Abstract
Macrophages are essential immune cells in all tissues and are vital for maintaining tissue homeostasis, immune surveillance, and immune responses. Considerable efforts have identified shared and tissue-specific gene programs for macrophages across organs during homeostasis. This information has dramatically enhanced the understanding of tissue-restricted macrophage programming and function. However, few studies have addressed the overlapping and tissue-specific responses of macrophage subsets following inflammation. One subset of macrophages observed across several studies, lipid-associated macrophages (LAMs), have gained interest due to their unique role in lipid metabolism and potential as a therapeutic target. LAMs are associated with regulating disease outcomes in metabolically related disorders including atherosclerosis, obesity, and metabolic dysfunction-associated steatotic liver disease. We utilized single-cell RNA sequencing datasets to profile LAM diversity across multiple tissues and inflammatory conditions in mice and humans, to define a shared LAM transcriptional profile, including Trem2 and Lpl, and sets of tissue-specific gene programs. Importantly, LAM markers were highly conserved with human LAM populations that emerge in inflammation. Overall, this analysis provides a detailed transcriptional landscape of tissue-restricted and shared LAM gene programs, data that may help instruct appropriate molecular targets for broad or tissue-restricted therapeutic interventions to modulate LAM populations in disease.
Similar content being viewed by others
Introduction
Macrophages are phagocytic immune cells, and are present in all tissues where they play diverse roles in tissue homeostasis, surveillance, and clearance of cellular debris1. While macrophages are recognized to have conserved transcriptional profiles, tissue-specific gene programs have been described in tissue resident macrophages across tissues2,3. Since foundational studies that described unique tissue-specific transcriptional programs, several groups have investigated the functional regulation of these genes and their contribution to promoting organ-specific functions4,5,6. As such, several recent studies have further resolved conserved macrophage profiles across tissues in the steady state7,8,9. Beyond these studies in homeostatic settings, it was suggested that conserved gene programs emerged following tissue injury. However, limited studies have investigated the conserved or tissue-specific programming of the macrophage subsets that emerge during tissue injury.
One macrophage subset recently recognized to expand following sterile tissue injury in several settings are lipid-associated macrophages (LAMs). LAMs are a unique subset of macrophages that play a pivotal role in lipid metabolism and tissue homeostasis10,11,12. These macrophages are increasingly appreciated in recent years for their influence in various diseases due to metabolic dysregulation, including atherosclerosis, obesity, metabolic dysfunction-associated steatotic liver disease (MASLD), and cancer13,14,15,16,17,18,19. Understanding the biological functions of LAMs is essential to developing therapeutic strategies for these conditions.
Multiple studies have shown that LAMs can originate from tissue resident macrophages or circulating monocytes that enter tissue in response to inflammatory cues, and differentiate into terminal cells with specific lipid-handling abilities, including sensing, engulfing, efflux, and breaking down of lipids11,20,21,22,23,24,25. These cells also exhibit unique lipid-associated transcriptomic profiles that distinguish them from other macrophage subsets. For instance, several studies have identified that LAMs are characterized by the expression of markers, such as triggering receptor expressed on myeloid cells 2 (Trem2) and Lipoprotein Lipase (Lpl), emphasizing their role in lipid sensing and metabolism. Specifically, LAMs help take up and break down excessive lipids to prevent accumulation that may lead to metabolic disorders. This homeostatic function contributes to metabolic and immune stability of tissues and ensures healthy energy storage and usage10,26,27,28.
LAMs are documented in multiple tissue sites, including adipose tissue, liver, adrenal gland, arterial plaques, and even the tumor microenvironment. In the adrenal gland, LAMs closely interact with lipid- and steroid hormone-producing cells under disease conditions21,22,29,30. In adipose tissue, LAMs possess dual functions. At homeostasis, LAMs facilitate tissue homeostasis by clearing cell debris from dead adipocytes and contribute to the remodeling of the extracellular matrix31,32. In contrast, under obesity conditions, the LAM population expands drastically, and they acquire a pro-inflammatory phenotype33,34,35. This alteration exacerbates local inflammation and insulin resistance, thereby promoting systemic dysregulation of metabolism. In the liver, LAMs are essential to the development and progression of MASLD15,36,37. These cells attempt to clear and digest excess lipid through the upregulation of cholesterol efflux and lipolysis pathways, but in the setting of obesity and atherosclerosis, these cells have been associated with excessive lipid overload and dysfunction. This lipid deposition causes the release of inflammatory mediators and promotes hepatic inflammation. Chronic hepatic inflammation eventually leads to liver fibrosis and irreversible damage. LAMs are also essential for the progression of atherosclerosis, a chronic inflammatory disease of the arterial wall. In the arterial intima, macrophages take up oxidized low-density lipoproteins and become dysfunctional once transformed into lipid-laden foamy macrophages17,38,39,40,41,42. Accumulation of foam cells in the intima ultimately drives atherosclerotic plaque progression. This process poses risks to cardiovascular health, and elevates the risk for myocardial infarction or stroke.
