Abstract
Pyruvate dehydrogenase B (PDHB) is an important component of the pyruvate dehydrogenase complex and is implicated in altering tumor metabolism and promoting malignancy. However, the specific impact of PDHB on hepatocellular carcinoma (HCC) metabolic reprogramming and its role in tumor progression remain to be elucidated. In our investigation, we have discerned a pronounced elevation in PDHB expression within HCC, intricately linked to delayed tumor staging, heightened tumor grading, and diminished prognostic outcomes. PDHB overexpression drives tumor growth and metastasis in vitro and in vivo. Mechanistically, PDHB mediates metabolic reprogramming by binding to the promoter regions of SLC2A1, GPI, and PKM2, promoting glycolysis-related gene transcription, contributes to HCC sorafenib resistance. In addition, Isoacteoside is a targeted inhibitor of PDHB and exert antitumor effects on HCC. In the mouse xenograft model, the combination of isoacteoside and sorafenib shows significantly better effects than sorafenib alone. In summary, our study validates PDHB as an oncogenic drug resistance-related gene capable of predicting HCC tumor progression. PDHB and Isoacteoside could be potential avenues for targeted and combination therapies in liver cancer.

Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) ranks among the most prevalent and lethal cancers globally, with China accounting for over half of the new cases and fatalities and posing a grave threat to public health1. HCC typically progresses insidiously and lacks effective and reliable screening methods, resulting in most patients being diagnosed at advanced or terminal stages. Early-stage HCC is amenable to radical treatments, including surgical resection, radiofrequency/microwave ablation, transarterial chemoembolization, liver transplantation, and, occasionally, systemic chemotherapy1,2. Nevertheless, these interventions carry a substantial risk of metastasis and recurrence3. For patients with advanced HCC, surgical options are not viable and usually require systemic therapy (e.g., chemotherapy)4. However, chemotherapeutic agents can severely affect patients’ normal physiological functions, inducing adverse reactions such as digestive issues, immune suppression, and organ damage, including pneumonia and esophagitis, thereby diminishing the quality of life and potentially exacerbating the disease5.
As the first multitargeted tyrosine kinase inhibitor approved by the FDA6, sorafenib is the cornerstone of first-line therapy for advanced-stage HCC, having demonstrated a significant survival benefit for patients with unresectable HCC6. Its effectiveness has been validated in two large-scale clinical trials, and showing superiority over dozens of other molecular drugs7,8. However, the emergence of sorafenib resistance has become a critical issue, undermining therapeutic outcomes and leading to recurrence and mortality9. The mechanisms underlying sorafenib resistance are multifaceted, encompassing not only inherent resistance but also acquired resistance factors such as dysregulation of the JAK-STAT and PI3K-AKT pathways, epithelial-mesenchymal transition (EMT), tumor hypoxia, and metabolic reprogramming10,11. Therefore, unraveling the mechanisms of acquired sorafenib resistance and developing novel combination therapy strategies to overcome this resistance remain imperative for enhancing the treatment efficacy for patients with advanced HCC.
Recent studies have unveiled that metabolic reprogramming is a hallmark of cancer cells, fostering growth and survival12,13. And there is a significant correlation between metabolic reprogramming and cancer drug resistance14, especially in the case of the glycolytic pathway dominated by aerobic glycolysis (the Warburg effect), which has been observed to significantly compromise the efficacy of chemo-radiotherapy treatments15. Pyruvate dehydrogenase E1 subunit β (PDHB), a key component of the pyruvate dehydrogenase (PDH) complex, catalyzes the decarboxylation of pyruvate, playing a pivotal role in the glycolytic pathway and cellular energy metabolism16,17. As a crucial part of the PDH, PDHB is implicated in altering tumor metabolism and promoting malignancy18. It has been recognized as an oncogenic factor that is overexpressed in various cancers, including those of the bladder, melanoma, ovary, and prostate19, and is known to spur tumor cell growth18,19. However, the specific impact of PDHB on HCC metabolic reprogramming and its role in sorafenib resistance and tumor progression remain to be elucidated.
In this study, we identified PDHB as a potential novel target and biomarker for HCC treatment. We found that PDHB expression was significantly elevated in HCC and correlated with advanced tumor stage, high grade, and poor prognosis. PDHB appears to play an oncogenic role, as its overexpression promotes tumor growth and metastasis both in vitro and in vivo. Mechanistically, we linked increased PDHB expression to sorafenib resistance in HCC, showing that PDHB promotes the transcription of glycolytic genes by binding to the promoters of the glycolysis-related genes glucose transporter protein 1 (GLUT-1, SLC2A1), glycosylphosphatidylinositol (GPI) and pyruvate kinase M2 (PKM2), thus influencing metabolism reprogramming and consequent HCC resistance and tumor progression. Furthermore, through targeted virtual docking to the PDHB protein structure, we identified isoacteoside (HY-N0022), a naturally occurring compound, as a selective inhibitor of PDHB. Isoacteoside, a dihydroxyphenylethyl glycoside isolated from various plants20, is known for its various pharmacological benefits, including immune modulation, antioxidant, endurance enhancement, and antifatigue effects21. Recently, its anti-inflammatory, neuroprotective, and antitumor activities have gained increasing amounts of attention22,23,24. However, its anticancer potential in HCC has not been well defined. Herein, we found that isoacteoside exerts antitumor effects on HCC by targeting and inhibiting PDHB. Moreover, compared to sorafenib monotherapy, the combination of isoacteoside and sorafenib significantly increased the therapeutic efficacy. In conclusion, our findings elucidate the critical role of PDHB in HCC sorafenib resistance and tumor progression, as well as the potential molecular mechanisms underlying its actions, and propose potential targeted treatments for sorafenib-resistant tumors. These insights will open new avenues for targeted and combination therapies in liver cancer.
Results
The expression of PDHB as a prognostic indicator in HCC progression
To evaluate the expression and clinical relevance of PDHB in HCC. We first analyzed the pancancer expression of PDHB using the TIMER 2.0 and TCGA databases. The results showed that PDHB was significantly overexpressed in a variety of cancers, including HCC (Supplementary Fig. 1A, B). Then, RT‒PCR was performed to compare PDHB expression between 30 HCC tissues and corresponding nontumor tissues. The results showed a significant increase in PDHB expression in HCC tissues compared to adjacent normal tissues, which was consistent with the results of bioinformatics analysis (Supplementary Fig. 1C). Additionally, Kaplan–Meier survival analysis revealed that elevated PDHB levels may be closely associated with unfavorable outcomes in individuals with HCC (Supplementary Fig. 1D). The analysis of the UALCAN (Supplementary Fig. 1E) and TIMER (Supplementary Fig. 1F) databases confirmed that high PDHB expression was significantly correlated with high HCC tumor stage and grade and a poor patient prognosis. Collectively, these findings indicate that PDHB expression is a potential prognostic biomarker for tumor progression and patient outcomes in HCC.
