Abstract
Inflammatory bowel disease (IBD) is a chronic, relapsing, and remitting disease characterized by chronic inflammation in the gastrointestinal tract. The exact etiology and pathogenesis of IBD remain elusive. Although ELF-1 has been known to be highly expressed in epithelial cells for past twenty years, little is known about its function in epithelial cells and epithelial-related IBD. Here, we demonstrated that ELF-1 deficiency in mouse lead to exacerbated DSS-induced colitis, marked by inflammation dominated by neutrophil infiltration and activation of IL-17 signaling pathways in various immune cells, including Th17, ILC3, γδT and NKT cells. Bone marrow transfer experiments confirmed ELF-1 deficiency in non-hematopoietic cells intrinsically worsened DSS-induced colitis. On one hand, ELF-1 deficiency enhanced the production of pro-inflammatory chemokines in colonic epithelial cells, leading to extensive infiltration of neutrophils and other immune cells into the colonic mucosal tissue. On the other hand, ELF-1 directly regulated the expression of the Rack1 gene in colonic epithelial tissue, which has been proved to play critical roles in maintaining intestinal homeostasis. Altogether, ELF-1 plays a protective role in colitis by maintaining intestinal epithelium homeostasis.
Similar content being viewed by others
Introduction
Inflammatory bowel disease (IBD) is a recurrent, chronic immune-mediated inflammatory disorder of the intestine recognized as a prevalent gastrointestinal problem globally1. IBD is characterized by two prominent clinical phenotypes: Crohn’s disease (CD) and ulcerative colitis (UC)2. Despite its high prevalence, the exact etiology of IBD remains inadequately understood due to its complex nature. Current evidence strongly supports that the pathophysiology of IBD arises from intricate interactions among environmental factors, microbial influences, and immune response disorders in genetic susceptible individuals1,3. It is currently believed that IBD results from multiple regulatory mechanisms being disturbed, disrupting intestinal homeostasis and leading to an abnormal innate and/or adaptive immune response to the intrinsic gut flora in genetically susceptible individuals4.
As the first line of defense, the colonic epithelium plays a crucial role in maintaining intestinal homeostasis by facilitating host-microorganism interactions to control mucosal immunity, coordinating nutrient recycling, and forming a mucus barrier. Once this barrier is compromised, a series of immune cells, receptors, and proteins are activated, ultimately promoting the development and progression of IBD5,6. Hence, understanding the complex relationships between the colonic epithelium and the host immune system may help to reveal the pathogenesis of IBD and provide new methods for its prevention and treatment.
ELF-1 (E74-Like Factor 1), a member of E26 transformation specific (ETS) family transcription factors, is most abundantly expressed in lymphoid and hematopoietic tissues7. It plays critical roles in immune system, being implicated in the transcriptional regulation of several T cell-specific molecules, such as CD3ζ8 and IL-2R9. ELF-1 is essential for the development and function of NKT cells, though it has minimal influence on the development of conventional T cells and NK cells10,11. As a key regulator of the hematopoietic system, ELF-1 directly regulates genes crucial for hematopoiesis, including Fli-1, Runx1, Lmo2 and Tie212,13,14,15. Additionally, ELF-1 has been studied as a transcription factor in diverse cancers, such as Hodgkin lymphoma, breast cancer and nasopharyngeal carcinoma16,17,18. Over twenty years ago, ELF-1 was found to be highly expressed in the epithelial cells of the gut and skin7. However, its function in epithelial cells and its role in epithelial-related IBD remain largely unknown.
In this study, we investigated the critical role of the transcription factor ELF-1 in colitis development and described a mechanism by which ELF-1 regulates colitis through maintaining intestinal epithelium homeostasis. We found that ELF-1 deficiency exacerbated DSS-induced colitis. Using single-cell RNA sequencing (scRNA-seq) transcriptional analysis, we characterized the intestinal cell landscape and discovered that colitis exacerbation caused by ELF-1 deficiency was marked by inflammation dominated by neutrophil infiltration and activation of IL-17 signaling pathways in various immune cells, including Th17, ILC3, γδT and NKT cells. Additionally, bone marrow transfer experiments demonstrated that ELF-1 deficiency in non-hematopoietic cells, but not hematopoietic-origin immune cells, intrinsically aggravated DSS-induced colitis. We also found that ELF-1 deficiency increased the production of inflammatory chemokines in colonic epithelial cells, which is supposed to lead to extensive infiltration of neutrophils and other immune cells. Furthermore, Chromatin immunoprecipitation sequencing (ChIP-seq) analysis revealed that ELF-1 could directly regulate the expression of the Rack1 gene, which plays critical roles in maintaining intestinal homeostasis.
Results
ELF-1-deficient mice developed severe DSS colitis
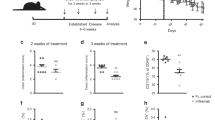
DSS-induced colitis, a commonly used mouse model to simulate the pathological characteristics of IBD19, was employed to investigate the role of ELF-1 in colitis. During the experiment, Elf-1−/− mice exhibited significantly more weight loss in the late phase (Fig. 1A) and a higher mortality rate than WT mice (Fig. 1B). Additionally, Elf-1−/− mice showed significantly higher disease activity index (DAI) scores compared to WT mice (Fig. 1C). The colon length of Elf-1−/− mice was considerably shorter (Fig. 1D), and they exhibited more severe splenomegaly than WT mice (Fig. 1E). Consistently, the colon tissues of Elf-1−/− mice were infiltrated with more inflammatory cells and showed more severe intestinal damage than those of WT mice (Fig. 1F, G). Furthermore, the expressions of inflammatory cytokines (Il1b and Il6) and chemokines (Cxcl1, Cxcl2, Ccl2 and Ccl3), which play critical roles in colitis, were markedly upregulated in the colonic tissues of Elf-1−/− mice (Fig. 1H). These results indicated that ELF-1 deficiency aggravated DSS-induced colitis, demonstrating that ELF-1 plays a protective role in restraining intestinal inflammation.
