Watson, P., Townley, A. K., Koka, P., Palmer, K. J. & Stephens, D. J. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic 7, 1678–1687 (2006).
Connerly, P. L. et al. Sec16 is a determinant of transitional ER organization. Curr. Biol. 15, 1439–1447 (2005).
Kurokawa, K., Okamoto, M. & Nakano, A. Contact of cis-Golgi with ER exit sites executes cargo capture and delivery from the ER. Nat. Commun. 5, 3653 (2014).
Tojima T., Suda Y., Jin N., Kurokawa K. & Nakano A. Spatiotemporal dissection of the Golgi apparatus and the ER-Golgi intermediate compartment in budding yeast. Elife 13, e92900 (2024).
Hughes, H. et al. Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER. J. Cell Sci. 122, 2924–2934 (2009).
Hammond, A. T. & Glick, B. S. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11, 3013–3030 (2000).
Weigel, A. V. et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429.e2416 (2021).
Kondylis, V. & Rabouille, C. A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 162, 185–198 (2003).
Takagi, J., Kimori, Y., Shimada, T. & Hara-Nishimura, I. Dynamic capture and release of endoplasmic reticulum exit sites by Golgi stacks in Arabidopsis. iScience 23, 101265 (2020).
Ito, Y. et al. cis-Golgi proteins accumulate near the ER exit sites and act as the scaffold for Golgi regeneration after brefeldin A treatment in tobacco BY-2 cells. Mol. Biol. Cell 23, 3203–3214 (2012).
daSilva, L. L. et al. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16, 1753–1771 (2004).
Yang, K. et al. ER exit sites in Drosophiladisplay abundant ER-Golgi vesicles and pearled tubes but no megacarriers. Cell Rep. 36, 109707 (2021).
Ito, Y., Uemura, T., & Nakano, A. The Golgi entry core compartment functions as a COPII-independent scaffold for ER-to-Golgi transport in plant cells. J. Cell Sci. 131, jcs203893 (2018).
Shimmen, T. The sliding theory of cytoplasmic streaming: fifty years of progress. J. Plant Res. 120, 31–43 (2007).
Shimmen, T. & Yokota, E. Cytoplasmic streaming in plants. Curr. Opin. Cell Biol. 16, 68–72 (2004).
Ueda, H. et al. Myosin-dependent endoplasmic reticulum motility and F-actin organization in plant cells. Proc. Natl. Acad. Sci. USA 107, 6894–6899 (2010).
Patel, S. et al. Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell 16, 3260–3273 (2004).
Jolliffe, N. A., Craddock, C. P. & Frigerio, L. Pathways for protein transport to seed storage vacuoles. Biochem. Soc. Trans. 33, 1016–1018 (2005).
Robinson, D. G., Oliviusson, P. & Hinz, G. Protein sorting to the storage vacuoles of plants: a critical appraisal. Traffic 6, 615–625 (2005).
Vitale, A. & Hinz, G. Sorting of proteins to storage vacuoles: how many mechanisms? Trends Plant Sci. 10, 316–323 (2005).
Shimada, T. et al. Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278, 32292–32299 (2003).
Shimada, T. et al. AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47, 1187–1194 (2006).
Yang, X., Liao, C. Y., Tang, J. & Bassham, D. C. Overexpression of trans-Golgi network t-SNAREs rescues vacuolar trafficking and TGN morphology defects in a putative tethering factor mutant. Plant J. 99, 703–716 (2019).
Kim, S. J. & Bassham, D. C. TNO1 is involved in salt tolerance and vacuolar trafficking in Arabidopsis. Plant Physiol. 156, 514–526 (2011).
Li, L. et al. MAG2 and three MAG2-INTERACTING PROTEINs form an ER-localized complex to facilitate storage protein transport in Arabidopsis thaliana. Plant J. 76, 781–791 (2013).
Li, L. et al. MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18, 3535–3547 (2006).
Takahashi, H. et al. MAG4/Atp115 is a golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant Cell Physiol. 51, 1777–1787 (2010).
Dhugga, K. S., Tiwari, S. C. & Ray, P. M. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: purification, gene cloning, and trans-Golgi localization. Proc. Natl. Acad. Sci. USA 94, 7679–7684 (1997).
Schaller, G. E. Isolation of endoplasmic reticulum and its membrane. Methods Mol. Biol. 1511, 119–129 (2017).
Nelson, B. K., Cai, X. & Nebenfuhr, A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007).
Geldner, N. et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178 (2009).
Uemura, T. et al. Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc. Natl. Acad. Sci. USA 109, 1784–1789 (2012).
Dettmer, J., Hong-Hermesdorf, A., Stierhof, Y. D. & Schumacher, K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18, 715–730 (2006).
Patel, S., Brkljacic, J., Gindullis, F., Rose, A. & Meier, I. The plant nuclear envelope protein MAF1 has an additional location at the Golgi and binds to a novel Golgi-associated coiled-coil protein. Planta 222, 1028–1040 (2005).
Brkljacic, J., Zhao, Q. & Meier, I. WPP-domain proteins mimic the activity of the HSC70-1 chaperone in preventing mistargeting of RanGAP1-anchoring protein WIT1. Plant Physiol. 151, 142–154 (2009).
Hughes, H. & Stephens, D. J. Sec16A defines the site for vesicle budding from the endoplasmic reticulum on exit from mitosis. J. Cell Sci. 123, 4032–4038 (2010).
Kondylis, V. & Rabouille, C. The Golgi apparatus: lessons from Drosophila. FEBS Lett. 583, 3827–3838 (2009).