PDHB promotes HCC cells proliferation and invasion in vitro
To investigate the oncogenic function of PDHB in HCC cells, we constructed PDHB overexpression and knockdown plasmids, and then overexpressed or knocked down PDHB in the HCC cell lines HepG2 and SMMC7721 (Fig. 1A, F, G and Supplementary Fig. 2A, F, G). The effects of PDHB overexpression and knockdown on the proliferation, colony formation, invasion and epithelial-mesenchymal transition (EMT) of HCC cell lines were detected. The results demonstrated that overexpression of PDHB significantly promoted cell proliferation (Fig. 1B and Supplementary Fig. 2B), colony formation (Fig. 1C and Supplementary Fig. 2C), invasion (Fig. 1D and Supplementary Fig. 2D), and EMT (Fig. 1E and Supplementary Fig. 2E) in HepG2 and SMMC7721 cells. Conversely, knockdown of PDHB expression markedly inhibited proliferation (Fig. 1H and Supplementary Fig. 2H), colony formation (Fig. 1I, J and Supplementary Fig. 2I, J), invasion (Fig. 1K and Supplementary Fig. 2K), and EMT (Fig. 1L and Supplementary Fig. 2L) of HCC cells.
A, F, G PDHB was either overexpressed or knocked down in HepG2 cells. B, H The CCK8 assay was conducted to evaluate the impact of PDHB overexpression or knockdown on HepG2 cell proliferation. C, I, J Colony formation assays were performed to assess the effect of PDHB overexpression or knockdown on the colony-forming capacity of HepG2 cells. D, K The Transwell assay was utilized to examine the influence of PDHB overexpression or knockdown on the invasion ability of HepG2 cells. Scale bar = 200 μm. E, L RT-PCR analysis was conducted to evaluate the effects of PDHB overexpression or knockdown on epithelial-mesenchymal transition (EMT) in HepG2 cells. Data are presented as means ± standard deviation from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
PDHB promotes HCC tumor growth and metastasis in vivo
To investigate the effects of PDHB on the tumor growth and metastasis of HCC, we first conducted a subcutaneous tumorigenesis experiment in nude mice by injecting HCC cells with overexpressed or knocked-down PDHB subcutaneously into the mice. The results showed that compared with the control group (pHBLV), the tumor growth in the PDHB overexpression group (PDHB) was significantly higher, with heavier tumor weight and larger tumor volume (Fig. 2A–C). In contrast, compared with the control group (pLKO.1), the tumor growth in the PDHB low-expression group (sh-PDHB) was significantly slower than the control group, with a noticeable reduction in tumor weight (Fig. 2H–J).
A–C Subcutaneous tumorigenesis assay was used to detect the effect of overexpression of PDHB on tumor growth in vivo, and the following are the statistics of tumor volume and body weight. D–G HepA1-6 cells were injected into mice via the tail vein after PDHB overexpression to establish an in vivo lung metastasis model (n = 3 mice/group). Bioluminescence was measured at designated time points, and representative images and quantifications are shown (D). Lungs were dissected and photographed (E), with representative HE-stained images of lung sections displayed (F, Scale bar = 2 mm) and immunohistochemical Ki67 staining images (G, Scale bar = 20 μm). H–J Subcutaneous tumorigenesis assay was used to detect the effect of knockdown of PDHB on tumor growth in vivo, and the following are the statistics of tumor volume and body weight. K–N HepA1-6 cells with PDHB knockdown were injected into mice via the tail vein to create the in vivo lung metastasis model (n = 3 mice/group). Bioluminescence was measured at designated time points, with representative images and quantifications shown (K). Lungs were dissected and photographed (L), along with representative HE-stained images of lung sections (M, Scale bar = 2 mm) and immunohistochemical Ki67 staining images (N, Scale bar = 20 μm). Data are presented as means ± standard deviation, with P values determined using a two-tailed t-test; *P < 0.05, **P < 0.01, ***P < 0.001.
Furthermore, we also injected the stably overexpressed or knocked-down mouse cell line HepA1-6-luci cells via the tail vein into mice and used a small animal live imaging instrument to dynamically monitor the growth and metastasis of tumors in mice. The results showed that compared with the control group (pCDH) mice, the lung metastasis in PDHB overexpression group (PDHB) mice was significantly enhanced, as shown by bioluminescence imaging (Fig. 2D). Notably, histological examination revealed that compared with the control group mice, the lung metastatic nodules in PDHB overexpression mice were larger and more numerous, with increased Ki67 staining (Fig. 2E–G). Conversely, compared with the control group mice (pLKO.1), the lung metastasis in PDHB knockdown group (sh-PDHB) was significantly reduced, with smaller and fewer lung metastatic nodules, and a noticeable decrease in Ki67 staining (Fig. 2K–N). In summary, these results indicate that PDHB can promote the proliferation, invasion, and metastasis of HCC cells both in vitro and in vivo suggest that PDHB is a potential oncogene, and high expression of PDHB is necessary for maintaining the occurrence and development of HCC tumors.
High PDHB expression correlates with sorafenib resistance in HCC
Given that sorafenib remains the standard therapy for patients with advanced HCC6 and its treatment effects are superior to those of dozens of other molecular drugs7,8. We probed the influence of PDHB expression on HCC cell sensitivity to sorafenib. The results showed that PDHB overexpression significantly increased the IC50 of sorafenib in HepG2 (Fig. 3A) and SMMC7721 (Fig. 3C) cells, while knockdown of PDHB decreased the IC50 of sorafenib in HepG2 (Fig. 3B) and SMMC7721 (Fig. 3D) cells.
A, C Overexpression of PDHB increases the IC50 of SMMC7721 and HepG2 cells to sorafenib. B, D Knockdown PDHB reduces the IC50 of SMMC7721 and HepG2 cells to sorafenib. E RT-PCR was used to detect the expression of PDHB in sorafenib-resistant strains of HCC (HepG2-R and SMMC7721-R). F, H Overexpression of PDHB increases the IC50 of HepG2-R and SMMC7721-R to sorafenib. G, I Knocking down PDHB reduces the IC50 of HepG2-R and SMMC7721-R to sorafenib. Data are presented as means ± standard deviation from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
Additionally, we developed sorafenib-resistant HepG2-R and SMMC7721-R cell lines by exposing HepG2 and SMMC7721 cells to gradually increasing concentrations of sorafenib over three months. Subsequently, we assessed PDHB expression and its effects on established HepG2-R and SMMC7721-R cells. The results showed that PDHB was significantly highly expressed in the HCC sorafenib-resistant strains HepG2-R and SMMC7721-R (Fig. 3E). Overexpression of PDHB significantly increased the IC50 in both HepG2-R (Fig. 3F) and SMMC7721-R (Fig. 3H) cells, while PDHB knockdown markedly decreased the IC50 of sorafenib (Fig. 3G, I).
In conclusion, these results indicate that overexpression of PDHB reduces the sensitivity of HCC to sorafenib, thereby promoting resistance. Conversely, downregulation of PDHB enhances sensitivity to the drug and significantly diminishes HCC resistance to sorafenib. The data thus suggest that PDHB expression is associated with sorafenib resistance in HCC and may contribute to the regulation of this resistance mechanism.