A The body weight of WT and Elf-1−/− mice under normal condition and DSS treatment (n = 8). The survival rate (B) and DAI scores (C) of WT and Elf-1−/− mice in DSS-induced colitis (n = 8). Colon lengths (D) and spleen weight (E) of WT and Elf-1−/− mice in DSS-induced colitis (n = 8). H&E staining (F) and H&E scores (G) of colon tissues of WT and Elf-1−/− mice under normal conditions and DSS treatment (n = 8). H The mRNA levels of Il1b, Il6, Cxcl1, Cxcl2, Ccl2 and Ccl3 in colonic tissues of WT and Elf-1−/− mice in DSS-induced colitis (n = 3). Data are shown as mean ± SD, representing one of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
ELF-1 deficiency led to an inflammatory condition characterized by predominant by neutrophil infiltration in DSS-induced colitis
To comprehensively explore how ELF-1 deficiency exacerbates colitis, we employed scRNA-seq to profile the colonic tissues of Elf-1−/− and WT mice treated with DSS. After quality filtering, a total of 16,831 cells were analyzed for transcriptional profiling. Based on known cell markers, we identified 12 cell types and states in colonic tissues, encompassing nearly every cell type in the colonic tissue, including fibroblasts, mural cells, endothelial cells, epithelial cells, erythrocytes, T cells and ILCs, B cells, plasma cells, mononuclear phagocytes, neutrophils, basophils and proliferating cells (Fig. 2A, B). To visualize changes in cell composition of the colon during inflammation, we counted the number and proportion of the identified clusters. Notably, ELF-1 deficiency resulted in a substantial increase in the percentage of neutrophils within the identified immune cell populations in the colonic tissues of DSS-induced colitis (Fig. 2A). This finding was further confirmed using fluorescence activated cell sorting (FACS) (Fig. 2C). Meanwhile, Gene Ontology (GO) enrichment analysis showed that the upregulated differentially expressed genes (DEGs) of neutrophils in DSS-treated Elf-1−/− mice were significantly involved in granulocyte migration (Fig. 2D). Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the upregulated DEGs of neutrophils in DSS-treated Elf-1−/− mice were significantly enriched in the IL-17 signaling pathway and TNF signaling pathway (Fig. 3E). The upregulated DEGs in neutrophils from DSS-treated Elf-1−/− mice that are involved in these signaling pathways included S100a8, S100a9, Mmp3, Lcn2, and Tnf (Fig. 2F). Further Uniform Manifold Approximation and Projection (UMAP) clustering analysis of neutrophils yielded six subclusters: neutrophils1 (Ngp+Hadha+Anxa1+), neutrophils2 (Crem+Thbs1+Anxa1+), neutrophils3 (Il23a+Ccl3+Ccl4+), neutrophils4 (Mrpl36+Clec4n+Kdm1a+), neutrophils5 (Ltf+Camp+Hnrnpll+), and neutrophils 6 (Fastk+Ciao1+Ankzf1+) (Fig. 2G, H). Notably, the neutrophils5 and neutrophils6 subclusters were almost exclusively present in colonic tissue of DSS-treated Elf-1−/− mice, but not in WT mice (Fig. 2G). GO enrichment analysis indicated that the upregulated DEGs of neutrophils5 compared to other neutrophil clusters were enriched in regulation of angiogenesis, neutrophil activation, granulocyte activation, and neutrophil degranulation (Fig. 2I). The upregulated DEGs of neutrophils6 compared to other neutrophil clusters were enriched in regulation of actin cytoskeleton organization, regulation of supramolecular fiber organization, protein polymerization, integrin-mediated signaling pathway and glucose-6-phosphate metabolic process (Fig. 2I). Meanwhile, HALLMARK analysis showed that the upregulated DEGs of neutrophils5 and neutrophils6 were co-enriched in pathways including oxidative phosphorylation, adipogenesis, apical junction, reactive oxygen species, allograft rejection, mTORC1, protein secretion and PI3K-AKT-mTOR (Fig. 2J). These results indicated that ELF-1 deficiency caused an inflammation situation dominated by neutrophil infiltration in DSS-induced colitis.
A UMAP plots and proportions of the identified cell types from the colonic tissues of DSS-treated WT and Elf-1−/− mice. B Dotplot of top 3 marker genes for the different cell subsets as indicated. C Percentages of CD11b+Ly6G+ neutrophils in colonic tissues of DSS-treated WT and Elf-1−/− mice (n = 3). GO (D) and KEGG (E) analyses of up-regulated DEGs in neutrophils of DSS-treated Elf-1−/− mice. F Expressions of representative genes of TNF and IL-17 signaling pathways in neutrophils of DSS-treated WT and Elf-1−/− mice. The intensity of expression is indicated as specified by the color legend. G UMAP plots and proportions of identified six subtypes in neutrophils of DSS-treated WT and Elf-1−/− mice. H Tracksplot of top marker genes for the subpopulations of neutrophil as indicated. I GO analysis of up-regulated DEGs in two identified subtypes of neutrophils (neutrophils5 and neutrophils6). J Differences in pathways scored by GSVA among different neutrophils subtypes.
A UMAP plots and proportions of the identified T cell types from the colonic tissues of DSS-treated WT and Elf-1−/− mice. B Heatmap of top marker genes for the subpopulations of T cells and ILCs group as indicated. C Percentages of CD4+IL-17+ Th17 cells and γδ T cells in colonic tissues of DSS-treated WT and Elf-1−/− mice (n = 3). KEGG (D) and GSEA (E) analyses of up-regulated DEGs in Th17, ILC3, NKT and γδT cells of DSS-treated Elf-1−/− mice. F Heatmap of representative genes of IL-17 signaling pathways in Th17, ILC3, NKT and γδT cells of DSS-treated WT and Elf-1−/− mice. The intensity of expression is indicated as specified by the color legend.
The IL-17 signaling pathway plays a vital role in the exacerbation of colitis caused by ELF-1 deficiency
In addition to neutrophils, expanded and dysregulated immune cells, including T cells, γδT cells, and ILCs, also play a critical role in driving gut inflammation20. Further UMAP clustering analysis of the T cells and ILCs group identified 10 subclusters with specific gene expression profiles: CD8+ T cells, γδT cells, Τh17 cells, ILC2 cells, ILC3 cells, NK cells, NKT cells, naïve T cells, Tfh cells, and Tregs (Fig. 3A, B). Notably, the proportions of Th17 cells, γδT cells and ILC3 cells in Elf-1−/− mice were much higher than those in WT mice in DSS-induced colitis (Fig. 3A). Furthermore, FACS assays also confirmed a significant increase in the percentages of Th17 cells and γδ T cells in the colonic tissues of DSS-treated Elf-1−/− mice (Fig. 3C). KEGG pathway and Gene Set Enrichment Analysis (GSEA) analyses revealed that the upregulated DEGs of Th17 cells, ILC3 cells, and NKT cells in DSS-treated Elf-1−/− mice were significantly enriched in the IL-17 signaling pathway (Fig. 3D, E), which is known to play critical roles in the pathogenesis of colitis20,21,22. The upregulated DEGs of Th17 cells, ILC3 cells, γδT cells, and NKT cells in DSS-treated Elf-1−/− mice included genes involved in IL-17 signaling pathway, such as Il17a, Il17f, Lcn2, S100a8, S100a9, Mmp3, Cxcl1, Cxcl2, Cxcl5, and Cebpb (Fig. 3F). These findings suggested that the IL-17 signaling pathway played a vital role in the exacerbation of colitis caused by ELF-1 deficiency.