The molecular mechanisms underlying the formation and function of LAMs are not fully elucidated, but it is proposed that adipose LAMs require Trem2 for LAM programming43. In recent atherosclerosis studies, conditional deletion of Trem2 in myeloid cells resulted in slowed atherosclerotic plaque growth41. Inversely, therapeutically agonizing Trem2 with an antibody stabilized plaque by promoting collagen accumulation and enhancing LAM survival. Furthermore, the expression of lipid-handling enzymes in LAMs, including LPL, facilitates their capability to perform lipolysis to break down and utilize the internalized lipids. Targeting LAMs offers potential therapeutic benefits, and in fact, many of these approaches are underway in the cancer and neuroinflammation fields. For example, peroxisome proliferator-activated receptor (PPAR) inhibition is used to regulate lipid metabolism in lipid and tumor-associated macrophages44. Fatty acid binding protein (FABP) inhibition intervenes in lipid transport proteins to reduce lipid handling in LAMs45. Similar inhibition approaches have also been developed to target other molecules regulating lipid uptake and accumulation in macrophages, including CD36, a fatty acid receptor and transporter on macrophages46, and ABC transporters, which play key roles in the efflux of cholesterol14,47,48. Overall, the field of LAM biology still lacks a universal documentation of LAM feature profile to potentially facilitate further therapeutic discovery.
In this study, we aimed to profile representative LAM signatures across chronic inflammatory diseases in mice, using scRNAseq data and integration approaches. We integrated macrophages from multiple tissues and diseases to identify gene programs that are conserved by LAMs across organs, as well as define genes that are unique to LAMs in specific disease settings. In addition, integration analysis of published human scRNAseq data revealed that LAMs are transcriptionally conserved between human and mouse. Overall, these data can be used as a valuable resource for defining LAM subsets, as well as providing insight into the tissue-restricted function of LAMs.
Results
scRNAseq integration reveals macrophage heterogeneity in response to chronic inflammation
Macrophages are appreciated for their functions in disease pathogenesis and resolution. Shared macrophage subpopulations are recognized to emerge following injury or chronic inflammation1,49,50, but the transcriptional profiles of these cells have not been thoroughly investigated. To computationally investigate the commonalities and differences of macrophage subsets from different murine tissues in response to metabolic dysregulation or chronic inflammation, we integrated scRNAseq data from atherosclerotic aorta, livers from MASLD, hearts with heart failure with preserved ejection fraction (HFpEF), white adipose tissue (WAT) under high-fat diet (HFD) fed obesity conditions, and adrenal glands under stress conditions (Table 1, Fig. 1A). We also included data from hearts bearing myocardial infarction to investigate macrophage responses to more acute sterile inflammation. Since we were particularly interested in cells from tissues under pathophysiological conditions, we did not include cells from healthy mice in the integration analysis. We first performed quality control to remove low-quality cells based on low read depth and high mitochondrial content. Next, monocyte and macrophages (MoMACs) were extracted from the datasets according to the original cell type annotation by the authors, after which post-processed cells underwent integration using Harmony and further downstream analysis (Fig. 1A). Clusters of monocytes and macrophages were subsequently projected to uniform manifold approximation (UMAP) space, where nine clusters were identified (Fig. 1B). All nine clusters were detected in different tissues and conditions, with cluster 0 and 1 being the dominant populations (Fig. 1C–E). The high uniformity of clusters between tissues may suggest MoMACs potentially undergo similar transcriptional programming in response to metabolic dysregulation or tissue injury, regardless of the homing site (Fig. 1E).
A Workflow for the design of the integration study. B Clusters of monocytes and macrophages shown in UMAP embedding. C Cluster composition of each tissue and condition. D Tissue and condition composition for each cluster. E UMAP cluster split by tissue and conditions. F Differentially expressed genes that represent each cluster, shown in a dot plot.