PDHB affects HCC sorafenib resistance by regulating glucose metabolism reprogramming
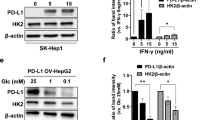
To further elucidate the molecular mechanisms underlying the pro-oncogenic role of PDHB in drug resistance, we conducted RNA sequencing on HepG2-R cells subjected to PDHB knockdown, followed by KEGG pathway, and differentially expressed gene (DEG) analyses (Fig. 4A–C and Supplementary Data 2 and 3). The analyses revealed that PDHB significantly influences metabolic pathways (Fig. 4A), especially oxidative phosphorylation and glycolysis/gluconeogenesis (Fig. 4B). PDHB also markedly affected the expression of pivotal metabolic genes such as PCK1, which is crucial for gluconeogenesis, and SLC2A1, essential for glycolysis (Fig. 4C). We first focused on the effect of PDHB on PCK1 expression. The results showed that overexpression of PDHB substantially suppressed PCK1 expression, while PDHB knockdown enhanced PCK1 expression (Fig. 4D, E and Supplement Fig. 3A, D), suggesting that PDHB may inhibit the gluconeogenesis pathway. Recognizing that gluconeogenesis is the reverse process of aerobic glycolysis25, we further explored the influence of PDHB on glycolysis. We found that overexpression of PDHB leads to increased expression of several glycolysis-related genes (including SLC2A1, HK2, GPI, PFKL, ALDOA, GAPDH, PGK1, ENO1, PKM2, and LDHA), whereas PDHB knockdown corresponded with a generalized reduction in the expression of these glycolysis-related genes (Fig. 4F, G and Supplementary Fig. 3B, E). To corroborate these results, we employed Western Blot analysis to evaluate the impact of PDHB on key enzymes involved in both gluconeogenesis and glycolysis at the protein level. Consistent with the RT-PCR findings, overexpression of PDHB inhibited gluconeogenesis and promoted glycolysis, whereas PDHB knockdown yielded opposite outcomes (Fig. 4H, I and Supplementary Fig. 3C, F).
A KEGG analysis of the effects of PDHB on physiological processes. B KEGG analysis of the effect of PDHB on metabolic pathways. C Volcano plot showing significantly differentially expressed genes affected by PDHB. D, E RT-PCR was used to detect the effect of overexpression or knockdown of PDHB on the expression of the gluconeogenic key enzyme PCK1. F, G RT-PCR was used to detect the effects of overexpression and knockdown of PDHB on the expression of glycolysis-related genes. H, I WB was used to detect the effects of overexpression and knockdown of PDHB on the expression of gluconeogenic and glycolysis-related genes. J Immunofluorescence was used to examine the intracellular distribution of PDHB following sorafenib treatment in HepG2 cells. PDHB (red) signals were observed under a laser microscope, while blue indicates the nuclei stained with 4’,6-diamidino-2-phenylindole (DAPI). Scale bar = 25 μm. K ChIP analysis of PDHB occupancy on the indicated gluconeogenic and glycolysis-related genes promoters in HepG2 cells. L Cytotoxicity analysis was performed to detect the role of glycolysis in PDHB-mediated resistance to sorafenib. M Cytotoxicity analysis was performed to detect the effect of glucose on PDHB-mediated resistance to sorafenib. Data are presented as means ± standard deviation from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
Additionally, GEPIA analysis revealed that the expression of PDHB in liver cancer tissue samples is significantly negatively correlated with the expression of the key gluconeogenic enzyme PCK1, significantly positively correlated with the expression of key glycolytic enzymes such as GPI, PFKL, ENO1, and weakly positively correlated with the expression of ALDOA, GAPDH, PGK1, LDHA, with no significant correlation with SLC2A1, HK2, PKM2, etc. (Supplementary Fig. 3G). STRING analysis also showed that among the proteins that have direct or indirect interactions with PDHB, there are many known glycolysis-related genes, such as GPI, LDHA, ENO1, PFKL, PKM2, etc. (Supplementary Fig. 3H). Overall, these findings suggest that PDHB may play an important role in sorafenib resistance in HCC by mediating metabolic reprogramming of glucose metabolism.
To further clarify the molecular mechanisms by which PDHB functions, we performed subcellular localization of PDHB. Immunofluorescence results showed that in HCC cells, PDHB is primarily localized in the cytoplasm (Fig. 4J). Interestingly, we also found that under the treatment of sorafenib, a portion of PDHB enters the nucleus (Fig. 4J). This finding suggests that PDHB may play a role in transcriptional regulation. Subsequently, we enhanced the expression of PDHB in HepG2 cells and performed ChIP-PCR. The results showed that PDHB can be recruited to the promoters of glycolytic enzymes SLC2A1, GPI, and PKM, but not to the upstream regions of other promoters (Fig. 4K). This indicates that PDHB can promote the transcription of glycolysis-related genes by binding to the promoters of glycolytic genes SLC2A1, GPI, and PKM2, thereby mediating the reprogramming of sugar metabolism.
Finally, based on the overexpression and knockdown of PDHB, we treated the cells with 2-DG (a glycolysis inhibitor that can reduce HCC resistance to sorafenib) to investigate the critical role of glycolysis in PDHB-induced HCC sorafenib resistance. The results showed that 2-DG could alleviate the sorafenib resistance induced by PDHB in HCC. Conversely, 2-DG treatment significantly enhanced the reduction of sorafenib resistance in HCC caused by sh-PDHB (Fig. 4L and Supplementary Fig. 3I). This indicates that glycolysis is a key factor in PDHB-induced HCC sorafenib resistance, and inhibiting glycolysis can relieve the sorafenib resistance induced by PDHB. Furthermore, we treated cells with high-glucose and low-glucose media based on PDHB overexpression, and used 2-DG to inhibit glucose metabolism to assess the IC50 value of sorafenib in HCC cells. The results showed that, compared to high-glucose conditions, low-glucose culture conditions enhanced the sensitivity to sorafenib mediated by PDHB, indicating that PDHB regulation of sorafenib resistance is glucose-dependent (Fig. 4M and Supplementary Fig. 3J). Moreover, the reduction of sorafenib resistance by 2-DG was reversed under the condition of PDHB overexpression (Fig. 4M and Supplementary Fig. 3J). Interestingly, we also observed that sorafenib treatment partially suppressed the expression of PDHB and glycolysis-related genes. Moreover, the effect of PDHB on glucose metabolism reprogramming was inhibited during sorafenib treatment, whereas the effects of PDHB knockdown on glucose metabolism were more pronounced during sorafenib treatment (Supplementary Fig. 3K, L). This suggests that the effect of PDHB on glucose metabolism may depend on its dynamic regulation and changes in the intracellular environment following sorafenib intervention. In conclusion, these data further indicate that PDHB can promotes sorafenib resistance in HCC by promoting glycolysis and affecting glucose metabolism reprogramming.
Isoacteoside inhibits HCC sorafenib resistance and tumor progression by targeting PDHB
Given that PDHB-mediated reprogramming of glucose metabolism plays a pivotal role in HCC sorafenib resistance and tumor progression, we sought to identify natural small molecule inhibitors that selectively target PDHB expression. Utilizing virtual docking techniques, multiple compounds were identified that interact with the active site of PDHB, exhibiting strong binding affinities. Among them, the five most potent compounds were HY-N0022, HY-101937C, Z1576583090, Z2436582684, and Z4480904659 (Supplementary Fig. 4).
Through a series of experimental verifications, we discovered that HY-N0022 (Isoacteoside) is one of the targeted inhibitors of PDHB (Fig. 5A, B). HY-N0022 significantly inhibits the expression of PDHB protein in a dose-dependent manners (Fig. 5C). Next, we evaluated the effect of HY-N0022 on HCC cell resistance and tumor progression. Initially, we determined the optimal dose of HY-N0022 through cytotoxicity assays (Fig. 5D, H). Subsequently, we examined the effect of HY-N0022 on HCC sorafenib resistance in HepG2 and SMMC7721 cells. After HY-N0022 treatment, the sensitivity of HCC cells to sorafenib significantly increased (Fig. 5E, I). Additionally, through in vitro and in vivo experiments, we observed that HY-N0022 can inhibit the proliferation (Fig. 5F, J), invasion (Fig. 5G, K), EMT (Fig. 5L, O), and tumor growth potential (Fig. 5M, N, P, Q) of HCC cells.