ELF-1 deficiency in non-hematopoietic cells, but not in hematopoietic-origin immune cells, intrinsically aggravated DSS-induced colitis
To determine whether the exacerbation of colitis due to ELF-1 deficiency is attributable to hematopoietic-origin immune cells or non-hematopoietic cells, we conducted reciprocal bone marrow transfer experiments between WT and Elf-1−/− mice, followed by DSS-induced colitis (Fig. 4A). Compared to WT recipient mice that received either WT or Elf-1−/− bone marrow cells (WT→WT, Elf-1−/−→WT), Elf-1−/− recipient mice that received either WT or Elf-1−/− mice bone marrow cells (WT→Elf-1−/−, Elf-1−/−→Elf-1−/−) exhibited significantly greater body weight loss and higher DAI scores (Fig. 4B, C). Additionally, Elf-1−/− recipient mice that received either WT or Elf-1−/− bone marrow cells (WT→Elf-1−/−, Elf-1−/−→Elf-1−/−) showed significantly shorter colon length and more severe splenomegaly than WT recipient mice that received WT or Elf-1−/− bone marrow cells (WT→WT, Elf-1−/−→WT) (Fig. 4D, E). Furthermore, the colon tissues of Elf-1−/− recipient mice that received WT or Elf-1−/− mice bone marrow cells (WT→Elf-1−/−, Elf-1−/−→Elf-1−/−) exhibited more severe histological inflammatory infiltration and intestinal damage compared to WT recipient mice that received WT or Elf-1−/− bone marrow cells (WT→WT, Elf-1−/−→WT) (Fig. 4F). There were nearly no differences between the two groups of WT recipient mice that received WT or Elf-1−/− bone marrow cells (WT→WT, Elf-1−/−→WT), and between the two groups of Elf-1−/− recipient mice that received WT or Elf-1−/− bone marrow cells (WT→Elf-1−/−, Elf-1−/−→Elf-1−/−) in terms of colitis indicators (Fig. 4B–F). Hence, ELF-1 deficiency aggravates colitis in the DSS-induced colitis model due to its deficiency in non-hematopoietic cells rather than hematopoietic-origin immune cells.
A The schematic diagram of construction of bone marrow chimera. The body weight (B) and DAI scores (C) of irradiated WT and Elf-1−/− mice transferred with WT or Elf-1−/− bone marrow in DSS-induced colitis (n = 8). Colon lengths (D) and spleen weight (E) of irradiated WT and Elf-1−/− mice transferred with WT or Elf-1−/− bone marrow in DSS-induced colitis (n = 8). F H&E staining and H&E scores of irradiated WT and Elf-1−/− mice transferred with WT or Elf-1−/− bone marrow in DSS-induced colitis (n = 8). Data are shown as mean ± SD, representing one of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
ELF-1 deficiency increased the production of inflammatory chemokines in colonic epithelial cells during DSS-induced colitis
Since it has been proven that ELF-1 deficiency exacerbated colitis due to its effects on non-hematopoietic cells, and given that the intestinal epithelium provides a critical physical barrier to prevent direct contact of luminal contents with subepithelial tissues, which is essential for maintaining intestinal homeostasis23, we further investigated the role of intestinal epithelial cells. In the colon tissues of DSS-treated WT and Elf-1−/− mice, epithelial cells were subdivided into six subclusters based on the transcriptional profiling: enterocytes, glial cells, goblet cells, neuroendocrine cells neurons, and transit amplifying cells (Fig. 5A, B). Goblet cells are known for their role in providing the protective mucus barrier that covers the intestine. Dysregulation of goblet cells has been associated with infectious and inflammatory conditions, as well as a predisposition to chronic disease later in life24. Compared to WT mice, the goblet cell subpopulation was absent in the colon tissues of DSS-treated Elf-1−/− mice (Fig. 5A), indicating that ELF-1 deficiency could lead to a severe DSS colitis. Cell-cell communication analyses revealed extensive interactions between epithelial cells and various immune cells through chemokines and their receptors during DSS colitis (Fig. 5C). Consistently, KEGG analyses showed that DEGs upregulated in colonic epithelial cells of DSS-treated Elf-1−/− mice were highly enriched in the cytokine-cytokine receptor interaction pathway, including many inflammatory chemokines such as Ccl2, Cxcl1, Cxcl2, Cxcl5, and Cxcl13 (Fig. 5D, E). The expression levels of these inflammatory chemokines and cytokines (Cxcl13, Ccl2, Cxcl2, Cxcl1, Ccl11, Ccl3 and Il1b) were significantly higher in the colonic epithelial tissues of DSS-treated Elf-1−/− mice compared to DSS-treated WT mice (Fig. 5F). Therefore, it is likely that ELF-1 deficiency increased the production of inflammatory chemokines in colonic epithelial cells, leading to an inflammatory state characterized by extensive infiltration of neutrophils and other immune cells.
A UMAP plots and proportions of the identified subtypes in colonic epithelial cells from the colonic tissues of DSS-treated WT and Elf-1−/− mice. B Tracksplot of top 3 marker genes for the subpopulations of colonic epithelial cells as indicated. C Ligand‒receptor interaction between colonic epithelial cell and immune cell types. D KEGG analysis of up-regulated DEGs in colonic epithelial cells of DSS-treated Elf-1−/− mice. E Expressions of representative genes of cytokine-cytokine receptor interaction signaling pathways in colonic epithelial cells of DSS-treated WT and Elf-1−/− mice. F The mRNA levels of Cxcl13, Cxcl2, Cxcl1, Ccl2, Ccl11, Ccl3 and Il1b in colonic epithelial tissues of normal and DSS-treated WT and Elf-1−/− mice (n = 4). Data are shown as mean ± SD, representing one of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
ELF-1 directly regulates the expression of Rack1 in DSS-induced colitis
Given that ELF-1 is a transcription factor, we performed ChIP-seq to identify the genes directly regulated by ELF-1 may play critical roles in DSS-induced colitis. ChIP-seq analysis revealed a total of 2391 peaks, with 5.23% were located at gene promoter regions (Fig. 6A). GO analysis of ELF-1-binding genes at promoter regions indicated they were primarily involved in biological adhesion, cell-cell adhesion and receptor tyrosine kinase binding (Fig. 6B). By integrating ChIP-seq data with scRNA-seq data, we identified Rack1 as the only one overlapping gene between ELF-1-binding genes (at promoter regions) and downregulated DEGs in epithelial cells of Elf-1−/− mice (Fig. 6C, D). ELF-1-binding peaks in the Rack1 gene promoter region were significantly enriched in GATA-rich consensus motifs (Fig. 6E). Additionally, Rack1 expressions were significantly downregulated in epithelial cells and colonic epithelial tissues of Elf-1−/− mice compared to WT mice (Fig. 6F, G). Given that RACK1 plays critical roles in maintaining intestinal homeostasis by protecting the integrity of the epithelial barrier25,26, so it is likely that ELF-1 protects against colitis by directly regulating the expression of the Rack1 gene.