Next, differential expression analysis was performed to identify the leading features of each cell cluster. Cluster 0 appeared to be classically activated macrophages as it yielded high expression of Tnf and pro-inflammatory chemokines, such as Ccl4, Ccl3, and Cxcl2 (Fig. 1F). Cluster 1 was enriched for Ly6c2 and Hp, and thus identified as classical monocytes. Notably, cluster 2 exhibited prominent expression of lipid-sensing molecules, such as Trem2 and Lpl, indicating these were LAMs. Major histocompatibility complex II (MHC-II) molecules, H2-Ab1 and H2-Eb1, and CD74 are high for cluster 3, suggesting these cells are prone to antigen presentation. Cluster 4 was distinguished by Ace and Adgre4. Interestingly, we were able to identify cluster 5 as Lyve1-resident macrophages, that also co-expressed Cd163, Mrc1, and Folr2, which has been documented in previous study8. Cluster 6 showed high expression of Top2a and Mki67, therefore distinguished as proliferating macrophages. According to our previous study, there was a smooth muscle cell derived macrophage population in atherosclerotic lesions that showed prominent expression of Acta241. Rather surprisingly, this subset of cells indeed appeared across all tissues in this integration analysis in cluster 7. In addition, collagen associated genes, Col1a2 and Col1a3, were differentially expressed in this cluster, suggesting these macrophages may be homeostatic by promoting collagen deposition. Interestingly, cluster 8 was distinguished by Ifit2 and Ifit3 expression, which are linked to type I interferon signaling, suggesting this population potentially responds to inflammatory stimuli51,52. Overall, this integration analysis across disease models and tissues suggested that macrophages may possess a conserved transcriptional response to cope with tissue injury.
LAMs transcriptional signatures are conserved across tissue and condition
LAMs are vital participants in metabolic homeostasis and regulation11,53,54,55. However, it is well recognized that LAMs play various roles in different tissues37,56,57,58,59,60. Whether LAMs from different tissue origins are transcriptionally conserved in mice is not well understood. To address this question, we first identified LAMs from the integrated meta-dataset. Since Trem2 and Lpl are well characterized LAM-associated genes13,14, we used these two molecules to first identify cluster 2 as the LAM population (Fig. 1F). Next, differential expression analysis between cluster 2 and all other cells was performed on each tissue independently. This step generated a set of genes for each tissue that represent LAM features in that tissue. This gene pool was integrated together, and we identified genes that were uniquely expressed by LAMs and their enrichment was shared by all tissues. Fourteen overlapping genes were identified, including Cd63, Cd68, Cstb, Ctsb, Ctsd, Fabp5, Gpnmb, Lgals3, Lipa, Lpl, Pld3, Plin2, Spp1, and Trem2 (Fig. 2A). Merging these genes into a pathway feature plot resulted in co-expression in cluster 2 (Fig. 2B, C), validating that cluster 2 is a conserved LAM cluster.
Of these 14 features, Cd63, Cd68, Ctsb, and Ctsd are associated with macrophage lysosomal functions61,62. Lipid sensing and signaling also appeared to be highlighted in LAMs as Fabp5, Lipa, Lpl, Trem2, and Pld3 are highly expressed26,43,63. In addition, the expression of Gpnmb and Spp1 may indicate LAMs participate in the regulation of inflammation64,65,66. Next, KEGG pathway analysis was performed to reflect the biological and cellular functions of genes. Notably, lipid utilization was reflected by PPAR signaling, steroid, and glycerolipid metabolism (Supplemental Fig. 1A). LAMs widely present lipid metabolism associated homeostatic characteristics as oxidative phosphorylation and lysosomal associated activities, including both proteosome and lysosome activation, are upregulated (Supplemental Fig. 1A). Together, the data support that LAMs possess complex functions to regulate lipid homeostasis, a feature that is conserved across tissues.
Furthermore, we investigated the potential developmental trajectory of LAMs from different tissue origins and conditions. According to previous studies in atherosclerosis, macrophages from tissue-resident and monocytic origins share a similar transcriptional profile67,68. Under HFD-induced obesity, WAT LAMs primarily derive from monocytic lineage69,70, but whether LAMs from other tissue niches follow the same differentiation pattern remains unclear. We hypothesized that the majority of LAMs arise from circulating monocytes to sustain local populations in different tissues during inflammatory diseases. We took a computational approach by inferring a pseudotime trajectory for each dataset to test this hypothesis. Cluster 1, classical monocytes, was set as the root of pseudotime trajectory. We observed a clear trend where pseudotime ends in cluster 2, LAM, and cluster 3, MHC-II high. This pattern was observed in all tissues and conditions, suggesting LAMs across tissue may depend, in part, on monocyte precursors (Supplemental Fig. 1B).