A Computer Virtual Docking Combination of Isoacteoside and PDHB. B Binding site of Isoacteoside to PDHB. C WB were used to detect the effects of Isoacteoside on PDHB expression in HCC cells. D, H Cytotoxicity assay was used to detect the IC50 of HepG2 and SMMC7721 after HY-N0022 treatment. E, I The effect of HY-N0022 on sorafenib resistance of HCC was detected. F, J CCK8 analysis was used to detect the effect of HY-N0022 on HCC cell proliferation. G, K Transwell assay was used to detect the effect of HY-N0022 on the invasion ability of HCC cells. Scale bar = 200 μm. L, O RT-PCR to detect the effect of HY-N0022 on the EMT ability of HCC cells. M, N, P, Q The effects of HY-N0022 on tumor growth was evaluated in vivo. R, S RT-PCR was used to detect the effects of HY-N0022 treatment on PDHB and glycolysis-related genes. Data are presented as means ± standard deviation from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
In addition, we also examined the effects of HY-N0022 on PDHB expression and glucose metabolism. The results showed that HY-N0022 could inhibit the expression of PDHB and glucose metabolism genes, and promote the expression of the key enzyme PCK1 in gluconeogenesis (Fig. 5R, S). These findings suggest that HY-N0022 exhibits anticancer effects on HCC, and its mechanism involves inhibiting HCC sorafenib resistance and progression by targeting PDHB to affect metabolic reprogramming.
The combination of isoacteoside and sorafenib significantly increased the therapeutic efficacy of HCC
Given the anticancer effects of HY-N0022, we explored the potential of combining isoacteoside with sorafenib to augment HCC therapeutic efficacy. Firstly, we established groups of HCC cells for a negative control, HY-N0022 treatment, sorafenib treatment, and a combination of HY-N0022 and sorafenib, and evaluated the impact of the various treatments on HCC cell growth using CCK-8 and apoptosis assays. The results showed that both monotherapy and combination therapy suppressed HCC cell proliferation (Fig. 6A, C) and induced tumor cell apoptosis (Fig. 6B, D). Notably, sorafenib monotherapy was more effective than HY-N0022 alone. And the combination therapy of HY-N0022 and sorafenib showed a significantly greater impact on tumor growth inhibition compared to sorafenib monotherapy. Moreover, we administered intraperitoneal injections of the four different treatments to an HCC xenograft mouse model. After a two-week treatment period, the mice were euthanized, and their tumor volumes were measured. The results corroborated the in vitro findings and showed that both monotherapy and combination therapy curtailed tumor growth in the mice, but sorafenib outperforming HY-N0022. Crucially, the combination therapy of HY-N0022 and sorafenib markedly enhanced the therapeutic effect compared to sorafenib alone (Fig. 6E, F).
A, C CCK8 analysis was used to detect the effects of HY-N0022, sorafenib monotherapy and combination treatment on HCC cell proliferation. B, D Flow cytometry was used to detect the effects of HY-N0022, sorafenib monotherapy and combination treatment on apoptosis of HCC cells. E, F The effects of HY-N0022, sorafenib monotherapy and combination treatment on tumor growth in vivo. Data are presented as means ± standard deviation from three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001.
Finally, we evaluated the drug toxicity of HY-N0022 in animal models. The results showed that HY-N0022 had no significant effect on body weight changes in mice at the recommended dosage, and no obvious toxic reactions were observed, as important organs such as the liver and kidneys showed no morphological abnormalities (Supplementary Fig. 5A–C). Additionally, blood biochemical tests indicated that HY-N0022 treatment did not disrupt electrolyte balance, nor did it affect glucose and lipid metabolism (Supplementary Fig. 5D–F). This suggests that HY-N0022 treatment has good safety and does not lead to hypokalemia, hyponatremia, or hyperuricemia commonly associated with traditional diuretics. It also does not impair normal kidney function or physiological processes related to glucose and lipid metabolism.
In summary, our study demonstrates that the PDHB-targeted inhibitor HY-N0022, when used in combination with sorafenib, can significantly enhance the therapeutic effect on HCC. Furthermore, HY-N0022 exhibits good safety in animal models, providing new insights and potential treatment options for the clinical management of HCC. These outcomes suggest that combining HY-N0022 with sorafenib can significantly boost the therapeutic efficacy in HCC treatment.
Discussion
HCC is one of the malignant tumors with a high mortality rate worldwide26,27. The optimization of treatment strategies and the discovery of new targets have always been hot topics in research. Sorafenib, as the first-line treatment drug for HCC at present, can significantly prolong the survival of patients28. However, the emergence of drug resistance severely affects the treatment effect, leading to an increased risk of recurrence and death for patients29. Therefore, exploring the molecular mechanisms of HCC resistance and seeking new therapeutic targets are of great significance for improving the prognosis of HCC patients.
This study first reveals that PDHB, an important component of the pyruvate dehydrogenase complex, is significantly upregulated in HCC, and its high expression is closely related to tumor staging, grading, and patient prognosis. This finding suggests that PDHB may play an important role in promoting the occurrence and development of HCC. Further in vitro and in vivo experiments confirm that PDHB promotes tumor growth and metastasis, indicating that PDHB is a potential oncogene. This suggests that PDHB is not only a biomarker for HCC progression but also a potential therapeutic target. Although few studies have pointed out that high expression of PDHB can inhibit the growth of tumor cells, such as Wang et al. finding that MEG3 can induce ER stress by upregulating PDHB, thereby inhibiting the proliferation and invasion of colorectal cancer cells30. Regardless, these studies indicate that PDHB may be a potential biomarker for pan-cancer prognosis, with different transcriptional and protein expression profiles, mechanisms, and functions in different types of tumors.
Mechanistically, our research has revealed a strong correlation between high expression of PDHB and the resistance of HCC to sorafenib. Specifically, overexpression of PDHB decreases the sensitivity of HCC to sorafenib and increases drug resistance. PDHB promotes the transcription of glycolysis-related genes SLC2A1, GPI, and PKM2 by binding to their promoters, leading to metabolic reprogramming in HCC. However, we also observed that sorafenib appears to inhibit the expression of PDHB and glycolysis-related genes to some extent, and the impact of PDHB overexpression on glucose metabolism is suppressed during sorafenib treatment, while the effect of PDHB knockdown on metabolism is more pronounced during sorafenib treatment. We guess that these different biological effects may be due to varying time points and drug dosages. At the initial stage of treatment, a certain duration and dose of sorafenib might inhibit the expression of oncogenes (such as PDHB) by regulating other regulatory factors or signaling pathways, altering the metabolism of tumor cells. With the accumulation of dosage and the extension of time, a subset of cancer cells may enter a “persistent state” under drug pressure. This process not only affects the energy metabolism of tumor cells but may also change the intracellular environment, affecting drug metabolism and mechanisms of action, further exacerbating the development of drug resistance. Alternatively, there may be compensatory mechanisms present in HCC, where the inhibitory effect of sorafenib on PDHB and glycolysis-related gene expression represents a complex feedback regulation mechanism aimed at suppressing the formation of resistance. Knockdown of PDHB may disrupt the metabolic balance within the cells, increasing their sensitivity to sorafenib and making the changes in glucose metabolism more pronounced. Additionally, the cellular environment and heterogeneity may also influence the experimental results. The HCC sorafenib-resistant cell lines and the non-induced HCC cells used in this study, as different cellular subpopulations, may exhibit varying responses to PDHB expression and sorafenib treatment. In summary, these seemingly conflicting experimental results may not be theoretically contradictory but rather reveal a more complex molecular regulatory network and drug action mechanisms that warrant further investigation.