A Distribution of ELF-1 binding sites from ChIP-Seq analysis of colonic tissues of WT mice. B KEGG pathway enrichment analysis of ELF-1 binding genes (binding peaks located in gene promoter regions). C Venn diagram showing overlapping gene between ELF-1 binding genes with down regulated DEGs in colonic epithelial cells of DSS-treated Elf-1−/− mice. D ChIP-seq peak for ELF-1 binding site in Rack1 gene. E Top 2 predicted ELF-1 binding motifs in Rack1 gene promoter with the most significant P values. F Rack1 expressions in colonic epithelial cells of DSS-treated WT and Elf-1−/− mice. G Rack1 expressions in colonic epithelial tissues of normal and DSS-treated WT and Elf-1−/− mice (n = 4). Data are shown as mean ± SD, representing one of at least three independent experiments. **P < 0.01.
Discussion
IBD is a chronic, relapsing, and remitting disease characterized by chronic inflammation in the gastrointestinal tract1. The exact etiology and pathogenesis of IBD remain elusive. ELF-1, an ETS family transcription factor, has been widely recognized for its role the transcriptional regulation of genes crucial for the immune and hematopoietic systems7. Although ELF-1 has been known to be highly expressed in epithelial cells for two decades, its functions in epithelial cells and epithelial-related IBD have been unclear. In the present study, we identified ELF-1 as a pivotal protective factor in IBD pathogenesis by maintaining intestinal homeostasis, suggesting that ELF-1 may serve as a novel risk gene for IBD.
DSS-induced colitis, which presents an inflammatory reaction morphologically and symptomatically similar to that observed in ulcerative colitis in humans27, was uesed to study the role of ELF-1 in IBD. Notably, we found that ELF-1 deficiency significantly exacerbated the symptoms of IBD in this model. Furthermore, the exacerbation of colitis due to ELF-1 deficiency was characterized by an inflammation condition dominated by neutrophil infiltration.
Neutrophils play a central role in the development and maintenance of intestinal inflammation. Once the epithelial barrier is disrupted, neutrophils are the first immune cells recruited to the site of inflammation. They recognize, phagocytose, and kill pathogens by producing reactive oxygen species with antimicrobial potential, releasing lytic enzymes from their granules, and liberating neutrophil extracellular traps28. Additionally, cytokines and mediators produced by neutrophils trigger feedback amplification, further promoting the recruitment and activation of neutrophils and other immune cells, leading to significant tissue damage and the progression toward chronic disease29. Neutrophil migration into the colon mucosa is considered a hallmark of inflammatory bowel diseases, especially in UC30. Bressenot et al. demonstrated that neutrophil infiltration was associated with disease severity31. The mucosal tissues of UC patients were characterized by an accumulation of immune cells, particularly mononuclear cells and neutrophils, which are associated with architectural distortion of tissue, crypt destruction and crypt abscess formation32. Increased neutrophils migration in the local colon could destroy junction proteins, such as E-cadherin and a junctional adhesion molecule-like factor, impairing the epithelial barrier and exacerbating intestinal inflammation in IBD33. Neutrophil depletion using anti-neutrophil antibodies has been shown to ameliorate DSS-induced colitis34. Therefore, the massive infiltration of neutrophils in the colonic mucosa may be one cause of colitis aggravation in Elf-1−/− mice. Furthermore, ELF-1 deficiency enhanced the activation of TNF signaling pathways and TNF-α production in neutrophils. Neutrophil-derived pro-inflammatory TNF-α played a critical role in IBD by increasing the recruitment and adhesion of leukocytes to vascular endothelial cells35. In IBD patients, TNF-α levels were elevated in the mucosa, and anti-TNF-α therapy is currently in used36.
It has been widely demonstrated that IL-17 and IL-17 signaling pathway play critical roles in IBD. The IL-17 family consists of six molecules, from IL-17A to IL-17F, with IL-17 being the key cytokine of Th17 cells. IL-17 can also be produced by various innate immune cells, including γδT, NKT, and ILC3 cells20,21,22. Dysregulation of the IL-23/IL-17 axis was associated with various genetic susceptibilities in CD and UC patients, due to dysfunctions in innate and adaptive immune responses37. As a pro-inflammatory cytokine, IL-17 promotes the expression of matrix metalloproteinases and proinflammatory cytokines such as IL-6, CXCL1, CXCL2, CCL20, and CSF3, which lead to neutrophil infiltration and tissue inflammation22,38. Consistently, we observed an increased infiltration of Th17, γδT and ILC3 cells in the colonic tissues of DSS-treated Elf-1−/− mice. Additionally, we found that the exacerbation of colitis due to ELF-1 deficiency was characterized by enhanced production of IL-17 and activation of IL-17 signaling pathways in various immune cells, including Th17, ILC3, γδT, and NKT cells. These findings suggest enhanced IL-17 production and activation of IL-17 signaling pathways contribute to the aggravation of colitis in DSS-treated Elf-1−/− mice.