Reynolds H. M., Zhang L., Tran D. T., Ten Hagen K. G. Tango1 coordinates the formation of ER/Golgi docking sites to mediate secretory granule formation. J. Biol. Chem. 294, 19498–19510 (2019).
Raote I. et al. TANGO1 builds a machine for collagen export by recruiting and spatially organizing COPII, tethers and membranes. Elife 7, e32723 (2018).
Liu, M. et al. Tango1 spatially organizes ER exit sites to control ER export. J. Cell Biol. 216, 1035–1049 (2017).
Saito, K. et al. cTAGE5 mediates collagen secretion through interaction with TANGO1 at endoplasmic reticulum exit sites. Mol. Biol. Cell 22, 2301–2308 (2011).
Raote, I. & Malhotra, V. Tunnels for protein export from the endoplasmic reticulum. Annu. Rev. Biochem. 90, 605–630 (2021).
Gallo, R., Rai, A. K., McIntyre, A. B. R., Meyer, K. & Pelkmans, L. DYRK3 enables secretory trafficking by maintaining the liquid-like state of ER exit sites. Dev. Cell 58, 1880–1897.e1811 (2023).
Mano, S. et al. Distribution and characterization of peroxisomes in Arabidopsis by visualization with GFP: dynamic morphology and actin-dependent movement. Plant Cell Physiol. 43, 331–341 (2002).
Heim, R., Cubitt, A. B. & Tsien, R. Y. Improved green fluorescence. Nature 373, 663–664 (1995).
Segami, S. et al. Vacuolar H(+)-pyrophosphatase and cytosolic soluble pyrophosphatases cooperatively regulate pyrophosphate levels in Arabidopsis thaliana. Plant Cell 30, 1040–1061 (2018).
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Karimi, M., Inze, D. & Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 (2002).
Nakamura, S. et al. Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74, 1315–1319 (2010).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Feldmann, K. A., Marks, M. D., Christianson, M. L. & Quatrano, R. S. A dwarf mutant of Arabidopsisgenerated by T-DNA insertion mutagenesis. Science 243, 1351–1354 (1989).
Shimada, T. et al. Vacuolar sorting receptor for seed storage proteins in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100, 16095–16100 (2003).
Ueda, H. et al. AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47, 164–175 (2006).
Hatano, K., Shimada, T., Hiraiwa, N., Nishimura, M. & Hara-Nishimura, I. A rapid increase in the level of binding protein (BiP) is accompanied by synthesis and degradation of storage proteins in pumpkin cotyledons. Plant Cell Physiol. 38, 344–351 (1997).
Tamura, K., Shimada, T., Kondo, M., Nishimura, M. & Hara-Nishimura, I. KATAMARI1/MURUS3 Is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17, 1764–1776 (2005).
Perkins, D. N., Pappin, D. J., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Rossignol, P., Collier, S., Bush, M., Shaw, P. & Doonan, J. H. Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120, 3678–3687 (2007).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Katoh, K. & Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinforma. 9, 286–298 (2008).
Steenwyk, J. L., Buida, T. J. 3rd, Li, Y., Shen, X. X. & Rokas, A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 18, e3001007 (2020).
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019).
Darriba, D. et al. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294 (2020).
Trigg, J., Gutwin, K., Keating, A. E. & Berger, B. Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS ONE 6, e23519 (2011).
Jeppe H. et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Preprint at bioRxiv (2022).
Hu, G. et al. flDPnn: accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat. Commun. 12, 4438 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Gilles, J. F., Dos Santos, M., Boudier, T., Bolte, S. & Heck, N. DiAna, an ImageJ tool for object-based 3D co-localization and distance analysis. Methods 115, 55–64 (2017).
Acknowledgements
We are grateful to T. Nakagawa (Shimane University), D. Bassham (Iowa State University), T. Uemura (Ochanomizu University), A. Nakano (RIKEN), K. Schumacher (Heidelberg University), N. Geldner (University of Lausanne), A. Nebenführ (University of Tennessee), and Plant System Biology (VIB) for sharing their materials and to the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre for providing seeds of A. thaliana T-DNA insertion mutants. This work was supported by Grants-in-Aid for Scientific Research to I.H.-N. (no. JP15H05776), to J.T. (nos. JP15J07657, JP18K14736, JP21K14825, and JP23K05143), to H.U. (nos. JP18H05496 and JP19K06732), to K.T. (nos. JP23H04205, and JP22K06269), and to T.S. (no. 18K06284) from the Japan Society for the Promotion of Science (JSPS), by the Human Frontier Science Program to K.T. (RGP0009/2018) from the International Human Frontier Science Program Organization; and by Academic Research to I.H.-N. and H.U. from the Hirao Taro Foundation of KONAN GAKUEN.
Author information
Authors and Affiliations
Contributions
Conceptualization; J.T., T.S., and I.H.-N. Methodology; J.T., H.U., and K.T. Investigation; J.T., H.T., Y.F., K.C.M., T.S., and M.N. Visualization; J.T. Supervision: J.T., T.S., and I.H.-N. Writing; J.T. and I.H.-N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Tomohiro Uemura and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Dr Gregory Lavieu and Dr Ophelia Bu.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Takagi, J., Takahashi, H., Moriya, K.C. et al. Plant-specific tail-anchored coiled-coil protein MAG3 stabilizes Golgi-associated ERESs to facilitate protein exit from the ER. Commun Biol 8, 358 (2025). https://doi.org/10.1038/s42003-025-07602-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07602-1