Notably, our study also found that PDHB is widely localized in the cytoplasm and translocates to the nucleus upon sorafenib stimulation. This phenomenon suggests that PDHB may play an important role in transcriptional regulation. Through CHIP experiments, we further confirmed that PDHB binds to the promoters of glycolytic genes SLC2A1, GPI, and PKM, promoting their transcription, which provides strong support for the role of PDHB as a transcription factor.
Moreover, studies have shown that the E1α subunit encoded by the PDHB gene is the active center of pyruvate dehydrogenase complex (PDHc), responsible for catalyzing the oxidative decarboxylation of pyruvate. The expression level and function of PDHB directly affect the overall catalytic activity of the PDH complex. The absence or dysfunction of PDHB can lead to reduced PDH activity, thereby affecting energy metabolism31. However, there is still controversy about the relationship between PDH activity and sugar metabolism and tumor progression. Some studies believe that PDH is a key enzyme in the glycolysis pathway, responsible for converting pyruvate into acetyl-CoA, which then enters the tricarboxylic acid (TCA) cycle. A decrease in PDH activity may promote glycolysis, providing energy and biosynthetic precursors for tumor cells. And reduced PDH activity may be related to tumor invasiveness, metastasis, and drug resistance32,33. Conversely, other studies indicate that PDH activity is higher in tumor cells, and PDH inhibitors could serve as new therapeutic targets for cancer treatment34. However, we are more interested in the new mechanism of PDHB, which acts as a transcription factor independently of PDH enzyme activity, affecting the transcription and expression of downstream genes. Overall, this discovery challenges the traditional understanding of PDHB function, suggesting its potential role in the regulation of tumor metabolism and the development of drug resistance. Of course, the broader functions and mechanisms of PDHB still need further exploration. Future research can focus on the role of PDHB in different types of cancer, its interaction with other metabolic pathways, and its regulatory mechanisms in the tumor microenvironment. This will help to deepen the understanding of the multiple roles of PDHB in cancer biology and provide a theoretical basis for the development of new treatment strategies.
Finally, we found that Isoacteoside can target and inhibit the expression of PDHB. Isoacteoside, a dihydroxyphenylethyl glycoside, is a major bioactive component found in plants such as Forsythia suspensa, Cistanche deserticola, and Plantago asiatica. Its primary pharmacological effects include enhancing physical strength, combating fatigue, immune regulation, antioxidant activity, antibacterial properties, and protective effects on the nervous and cardiovascular systems. Its functions involve a variety of complex mechanisms such as free radical scavenging, modulation of inflammatory pathways, regulation of energy sensing pathways, and cell cycle control22,23,35. In recent years, studies have found that isoacteoside has the ability to inhibit the proliferation of tumor cells, such as ovarian cancer24. However, the efficacy and mechanism of Isoacteoside in the treatment of HCC are rarely explored. Here, we propose the antitumor activity of Isoacteoside in HCC and preliminarily explore its therapeutic effects when used in combination with sorafenib. We found that HY-N0022 can target PDHB to inhibit sorafenib resistance and tumor progression in HCC. Moreover, the combination of HY-N0022 with sorafenib significantly enhances the therapeutic effect of sorafenib monotherapy. Additionally, HY-N0022 demonstrates good biological safety. These findings suggest that Isoacteoside holds great potential in the treatment of HCC, both as a standalone therapy and in combination with other treatments, warranting further investigation. Therefore, future research should delve into the biological activity of Isoacteoside in targeting PDHB and its antitumor activity in tumor treatment, including HCC, providing new insights for the development of targeted and combination therapies for HCC. This research may also lay the groundwork for clinical trials and the promotion of Isoacteoside.
In summary, tumor drug resistance is currently one of the most important and challenging issues in cancer treatment. Understanding the mechanisms of tumor drug resistance is crucial for overcoming drug resistance and exploring new treatment strategies. This study reveals the important role of PDHB in sorafenib resistance and tumor progression in HCC, and proposes PDHB-based targeted therapy and combination antagonism strategies. These findings not only provide new molecular targets and biomarkers for the treatment of HCC, but also provide new ideas and pathways for clinical diagnosis and treatment.
Methods
Clinical tissue specimens
HCC cancer tissues and adjacent tissues from 30 HCC patients (from August 2020 to August 2022) who underwent surgical resection at the First Affiliated Hospital of Xinxiang Medical College were collected. None of the patients received chemotherapy or radiotherapy before surgery, and the excised tumor tissues were confirmed by at least two pathologists. Informed consent was obtained from each patient, and the study protocol was approved by the Ethics Committee of Xinxiang Medical University (Number: XYLL-2020653, Date: 2020.08.01). All ethical regulations relevant to human research participants were followed.
Cell lines
The human hepatocellular carcinoma cell lines HepG2 (#CBP60199, COBIOER) and SMMC-7721 (#CBP60210, COBIOER) and the mouse hepatoma cell line HepA1-6 (#CBP60574, COBIOER) were purchased from Nanjing Cobioer Biosciences Co., Ltd. Cells were authenticated by STR profiling and tested for mycoplasma contamination. The HCC cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM, #12100, Solarbio) supplemented with 10% fetal bovine serum (FBS, #P08X20, Kamplai) and 100 U/ml of penicillin/streptomycin (#C100C5, New Saemai Biotechnology Co., Ltd.). The cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2.
RNA extraction and real-time quantitative PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (# R401-01, Vazyme), and the reverse transcription reaction was performed with HiSCRIPTR II Q RT SuperMix (#R223-01, Vazyme). qRT-PCR analysis was performed using HiSCRIPTR Universal SYBR qPCR Master Mix (#Q511-02, Vazyme) on a Pikorea196 Real-Time PCR machine (Thermo). Gene expression levels were normalized to β-actin, and the 2−ΔΔCt method was employed for calculating fold changes. Each experiment was independently replicated three times independently. The sequences of primers used are listed in Supplementary Information Table 1A.
Western blot (WB)
Total protein extraction was performed using RIPA buffer (#R0020, Solarbio). The lysates were then mixed with 5x protein loading buffer (#WB2001, New Saimet Biotech LTD.) and denatured at 95 °C for 10 min. Proteins were separated on an 8% to 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel and subsequently transferred onto a PVDF membrane (#IPVH00010, Millipore). The membrane was blocked with blocking solution (#P0252, Beyotime) for 2 h at room temperature. Primary antibodies were applied and incubated overnight at 4 °C according to the provided instructions. Following this, the membrane was treated with an appropriate HRP-conjugated secondary antibody and developed using an enhanced chemiluminescence kit (#KE0101, Kemix). ECL signals were detected with a GE Amersham Imager 680. Antibodies used included anti-β-actin (AB0035, 1:1000, Abways), anti-PDHB (#14744-1-AP, 1:1000, Proteintech), anti-PCK1(CY8626, 1:1000, Abways), anti-SLC2A1(CY8069, 1:1000, Abways), anti-GPI (CY8078, 1:1000, Abways), anti-PFKL (CY8047, 1:1000, Abways), anti-ALDOA (CY7206, 1:1000, Abways), anti-GAPDH (AB0037, 1:1000, Abways), anti-PGK1(CY5536, 1:1000, Abways), anti-ENO1(CY5106, 1:1000, Abways), anti-HK2 (HA500186-50, 1:1000, HuaBio), anti-PKM2 (CY5764, 1:1000, Abways), anti-LDHA(3582S, 1:2000, Cell Signaling Technology), and anti-rabbit IgG (#P03802M, 1:5000, Kimplai).