Bone marrow chimeras experiments were constructed to further investigate whether the exacerbation of colitis due to ELF-1 deficiency is attributed to hematopoietic-origin immune cells or non-hematopoietic cells. The results revealed that ELF-1 deficiency in non-hematopoietic cells intrinsically aggravated DSS-induced colitis. Intestinal homeostasis is maintained by intestinal barrier, with the intestinal epithelium forming a protective physical barrier that separates intestinal contents from host tissues, regulates nutrient absorption, and allows interactions between resident bacterial flora and the mucosal immune system39. The current understanding of IBD etiology involves the interplay of host genetics, host immunity, gut microbiota, and environmental exposures40. The intestinal epithelium, as the interface between these factors, is crucial in governing these interactions41. An array of pro-inflammatory chemokines, such as CXCL8, CXCL1, CXCL5, CCL2, CCL3, and CCL5, have been found to be elevated in the intestinal mucosa of IBD patients resulting in increased recruitment and localization of neutrophils, monocytes, dendritic cells, natural killer cells, and T lymphocytes to the gut mucosa42,43. CXCL1, CXCL2, and CXCL5, which are ligands for the chemokine receptor CXCR2, play a vital role in the recruitment of neutrophils during inflammation. Blocking CXCR2 with its inhibitor SB225002 has been shown to markedly ameliorate DSS-induced colitis in mice44. Intestinal epithelial cell-derived CXCL5 also participated in the colonic epithelial cell inflammatory response by enabling neutrophil recruitment in UC, and BATF3 promoted colitis-associated colon cancer through facilitating CXCL5-mediated neutrophil recruitment in the intestinal mucosa45,46. Meanwhile, CCL5 could induce the massive recruitment of a variety of innate and adaptive immune cells carrying CCR1, CCR3, CCR4, and CCR547. Elinav et al. found that gut microbiota from NLRP6 inflammasome-deficient mice induced CCL5 production and immune cell recruitment, leading to spontaneous inflammation48. Consistently, we observed marked increase in the production of these pro-inflammatory chemokines in the colonic epithelial cells of DSS-treated Elf-1−/− mice. Additionally, scRNA-seq data showed a marked increase in the fibroblast population in DSS-treated Elf-1−/− mice. However, the effects of fibroblasts in IBD seem to manifest in the later stages, and the increased fibroblast population in DSS-treated Elf-1−/− mice was likely a consequence of DSS colitis rather than a cause of its onset. The role of intestinal fibroblasts in IBD has primarily been studied concerning their involovement in extracellular matrix (ECM) remodeling, particularly through the expression of matrix metalloproteinases (MMPs)49. Fibroblasts and myofibroblasts are the primary sources of excessive ECM, which accumulates in a time-dependent manner50. Excessive ECM deposition is a hallmark feature of IBD51, leading to tissue stiffness and functional impairment52. Imbalance in expression of MMPs and tissue inhibitors of metalloproteinases, along with dysregulation in the production and degradation of ECM proteins under chronic inflammatory conditions may lead to the development of intestinal fibrosis53,54. Thus, Fibroblasts are regarded as the main effector cells in the pathophysiology of Intestinal fibrosis55, which is recognized as a late-stage phenotype of IBD56. Therefore, ELF-1 deficiency enhanced the production of pro-inflammatory chemokines in colonic epithelial tissues was suspected to lead to the inflammation state characterized by extensive infiltration of neutrophils and other immune cells in DSS-treated Elf-1−/− mice.
Additionally, the Rack1 gene was identified as a target gene of ELF-1, with its expression downregulated in the colonic epithelial tissues and cells of DSS-treated Elf-1−/− mice. Cheng et al. demonstrated that RACK1 deficiency in intestinal epithelium promoted the proliferation of crypt cells and hindered their differentiation into enterocytes, goblet cells, and enteroendocrine cells25. Furthermore, they established that RACK1 plays a protective role in maintaining the integrity of the intestinal epithelium. Mice with RACK1 deficiency in their intestinal epithelium, including the small intestine and colon, developed inflammatory enterocolitis characterized by denuded and sloughed-off surface epithelium and crypt hyperplasia26. RACK1 has also been shown to promote epithelial cell-cell adhesion by stabilizing E-cadherin and catenins at cell-cell contacts57. Therefore, the exacerbation of colitis due to ELF-1 deficiency is partly mediated through its direct regulation of the Rack1 gene, which is crucial for maintaining intestinal homeostasis.
In summary, our data provide direct evidence that the transcription factor ELF-1 is a novel protective regulator in colitis development and elucidate how ELF-1 maintains intestinal epithelial homeostasis to regulate colitis. On one hand, ELF-1 deficiency increases the production of pro-inflammatory chemokines and cytokines in colonic epithelial cells, leading to an inflammation state characterized by extensive infiltration of neutrophils and other immune cells, as well as activation of IL-17 signaling pathways in Th17, ILC3, γδT and NKT cells during DSS colitis. On the other hand, ELF-1 directly regulates the expression of the Rack1 gene, which is critical for maintaining intestinal homeostasis in colonic epithelial cells. Our results suggest that ELF-1 plays a crucial protective role in colitis by maintaining intestinal homeostasis, and it may be a potential candidate risk gene for colitis.
Materials and methods
Mice
Elf-1−/− mice (Saiye, Guangzhou, China) were crossed with C57BL/6 J (B6) (Huafukang Bioscience, Beijing, China). Sex-matched littermate WT and Elf-1−/− mice of 8 to 10-week were used for experiments. All mice were bred and maintained under specific pathogen-free condition in the animal facility of Zhengzhou University. All animal studies were approved by the Animal Ethics Committee of Zhengzhou University (Zhengzhou, China). We have complied with all relevant ethical regulations for animal use.
DSS colitis
Mice were fed with 2.5% (w/v) DSS (M.W. = 36,000–50,000 Da; MP Biomedicals) dissolved in drinking water for 7 or 8 consecutive days to induce DSS colitis. Body weight, and DAI were assessed daily. DAI scores were calculated according to weight loss, stool consistency, and stool blood content/rectal bleeding, details are as follows: Score = 0, Weight loss: none, Stool consistency: normal, Blood in stool: none; Score = 1, Weight loss: 1–5%, Stool consistency: loose stools, Blood in stools: present; Score = 2, Weight loss: 5–10%, Stool consistency: watery diarrhea, Blood in stools: present; Score = 3, Weight loss: 10–20%, Stool consistency: slimy diarrhea, little blood, Blood in stools: present; Score = 4, Weight loss: > 20%, Stool consistency: severe watery diarrhea with blood, Blood in stools: gross bleeding. The colon tissue was harvested for scRNA-seq, histology and RT-qPCR.
Histopathology
Colon tissues were fixed in 4% paraformaldehyde and then embedded in paraffin. Subsequently, 5 μm sections were stained with hematoxylin and eosin (H&E). Histological sections were scored as follows58,59: Epithelium: normal morphology (0), loss of goblet cells (1), loss of goblet cells in large areas (2), loss of crypts (3), and loss of crypts in large areas (4); and infiltration: no infiltrate (0), infiltrate around crypts (1), infiltrate reaching the lamina muscularis mucosae (2), extensive infiltration reaching the lamina muscularis mucosae and thickening of the mucosa (3), and infiltration of the submucosal layer (4). A total histological score equals to the sum of both scores.
Single-cell preparation and flow cytometry assay
For the isolation of colon lamina propria, residual mesenteric fat tissue and Peyer’s patches were removed, and the colon was then opened longitudinally. The colon was thoroughly washed with ice-cold PBS and cut into 0.5 - 1 cm pieces. These pieces were incubated in 3 ml PBS containing 5 mM EDTA for 20 min at 37 °C with shaking at 220 rpm. Following intensive vortexing, the pieces were collected and washed once with complete RPMI medium. The tissue was then shredded and digested in complete RPMI medium containing DNase I (40 μg/mL, ACMEC) and collagenase IV (1 mg/mL, BioFroxx) at 37 °C for 1 h. For the analysis of surface markers, prepared single cell suspension (100 μL, 5 × 106 cells/ml) was stained with anti-mouse CD11b (M1/70, Biolegend), anti-mouse Ly6G (1A8, Biolegend), anti-mouse CD45 (30-F11, Tonbo), anti-mouse CD4 (RM4-5, Biolegend), anti-mouse TCR γ/δ(GL3, Biolegend). Intracellular cytokine staining was conducted using the Fixation/Permeabilization buffer solution in accordance to the manufacturer’s instructions (Invitrogen). Fluorochrome-conjugated anti-mouse IL-17A (Miltenyi) was employed for staining. The stained cells were analyzed using FACSCelesta (BD Biosciences), and the acquired data were analyzed with FlowJo_V10 software.