Plasmid construction and cell transfection
The PDHB coding sequence was cloned into the pHBLV vector to construct the PDHB overexpression plasmid (pHBLV-PDHB) (Supplementary Information Table 1B). The mouse PDHB overexpression plasmid (M-pCDH-PDHB) and the human PDHB overexpression plasmid with a GFP tag (H-pCDH-EGFP-PDHB) were purchased from YiXueSheng Biosciences Inc (Shanghai, China). Using OligoEngine software, we designed three shRNAs targeting PDHB for both human and mouse (Supplementary Information Table 1C) and cloned into the pLKO.1 vector to create the PDHB knockdown plasmids (pLKO.1-sh-PDHB) for human and mouse. All plasmid sequences were confirmed by DNA sequencing. Plasmids were transfected into cells using PEI Transfection Reagent (#23966-1, Polysciences) according to the manufacturer’s protocol. Subsequently, transduced cells were treated with 2 µg/ml puromycin (#ST551, Beyotime) to generate stable PDHB knockout cell lines. At 48 h post-transfection, gene overexpression or silencing effects were assessed at both the mRNA and protein levels.
Cell proliferation and colony formation assays
Cell proliferation was assessed using the CCK-8 colorimetric assay (#B34304, Bimake). Cells were seeded at 1 × 103 cells/well in 96-well plates and incubated at 37 °C with 5% CO2 for 24, 48, 72, or 96 h. At specific time points, 10 μL of CCK-8 solution was added to each well and further incubated for 2 h. Absorbance was measured at 450 nm using a Multiskan MK3 microplate reader (Thermo).
For the colony formation assay, cells from various groups were counted and seeded at 500 cells per well in 6-well plates with complete medium containing 10% FBS. After 14 days of culture, the cells were fixed with 4% paraformaldehyde (#SLI830, Coolaber) and stained with 0.1% crystal violet (#G1062, Solarbio). Images were captured using a Leica DMI8 light microscope, and colonies in 5 randomly selected fields were counted for quantification.
Immunofluorescence (IF)
After a 48-hour treatment with sorafenib, the round coverslip was performed. The following day, the culture medium was discarded, and the cells were washed three times with PBS. They were then fixed with 4% paraformaldehyde for 20 min. After removing the fixation solution, the cells were washed three times and permeabilized with 0.3% Triton X-100 for 2 min, followed by another wash. The cells were then blocked with 3% BSA at room temperature for 1 h before being incubated overnight at 4 °C with anti-PDHB antibody (#14744-1-AP, Proteintech, 1:300). After the incubation, the cells were washed with PBST and incubated with a Cy3 conjugated Donkey Anti-Rabbit IgG (#GB21403, Servicebio, 1:800) at room temperature for 1 h. Following another wash, the cells were stained with DAPI and images were taken using a Leica DMI8 fluorescence microscope. Finally, the images were merged using ImageJ software.
Transwell assay
Transwell assay was used to detect the invasion of HCC cells according to the manufacturer’s instructions. In brief, 1 × 105 cells were suspended in serum-free medium and placed into the upper compartment of the Transwell chamber (#3422, Costar). The lower chamber was filled with culture medium containing 20% FBS to serve as a chemoattractant. Following a 28-h incubation at 37 °C, non-invading cells were removed from the upper surface of the membrane using a cotton swab. Cells that had migrated to the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Images were captured with a Leica DMI8 light microscope, and invaded cells were counted in five randomly selected fields for quantification.
RNA sequencing
HepG2 Sorafenib-resistant strain (HepG2-R) were transiently transfected with the sh-PDHB plasmid. 48 h post-transfection, the cells were harvested, and total RNA was extracted using Trizol reagent (Invitrogen, MA, USA), followed by quantification with a NanoDrop (Thermo Fisher Scientific, USA). Then, the RNA samples were sent to OE Biotech Co., Ltd. (Shanghai, China) for library construction and sequencing, followed by bioinformatics analysis on the Illumina HiSeq X Ten platform. Raw data in fastq format were processed using Trimmomatic to remove low-quality reads and generate clean reads, which were then mapped to the human genome (GRCh38) using HISAT2. FPKM for each gene was calculated using cufflinks, and gene read counts were obtained through HTSeq-count. Differential expression analysis was performed using the DESeq R package, with a significance threshold of P < 0.05 and a fold change cutoff of ≥2 or ≤0.5. Hierarchical clustering analysis revealed expression patterns of differentially expressed genes (DEGs) across the samples. GO term and KEGG pathway enrichment analysis for DEGs was conducted using R, based on a hypergeometric distribution.
Chromatin immunoprecipitation (ChIP)
ChIP assays were carried out using the Hyperactive pG-MNase CUTandRUN Assay Kit for PCR/qPCR (#HD101-01, Vazyme). Cell chromatin complexes were collected as per the manufacturer’s guidelines and analyzed by qRT-PCR. The primer sequences are provided in Supplementary Information Table 1D.
Virtual screening of small molecule drugs targeting PDHB
Virtual screening was performed using Schrödinger Maestro software (version 11.4) to identify potential small molecule inhibitors targeting the active sites of PDH β subunits. The PDH crystal structure (PDB ID: 3EXE) was obtained from the RCSB Protein Data Bank (http://www.rcsb.org/). The PDHB protein structure was optimized using the Protein Preparation Wizard module. Subsequently, Discovery Diversity Set 50 (DDS-50, comprising 50.2 K compounds) and MCE Bioactive Compound Library Plus (12.2 K compounds) were screened against PDHB using the Virtual Screening Workflow module in High-Throughput Virtual Screening (HTVS) mode. Compounds demonstrating the highest binding affinity to the PDHB active site were shortlisted as candidate inhibitors.
Drug, cytotoxicity, and sorafenib resistance testing
Isoacteoside (#HY-N0022, MCE), Sorafenib (#HY-10201, MCE), and 2-deoxy-D-glucose (2-DG, #HY-13966, MCE) were purchased from MCE and dissolved in dimethylsulfoxide (DMSO, #D8371, Solarbio). These were then further diluted to various concentrations in the medium. Cells, numbering ≥5 × 104, were seeded into each well of a 96-well plate with approximately 100 μl of cell suspension. Replicates of 4–6 were prepared for each sample. To perform the cytotoxicity analysis, after cell attachment, varying concentrations of drugs were administered. For detecting sorafenib resistance, upon cell attachment, the medium was substituted with one containing different concentrations of sorafenib. The cells were then incubated at 37 °C. Post-treatment for the specified duration, 10 μL of CCK-8 solution was added to each well, and the plates were incubated at 37 °C for 2 h. Absorbance was measured at 450 nm using a Multiskan MK3 microplate reader (Thermo).
Apoptosis analysis
An apoptosis assay kit (#E-CK-A211, Elabscience) was utilized for apoptosis detection. Cells, prepared as single-cell suspensions, underwent FITC/PI double staining for 15 min at room temperature (25 °C). Analysis was conducted using a BD FACS Calibur™ Flow Cytometer (#342975, BD Biosciences), with FlowJo software (Tree Star, Ashland, OR) for data analysis.