RT-qPCR
Total RNAs were extracted from colonic tissues or colonic epithelial tissues by using total RNA kit (Promega) followed by cDNA synthesis with cDNA Reverse Transcription Kit (Takara Bio). RT-qPCR was conducted on the QuantStudio 7 Flex System (Applied Biosystems) by using Power SYBR PCR Master Mix (Applied Biosystems). Every sample was run in triplicates. The primers used are listed in Table 1. RT-qPCR data were analyzed by using the comparative cycle threshold method and were normalized by using the housekeeping gene Hprt or Gapdh60.
Bone marrow transplantation
Eight to 10-week WT and Elf-1−/− mice were used as donor of bone marrows cells. Bone marrow was extracted from the femurs of hind legs. After red blood cell lysis and strained through cell strainer (40 µm), cell pellets were washed three times with sterile PBS, counted and stored on ice for later injection. Recipient WT and Elf-1−/− mice (8 to 10-week) were sublethally irradiated with 9 Gy and received 1 × 107 bone marrow cells prepared above 4 hours later. Chow feed for 8 weeks to reconstitute the immune system and then processed for DSS treatment.
scRNA-seq and data analysis preparation of single cell suspensions from colon tissue
For isolation of colon cells, proximal, distal colon, and rectum were collected from DSS-treated WT and Elf-1−/− mice. Three biological replicates were pooled in each group. Intestines were opened longitudinally and washed to remove fecal content with cold PBS, then placed into the GEXSCOPE Tissue Preservation Solution (Singleron Biotechnologies) and transported at 2–8 °C. All tissues were cut into small pieces (1–2 mm3) and digested for 15 min at 37 °C in 2 mL GEXSCOPE Tissue Dissociation Solution (Singleron Biotechnologies). Subsequently, cells were passed through a 40-μm cell strainer and washed in PBS. Library preparation: Colonic cells from three DSS-treated WT mice were mixed and pooled as one sample/group designated as WT-DSS, and colonic cells from three DSS-treated Elf-1−/− mice were mixed and pooled as one sample/group designated as Elf-1−/−-DSS for scRNA-seq. Cells were loaded into GEXSCOPE Single Cell Microchip (Singleron) and scRNA-seq libraries were constructed according to protocol from GEXSCOPE Single-Cell RNA Library Kit (Singleron). The libraries were finally sequenced with 150 bp paired end on the Illumina Novaseq 6000 platform with a sequencing depth of at least 100,000 reads per cell. Quality control, dimension-reduction and clustering: Scanpy v1.8.2 was used for quality control, dimensionality reduction and clustering under Python 3.7. For each sample dataset, we filtered expression matrix by the following criteria: 1) cells with gene count less than 200 or with top 2% gene count were excluded; 2) cells with top 2% UMI count were excluded; 3) cells with mitochondrial content > 20% were excluded; 4) genes expressed in less than 5 cells were excluded. After filtering, 16831 cells were retained for the downstream analyses, with on average 1175 genes and 3424 UMIs per cell. The raw count matrix was normalized by total counts per cell and logarithmically transformed into normalized data matrix. Top 2000 variable genes were selected by setting flavor = ‘seurat’. Principal component analysis was performed on the scaled variable gene matrix, and top 12 principal components were used for clustering and dimensional reduction. Cells were classified into 12 clusters using Louvain algorithm with a resolution parameter of 1.2 and were subsequently manually annotated with specific cell types based on specific cell markers (Supplementary Table 1). Within the annotated clusters of T cells and innate lymphoid cells (ILCs), cells were further divided into 10 subclusters using the same Louvain algorithm (resolution parameter: 1.2) and were annotated according to specific cell markers (Supplementary Table 2). In the annotated epithelial cells cluster, cells were categorized into 6 subclusters using the Louvain algorithm (resolution parameter: 1.2) and annotated as specific epithelial subtypes based on designated cell markers (Supplementary Table 3).
Cell clusters were visualized by using UMAP. DEGs analysis: To identify DEGs, we used the scanpy.tl.rank_genes_groups function based on Wilcoxon rank sum test with default parameters, and selected the genes expressed in more than 10% of the cells in either of the compared groups of cells and with an average log (Fold Change) value greater than 0.25 as DEGs. Adjusted p value was calculated by benjamini-hochberg correction and the value 0.05 was used as the criterion to evaluate the statistical significance. Pathway enrichment analysis: To investigate the potential functions of identified cell types. GO, KEGG and GSEA analyses were used with the “clusterProfiler” R package v 3.16.1. Pathways with p_adj value less than 0.05 were considered as significantly enriched. Selected significant pathways were plotted as bar plots. Hallmark gene sets were obtained from the Molecular Signature Database (http://gsea-msigdb.org). Finally, the limma R package was used to determine the p values. Statistical significance was defined as p value < 0.05. Cell-cell interaction analysis: Receptor-ligand interactions analysis between cell clusters was performed using CellphoneDB60.
ChIP-seq and data analysis
Colon tissues from DSS-treated WT mice were obtained to perform ChIP-seq assay. The ChIP-seq assay was performed by Wuhan Igenebook Biotechnology Co. Ltd (China). Briefly, 2 g tissue was washed twice in cold PBS buffer and cross-linked with 1% formaldehyde for 10 minutes at room temperature and then quenched by addition of glycine (125 mmol/L final concentration). Afterwards, samples were lysed with 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS, 1× protease inhibitor cocktail, and chromatins were obtained on ice. Chromatins were sonicated to get soluble sheared chromatin (average DNA length of 200-500 bp). 20 uL chromatin was saved at −20 °C for input DNA, and 100ul chromatin was used for immunoprecipitation by anti-mouse ELF-1 antibodies (sc-133096 X, Santa Cruz). 10 μg of antibody was used in the immunoprecipitation reactions at 4 °C overnight. The next day, 30 μL of protein beads was added and the samples were further incubated for 3 h. The beads were next washed once with 20 mM Tris/HCL (pH 8.1), 50 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS; twice with 10 mM Tris/HCL (pH 8.1), 250 mM LiCl, 1 mM EDTA, 1% NP-40, 1% deoxycholic acid; and twice with 1× TE buffer (10 mM Tris-Cl at pH 7.5, 1 mM EDTA). Bound material was then eluted from the beads in 300 μL of elution buffer (100 mM NaHCO3, 1% SDS), treated first with RNase A (8 μg/mL) during 6 h at 65 °C and then with proteinase K (345 μg/mL) overnight at 45 °C. Immunoprecipitated DNA was used to construct sequencing libraries following the protocol provided by the I NEXTFLEX ChIP-seq Library Prep Kit for Illumina Sequencing (NOVA-5143-02, Bio Scientific) and sequenced on Illumina Novaseq 6000 with PE 150 method.
Two biological replicates were performed in ChIP-seq analysis. Trimmomatic (version 0.36) was used to filter out low-quality reads. Clean reads were mapped to the GRCm39 genome by Bwa (version 0.7.15). Samtools (version 1.3.1) was used to remove potential PCR duplicates. MACS2 software (version 2.1.1.20160309) was used to call peaks by default parameters (bandwidth, 300 bp; model fold, 5, 50; q value, 0.05). If the midpoint of a peak located closest to the TSS of one gene, the peak will be assigned to that gene. HOMER (version3) was used to predict motif occurrence within peaks with default settings for a maximum motif length of 12 base pairs. ClusterProfiler in R package was employed to perform GO analysis. The GO analysis was calculated using hypergeometric distribution with a q value cutoff of 0.05.
Statistics and reproducibility
At least three independent experiments were carried out unless otherwise stated. All data were presented as mean ± SD, sample sizes and number of biological replicates are given in figures and figure legends. Unpaired two-tailed Student’s t-test was used for data from two groups. Data from more than two groups were subjected to a one-way analysis of variance with GraphPad Prism v10.0. Comparisons were considered statistically significant when the P value was less than 0.05. * P < 0.05, ** P < 0.01, *** P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors confirm that all relevant data are included in the paper and/or its supplementary information files. The scRNA-seq and ChIP-seq data has been submitted to the GEO database (GSE269385 and GSE289183). Numerical source data for all graphs in the manuscript can be found in Supplementary data 1 file.
References
de Souza, H. S. P., Fiocchi, C. & Iliopoulos, D. The IBD interactome: an integrated view of aetiology, pathogenesis and therapy. Nat. Rev. Gastroenterol. Hepatol. 14, 739–749 (2017).
Flynn, S. & Eisenstein, S. Inflammatory Bowel Disease Presentation and Diagnosis. Surg. Clin. North Am. 99, 1051–1062 (2019).
de Souza, H. S. & Fiocchi, C. Immunopathogenesis of IBD: current state of the art. Nat. Rev. Gastroenterol. Hepatol. 13, 13–27 (2016).
Mayer, L. Evolving paradigms in the pathogenesis of IBD. J. Gastroenterol. 45, 9–16 (2010).
Parikh, K. et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 567, 49–4 (2019).
Hayashi, S. Unraveling the Pathogenesis of Inflammatory Bowel Disease and Search for New Therapeutic Medicines. Yakugaku Zasshi 140, 123–128 (2020).
Bassuk, A. G., Barton, K. P., Anandappa, R. T., Lu, M. M. & Leiden, J. M. Expression pattern of the Ets-related transcription factor Elf-1. Mol. Med. 4, 392–401 (1998).
Juang, Y. T. et al. PP2A dephosphorylates Elf-1 and determines the expression of CD3ζ and FcRγ in human systemic lupus erythematosus T cells. J. Immunol. 181, 3658–3664 (2008).
Serdobova, I. et al. Elf-1 contributes to the function of the complex interleukin (IL)-2-responsive enhancer in the mouse IL-2 receptor alpha gene. J. Exp. Med. 185, 1211–1221 (1997).
Garrett-Sinha, L. A., Dahl, R., Rao, S., Barton, K. P. & Simon, M. C. PU.1 exhibits partial functional redundancy with Spi-B, but not with Ets-1 or Elf-1. Blood 97, 2908–2912 (2001).
Choi, H. J. et al. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 117, 1880–1887 (2011).
Landry, J. R. et al. Fli1, Elf1, and Ets1 regulate the proximal promoter of the gene in endothelial cells. Blood 106, 2680–2687 (2005).
Nottingham, W. T. et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110, 4188–4197 (2007).
Nowling, T. K., Fulton, J. D., Chike-Harris, K. & Gilkeson, G. S. Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes. Mol. Immunol. 45, 1–12 (2008).
Dube, A. et al. ELF-1 is a transcriptional regulator of the Tie2 gene during vascular development. Circ. Res. 88, 237–244 (2001).
Paczkowska, J. et al. Expression of ELF1, a lymphoid ETS domain-containing transcription factor, is recurrently lost in classical Hodgkin lymphoma. Brit J. Haematol. 185, 79–88 (2019).
Gerloff, A., Dittmer, A., Oerlecke, I., Holzhausen, H. J. & Dittmer, J. Protein expression of the Ets transcription factor Elf-1 in breast cancer cells is negatively correlated with histological grading, but not with clinical outcome. Oncol. Rep. 26, 1121–1125 (2011).
Chen, C. H., Su, L. J., Tsai, H. T. & Hwang, C. F. ELF-1 expression in nasopharyngeal carcinoma facilitates proliferation and metastasis of cancer cells via modulation of CCL2/CCR2 signaling. Cancer Manag Res. 11, 5243–5254 (2019).
Chassaing, B., Aitken, J. D., Malleshappa, M. & Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Prot. Immunol. 104, 15 25 11–15 25 14 (2014).
Cua, D. J. & Tato, C. M. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 10, 479–489 (2010).
Sutton, C. E. et al. Interleukin-1 and IL-23 Induce Innate IL-17 Production from γδ T Cells, Amplifying Th17 Responses and Autoimmunity. Immunity 31, 331–341 (2009).
Zeng B. N. et al. ILC3 function as a double-edged sword in inflammatory bowel diseases. Cell Death Dis. 10, 315 (2019).
Soderholm, A. T. & Pedicord, V. A. Intestinal epithelial cells: at the interface of the microbiota and mucosal immunity. Immunology 158, 267–280 (2019).
Gustafsson, J. K. & Johansson, M. E. V. The role of goblet cells and mucus in intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 19, 785–803 (2022).
Cheng, Z. F., Pai, R. K. & Cartwright, C. A. Rack1 function in intestinal epithelia: regulating crypt cell proliferation and regeneration and promoting differentiation and apoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G1–G13 (2018).
Cheng, Z. F. & Cartwright, C. A. Rack1 maintains intestinal homeostasis by protecting the integrity of the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 314, G263–G274 (2018).
Wéra, O., Lancellotti, P. & Oury, C. The Dual Role of Neutrophils in Inflammatory Bowel Diseases. J. Clin. Med. 5, 118 (2016).
Biasi, F., Leonarduzzi, G., Oteiza, P. I. & Poli, G. Inflammatory Bowel Disease: Mechanisms, Redox Considerations, and Therapeutic Targets. Antioxid. Redox Sign 19, 1711–1747 (2013).