Animal experiments
BALB/c nude male mice and C57BL/6 black male mice, both 5 weeks old, were sourced from Beijing Vital River Laboratory Animal Technology Co., Ltd. and housed under specific pathogen-free (SPF) conditions. All animal experimental protocols were reviewed and approved by the Animal Care and Use Committee of Xinxiang Medical University (approval number: XYLL-2020653, date: 2020-08-01), and we have complied with all relevant ethical regulations for animal use. In animal experiments, the tumor was excised before reaching a maximum volume of 2000 mm³. We ensured that the maximum allowed tumor volume was not exceeded during the experiment.
In subcutaneous tumorigenesis experiments, 5 × 106 SMMC7721 cells with either control vectors (pHBLV or pLKO.1) or modified for PDHB overexpression (PDHB) or knockdown (sh-PDHB) were subcutaneously injected into BALB/c nude mice (n = 6). After 4–6 weeks, the mice were euthanized under deep anesthesia, and the tumor were excised, weighed, measured, and photographed.
For the in vivo metastasis experiment, 2.0 × 105 HepA1-6-luciferase cells with stable overexpression of PDHB (PDHB) or knockdown (sh-PDHB), or control groups (pCDH or pLKO.1) were injected into mice via the tail vein. After injecting the substrate (D-arginine) and anesthetizing with isoflurane, the fluorescence detection of mice bearing HepA1-6 tumors was monitored using an in vivo imaging system. Data were analyzed using LivingImage software. Finally, the tumor metastatic tissues were fixed with 10% neutral formalin or stored at −80 °C for subsequent analysis.
For HY-N0022 drug toxicity assessment experiment, C57BL/6 mice were divided into 2 groups (n = 4) to evaluate the toxic side effects of the small molecule drug. One group received saline solution, while the other group received HY-N0022 at a dose of 30 mg/kg via intraperitoneal injection for two weeks, with the initial dose being doubled. Body weight was recorded every two days, and the overall condition of the mice (including food intake, water consumption, and activity level) was monitored daily. After 2 weeks, the mice were euthanized under deep anesthesia, and liver and kidney tissues were fixed in 4% paraformaldehyde, while plasma samples were quickly frozen at −80 °C for subsequent biochemical analysis.
For the antitumor activity evaluation of small molecule drugs, 1.2 × 106 HepA1-6 cells were injected subcutaneously into C57BL/6 mice (n = 6). Upon reaching a tumor volume of ~50 mm3, mice were allocated into four groups for a 2-week treatment with either normal saline, HY-N0022 (30 mg/kg), sorafenib (30 mg/kg), or a combination of both drugs, all administered via intraperitoneal injection. Tumor volume and body weight were recorded every 2 days. After 2 weeks, the mice were euthanized under deep anesthesia, and the tumors were processed as described above. Tumor volume (V) was calculated using the formula: V = (Length × Width2) × 0.5. Tumor tissues were either fixed with 4% paraformaldehyde or snap-frozen at −80 °C for further analysis.
Hematoxylin and Eosin staining (HE) and immunohistochemistry (IHC)
The procedure was performed as previously described36. Briefly, tissues obtained from animal experiments (lung tissues with tumor metastasis and liver and kidney tissues from drug treatment) were fixed in formalin, processed through a graded series of ethanol and xylene, and then paraffin-embedded. Finally, sections (3 μm) were cut and mounted on slides. The sections were dewaxed, rehydrated, and treated with 3% H2O2.
For HE staining, the sections were stained with hematoxylin (Servicebio, G1076) for 5–10 min, rinsed, and quickly differentiated in acidic alcohol. They were then stained with eosin for 2–5 min, dehydrated through a series of ethanol washes, and treated with xylene.
For IHC staining, the sections were incubated with 5% bovine serum albumin (BSA) blocking solution at room temperature for 30 min. Then, a diluted primary antibody was added and incubated overnight at 4 °C. After washing the sections three times with PBS, a secondary antibody was added and incubated at room temperature for 1 h. The sections were washed three times with PBS again. After adding the chromogenic substrate, a mounting medium was applied, and coverslips were placed on top, allowing them to dry. Finally, a neutral mounting medium was applied, and the sections were visualized, and images were captured using an Olympus microscope.
Biochemical tests
Blood specimens were collected using heparin anticoagulant tubes and centrifuged at 3000 rpm for 15 min within 30 min of collection, maintaining a temperature of 2–8 °C. The supernatant was then promptly analyzed using the Chemray 800.
Bioinformatics analysis
The analysis of PDHB expression between normal tissues and tumor tissues was completed by tumor immune estimation resource, version 2 (TIMER2.0, http://timer.cistrome.org/)37. We also integrated the normal samples from GTEx with tumor sample from TCGA38 to confirm PDHB expression in several tumors. The gene expression profile and correlation analysis of PDHB, and their correlation with prognosis in HCC patients was predicted with the help of the GEPIA database (http://www.gepia.cancer-pku.cn/)39 and UALCAN40 database. The Protein-protein interaction network (PPI) is constructed using STRING41.
Statistics and reproducibility
All statistical analyses were performed using Prism 9.0 (GraphPad Software, La Jolla, CA, USA) software. The two-tailed students t-test was used for comparison between the two groups. Kaplan–Meier analysis was used for survival analysis. Spearman rank correlation analysis was used for correlation between genes. All experiments were repeated 3 times, and the data were expressed as mean ± standard deviation (SD), P < 0.05 was considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001. All data were collected from at least three independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data of the sorafenib-resistant cells are available at NCBI, Biological Project Entry number (PRJNA1202723). The raw data for all the figures in the manuscript can be found as Supplementary Data 1. Uncropped Western blot images are available in the Supplementary Information. The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. A report summary of the study is also provided as a supplementary file.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019).
Yang, J. D. et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 16, 589–604 (2019).
Simile, M. M. et al. Targeted therapies in cholangiocarcinoma: emerging evidence from clinical trials. Medicina 55, 42 (2019).
Bruix, J., da Fonseca, L. G. & Reig, M. Insights into the success and failure of systemic therapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 16, 617–630 (2019).
Kumari, R., Sahu, M. K., Tripathy, A., Uthansingh, K. & Behera, M. Hepatocellular carcinoma treatment: hurdles, advances and prospects. Hepat. Oncol. 5, HEP08 (2018).
Bangaru, S., Marrero, J. A. & Singal, A. G. Review article: new therapeutic interventions for advanced hepatocellular carcinoma. Aliment Pharm. Ther. 51, 78–89 (2020).
Zhu, A. X. et al. Ramucirumab versus placebo as second-line treatment in patients with advanced hepatocellular carcinoma following first-line therapy with sorafenib (REACH): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 16, 859–870 (2015).
El-Khoueiry, A. B. et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet 389, 2492–2502 (2017).
Jindal, A., Thadi, A. & Shailubhai, K. Hepatocellular carcinoma: etiology and current and future drugs. J. Clin. Exp. Hepatol. 9, 221–232 (2019).
Xu, J. et al. UBQLN1 mediates sorafenib resistance through regulating mitochondrial biogenesis and ROS homeostasis by targeting PGC1beta in hepatocellular carcinoma. Signal Transduct. Target Ther. 6, 190 (2021).
Mendez-Blanco, C., Fondevila, F., Garcia-Palomo, A., Gonzalez-Gallego, J. & Mauriz, J. L. Sorafenib resistance in hepatocarcinoma: role of hypoxia-inducible factors. Exp. Mol. Med. 50, 1–9 (2018).
Jeong, H., Lee, B., Han, S. J. & Sohn, D. H. Glucose metabolic reprogramming in autoimmune diseases. Anim. Cells Syst.27, 149–158 (2023).
Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science 368, eaaw5473 (2020).
Bao, M. H. R. & Wong, C. C. L. Hypoxia, metabolic reprogramming, and drug resistance in liver cancer. Cells 10, 1715 (2021).
Rahman, M. & Hasan, M. R. Cancer metabolism and drug resistance. Metabolites 5, 571–600 (2015).
Kikuchi, D., Minamishima, Y. A. & Nakayama, K. Prolyl-hydroxylase PHD3 interacts with pyruvate dehydrogenase (PDH)-E1beta and regulates the cellular PDH activity. Biochem. Biophys. Res. Commun. 451, 288–294 (2014).
Larrieu, C. M. et al. Refining the role of pyruvate dehydrogenase kinases in glioblastoma development. Cancers 14, 3769 (2022).
Yonashiro, R., Eguchi, K., Wake, M., Takeda, N. & Nakayama, K. Pyruvate dehydrogenase PDH-E1beta controls tumor progression by altering the metabolic status of cancer cells. Cancer Res. 78, 1592–1603 (2018).
Echeverri Ruiz, N. P. et al. Dynamic regulation of mitochondrial pyruvate metabolism is necessary for orthotopic pancreatic tumor growth. Cancer Metab. 9, 39 (2021).
Sannigrahi, S., Mazumder, U. K., Pal, D. & Mishra, S. L. Terpenoids of methanol extract of Clerodendrum infortunatum exhibit anticancer activity against Ehrlich’s ascites carcinoma (EAC) in mice,. Pharm. Biol. 50, 304–309 (2012).
Chae, S. et al. Antioxidant activity of isoacteoside from Clerodendron trichotomum. J. Toxicol. Environ. Health A 68, 389–400 (2005).
Gao, H. et al. Isoacteoside, a dihydroxyphenylethyl glycoside, exhibits anti-inflammatory effects through blocking toll-like receptor 4 dimerization. Br. J. Pharm. 174, 2880–2896 (2017).
Ji, S. L. et al. Antioxidant activity of phenylethanoid glycosides on glutamate-induced neurotoxicity. Biosci. Biotechnol. Biochem. 83, 2016–2026 (2019).
Yang, X., Guo, F., Peng, Q., Liu, Y. & Yang, B. Suppression of in vitro and in vivo human ovarian cancer growth by isoacteoside is mediated via sub-G1 cell cycle arrest, ROS generation, and modulation of AKT/PI3K/m-TOR signalling pathway. J. BUON 24, 285–290 (2019).
Wang, Z. & Dong, C. Gluconeogenesis in cancer: function and regulation of PEPCK, FBPase, and G6Pase. Trends Cancer 5, 30–45 (2019).
Chakraborty, E. & Sarkar, D. Emerging therapies for hepatocellular carcinoma (HCC). Cancers 14, 2798 (2022).
Asafo-Agyei, K. O. & Samant, H. Hepatocellular carcinoma. StatPearls, Treasure Island (FL) ineligible companies. Disclosure: Hrishikesh Samant declares no relevant financial relationships with ineligible companies (2023).
Cheng, A. L. et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 10, 25–34 (2009).
Huang, A., Yang, X. R., Chung, W. Y., Dennison, A. R. & Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target Ther. 5, 146 (2020).
Wang, G. et al. LncRNA MEG3 promotes endoplasmic reticulum stress and suppresses proliferation and invasion of colorectal carcinoma cells through the MEG3/miR-103a-3p/PDHB ceRNA pathway. Neoplasma 68, 362–374 (2021).
Patel, M. S. & Korotchkina, L. G. Regulation of the pyruvate dehydrogenase complex. Biochem. Soc. Trans. 34, 217–222 (2006).
Wang, H. et al. Metabolic and oncogenic adaptations to pyruvate dehydrogenase inactivation in fibroblasts. J. Biol. Chem. 294, 5466–5486 (2019).
Gray, L. R., Tompkins, S. C. & Taylor, E. B. Regulation of pyruvate metabolism and human disease. Cell Mol. Life Sci. 71, 2577–2604 (2014).
Stacpoole, P. W. Therapeutic targeting of the pyruvate dehydrogenase complex/pyruvate dehydrogenase kinase (PDC/PDK) axis in cancer. J. Natl Cancer Inst. 109, 11 (2017).
Shiao, Y. J., Su, M. H., Lin, H. C. & Wu, C. R. Acteoside and isoacteoside protect amyloid beta peptide induced cytotoxicity, cognitive deficit and neurochemical disturbances in vitro and in vivo. Int. J. Mol. Sci. 18, (2017).
Wang, L. et al. c-Myc-mediated epigenetic silencing of MicroRNA-101 contributes to dysregulation of multiple pathways in hepatocellular carcinoma. Hepatology 59, 1850–1863 (2014).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020).
The TCGA legacy. Cell 173, 281–282 (2018).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47, W556–W560 (2019).
Chandrashekar, D. S. et al. UALCAN: an update to the integrated cancer data analysis platform. Neoplasia 25, 18–27 (2022).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49, D605–D612 (2021).
Acknowledgements
We would like to thank Director Zhao Baosheng of the Department of Thoracic Surgery of the First Affiliated Hospital of Xinxiang Medical University for providing HCC clinical samples. This work was supported by Key Research Projects of Higher Education Institutions in Henan Province (25A180011 and 25A320018) and Key Science and Technology Program of Henan Province (222102310688 and 232102311062).
Author information
Authors and Affiliations
Contributions
Lijun Zhao, PhD (Data curation: Lead; Formal analysis: Equal; Investigation: Lead; Methodology: Equal; Funding acquisition: Equal; Project administration: Equal; Software: Lead; Supervision: Equal; Writing-original draft: Lead; Writing-review & editing: Equal). Haonan Qi, Master (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Project administration: Equal; Writing-original draft: Equal; Writing-review & editing: Equal). Weiting Liu, bachelor (Data curation: Equal; Investigation: Equal; Methodology: Equal; Project administration: Equal; Writing-original draft: Equal). Huiying Lv, Master (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting). Peixian Li, Master (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting). Wenyue Liu, Master (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting). Ruili Sun, PhD (Formal analysis: Equal; Methodology: Equal; Resources: Equal, Project administration: Equal; Supervision: Lead; Writing-review & editing: Equal). Qiongzi Wang, PhD (Formal analysis: Equal; Methodology: Equal; Funding acquisition: Equal; Project administration: Equal; Resources: Equal, Supervision: Equal; Writing-review & editing: Equal). Xiangpeng Wang, PhD (Conceptualization: Lead; Formal analysis: Lead; Funding acquisition: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing-review & editing: Equal). The work reported in the paper has been performed by the authors, unless clearly specified in the text.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics statement
All animal care and experimental studies were performed according to the guidelines and approval of the Ethics Committee of Xinxiang Medical University (Approval Number: XYLL-20200653; Date: 2020.08.01). This study included 30 patients was conducted in accordance with the Declaration of Helsinki, and all patients provided written informed consent. Patient information was completely anonymized. The study protocol was approved by the Ethics Committee of Xinxiang Medical University (Approval Number: XYLL-20200653; Date: 2020.08.01).
Peer review
Peer review information
Communications Biology thanks Kui Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Johannes Stortz and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, L., Qi, H., Liu, W. et al. Isoacteoside alleviates hepatocellular carcinoma progression by inhibiting PDHB-mediated reprogramming of glucose metabolism. Commun Biol 8, 205 (2025). https://doi.org/10.1038/s42003-025-07622-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07622-x