Németh, T. & Mócsai, A. Feedback Amplification of Neutrophil Function. Trends Immunol. 37, 412–424 (2016).
Muthas, D. et al. Neutrophils in ulcerative colitis: a review of selected biomarkers and their potential therapeutic implications. Scand. J. Gastroentero 52, 125–135 (2017).
Bressenot, A. et al. Comparing histological activity indexes in UC. Gut 64, 1412–1418 (2015).
Friedrich, M., Pohin, M. & Powrie, F. Cytokine Networks in the Pathophysiology of Inflammatory Bowel Disease. Immunity 50, 992–1006 (2019).
Fournier, B. M. & Parkos, C. A. The role of neutrophils during intestinal inflammation. Mucosal Immunol. 5, 354–366 (2012).
Natsui, M. et al. Selective depletion of neutrophils by a monoclonal antibody, RP-3, suppresses dextran sulphate sodium-induced colitis in rats. J. Gastroen Hepatol. 12, 801–808 (1997).
Danese, S. & Fiocchi, C. MEDICAL PROGRESS Ulcerative Colitis. N. Engl. J. Med. 365, 1713–1725 (2011).
Neurath, M. F. Strategies for targeting cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 24, 559–576 (2024).
Moschen, A. R., Tilg, H. & Raine, T. IL-12, IL-23 and IL-17 in IBD: immunobiology and therapeutic targeting. Nat. Rev. Gastro Hepat. 16, 185–196 (2019).
Iwakura, Y., Ishigame, H., Saijo, S. & Nakae, S. Functional Specialization of Interleukin-17 Family Members. Immunity 34, 149–162 (2011).
Roda, G. et al. Intestinal epithelial cells in inflammatory bowel diseases. World J. Gastroentero 16, 4264–4271 (2010).
Plichta, D. R., Graham, D. B., Subramanian, S. & Xavier, R. J. Therapeutic Opportunities in Inflammatory Bowel Disease: Mechanistic Dissection of Host-Microbiome Relationships. Cell 178, 1041–1056 (2019).
Okamoto, R. & Watanabe, M. Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. J. Gastroenterol. 51, 11–21 (2016).
Zimmerman, N. P., Vongsa, R. A., Wendt, M. K. & Dwinell, M. B. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm. bowel Dis. 14, 1000–1011 (2008).
Egesten, A. et al. The proinflammatory CXC-chemokines GRO-α/CXCL1 and MIG/CXCL9 are concomitantly expressed in ulcerative colitis and decrease during treatment with topical corticosteroids. Int J. Colorectal Dis. 22, 1421–1427 (2007).
Zhu, F. Q. et al. Blockade of CXCR2 suppresses proinflammatory activities of neutrophils in ulcerative colitis. Am. J. Transl. Res. 12, 5237–5251 (2020).
Koukos, G. et al. A MicroRNA Signature in Pediatric Ulcerative Colitis: Deregulation of the miR-4284/CXCL5 Pathway in the Intestinal Epithelium. Inflamm. bowel Dis. 21, 996–1005 (2015).
Lin, Y. et al. Intestinal epithelium-derived BATF3 promotes colitis-associated colon cancer through facilitating CXCL5-mediated neutrophils recruitment. Mucosal Immunol. 14, 187–198 (2021).
Mantovani, A. et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686 (2004).
Elinav, E. et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell 145, 745–757 (2011).
Roulis, M. & Flavell, R. A. Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation 92, 116–131 (2016).
Rieder, F., Mukherjee, P. K., Massey, W. J., Wang, Y. & Fiocchi, C. Fibrosis in IBD: from pathogenesis to therapeutic targets. Gut 73, 854–866 (2024).
Prockop, D. J. Inflammation, fibrosis, and modulation of the process by mesenchymal stem/stromal cells. Matrix Biol. 51, 7–13 (2016).
Li, J. et al. Pathogenesis of fibrostenosing Crohn’s disease. Transl. Res. 209, 39–54 (2019).
Mortensen, J. H. et al. The intestinal tissue homeostasis ? the role of extracellular matrix remodeling in inflammatory bowel disease. Expert Rev. Gastroent 13, 977–993 (2019).
Latella, G., Di Gregorio, J., Flati, V., Rieder, F. & Lawrance, I. C. Mechanisms of initiation and progression of intestinal fibrosis in IBD. Scand. J. Gastroenterol. 50, 53–65 (2015).
Rieder, F. & Fiocchi, C. Intestinal fibrosis in IBD–a dynamic, multifactorial process. Nat. Rev. Gastroenterol. Hepatol. 6, 228–235 (2009).
Bamias, G., Pizarro, T. T. & Cominelli, F. Immunological Regulation of Intestinal Fibrosis in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 28, 337–349 (2022).
Swaminathan, G. & Cartwright, C. A. Rack1 promotes epithelial cell-cell adhesion by regulating E-cadherin endocytosis. Oncogene 31, 376–389 (2012).
Wang Y. S. et al. Ring1a protects against colitis through regulating mucosal immune system and colonic microbial ecology. Gut Microbes 15, 2251646 (2023).
Hu, X. et al. Phosphatase Wip1 Masters IL-17-producing Neutrophil-mediated Colitis in Mice. Inflamm. bowel Dis. 22, 1316–1325 (2016).
Wang, P. et al. The amino acid sensor general control nonderepressible 2 (GCN2) controls TH9 cells and allergic airway inflammation. J. Allergy Clin. Immun. 144, 1091–1105 (2019).
Acknowledgements
This work is supported by the Natural Science Foundation of Henan Province (232300421180), the National Natural Science Foundation of China (811900024), Key scientific research projects of colleges and universities in Henan Province (23A310024, 20A180030), China Postdoctoral Science Foundation (2019M662542, 2020T130606), Young Talent Lift Engineering Project in Henan Province (2020HYTP057), Scientific and technological research projects in Henan Province (212102310747) and Zhongyuan Thousand Talents Program (ZYQR201912186).
Author information
Authors and Affiliations
Contributions
G.H. and X.X. carried out the experiments and analyzed data; P.L. carried out the scRNA-seq experiments, analyzed data, and uploaded data; M.Z. carried out the RT-qPCR experiments; H.Z. (Hongxia Zhang), K.Y., Y.L., Q.Y., and J.Y. breed mice and genotyped the genetically modified mice; Q.L. and H.Z. (Huaixin Zheng) revised the manuscript; P.W. conceived and designed the study, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Joao Valente. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
He, G., Liu, P., Xuan, X. et al. Transcription factor ELF-1 protects against colitis by maintaining intestinal epithelium homeostasis. Commun Biol 8, 395 (2025). https://doi.org/10.1038/s42003-025-07742-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07742-4