LAMs exhibit tissue-specific characteristics
Despite the shared features across tissues, we next sought to investigate tissue-specific markers that may represent LAM population heterogeneity across tissues. First, we performed re-clustering of MoMAC cluster 2. Four LAM subclusters were generated (Fig. 3A). Subcluster 0 appeared to be the dominant cluster. Interestingly, LAM subcluster 3 was unique to mouse aorta bearing atherosclerosis, and it was not present in other conditions and tissues (Fig. 3B). This was shown more clearly in cluster split by tissue, where subcluster 3 only appeared in the aorta dataset (Fig. 3C, D). Next, differential expression was performed to calculate features representing each subcluster. The canonical LAM markers, such as Lpl, Mmp12, and Spp1, were expressed by all cells, although the average expression for these features varied (Fig. 3E). Subcluster 0 and 1 were particularly high for these markers. Subcluster 2 showed relatively high expression of classical monocyte markers, such as Ly6c2, and was therefore identified as newly differentiated LAMs deriving from recently recruited monocytes. Notably, subcluster 3 was distinguished by high Gab2, Slc8a1 and Tanc2, which was specific to the aorta. To investigate the functional properties of LAM subpopulations, we performed gene set enrichment analysis (GSEA). First, we identified pathways highly enriched across all LAMs. As expected, these cells exhibited strong enrichment in lipid-related processes, including lipid storage, lipid clearance, low-density lipoprotein receptor (LDLR) activity, and response to LDL (Supplemental Fig. 2A). Despite the shared enrichment in lipid-associated pathways, LAM subpopulations displayed notable heterogeneity. LAM subcluster 1 showed a strong association with cholesterol efflux and catabolism (Supplemental Fig. 2B, C). In contrast, subcluster 2 exhibited a more pronounced pro-inflammatory profile relative to other LAM subpopulations (Supplemental Fig. 2D). Subcluster 3 was specifically characterized by enrichment in lipid translocation pathways (Supplemental Fig. 2E). Lastly, subcluster 0 was predominantly enriched in lipid metabolic processes (Supplemental Fig. 2F). Together, these findings highlight the overall lipid-associated nature of LAMs, with specialized lipid and cholesterol handling capacity, while also underscoring their transcriptional heterogeneity across tissues.
A UMAP embedding showing four LAM subclusters. B Cluster composition for each tissue. C Tissue/condition composition for each LAM cluster. D UMAP cluster split by tissue. E Top ten signature genes for each LAM cluster. F Pseudotime trajectory involving all monocytes and macrophage in UMAP space. G LAM subclusters shown in previous total MoMAC UMAP space.
To computationally develop a more defined differentiation trajectory within the LAM subpopulations, we reintroduced pseudotime to the new clusters. Interestingly, subcluster 2 was upstream of trajectory pseudotime, suggesting this is the population that first to differentiate and adopt LAM phenotype (Fig. 3F, G). Since subcluster 2 expresses lipid-associated genes while maintaining genes such as Ly6c2, reflecting a monocyte phenotype, we argued that this stage of cells could be indicative of early events leading to LAM differentiation. To investigate this idea, we performed differential gene expression analysis between monocyte cluster 1 and LAM subcluster 2 (Supplemental Fig. 3A). Notably, we observed prominent upregulation of LAM features in subcluster 2, including Fabp5, Gpnmb, Ctsd, Cd63, Cstb, Lgals3, Plin2, Cd68, Trem2, Cd9, Spp1, Lpl, and Mmp12. Of these markers, Fabp5, Gpnmb, Spp1, and Mmp12 were at a more significantly upregulated state (Supplemental Fig. 3B), suggesting LAMs may initially turn on these genes during differentiation toward mature LAMs. Subcluster 0 subsequently emerged from cluster 2. Compared with cluster 1, cluster 0 showed lower expression of Apoe, which may suggest that this population has already undergone LAM transcriptional programming but is not at a fully mature differentiation state (Fig. 3F, G). This reclustering approach revealed unique heterogeneity and composition of LAMs during disease and proposes the co-existence of several LAM populations in tissues, likely with common origins from monocyte precursors.
Next, we specifically investigated the differences of LAMs across tissues. Although the majority of differentially expressed genes (DEGs) are not tissue-specific, aortic and HFpEF LAMs exhibited the most segregated gene expression patterns, where 447 and 500 features were differentially expressed, respectively (Fig. 4A). Liver, MI heart, WAT, and adrenal LAMs showed 97, 71, 240, and 191 DEGs, respectively, suggesting LAMs indeed possess tissue-specific characteristics (Fig. 4A). Aortic LAMs, in particular, showed high expression of Abca1, consistent with aortic LAMs promoting cholesterol efflux pathways during atherosclerosis (Fig. 4B, C). Lyz1 was exclusively expressed in liver LAMs. In addition, these cells showed distinguishable Id3, Ear2, and Apoc1 expression (Fig. 4B, C). As for hearts bearing MI condition, Arg1 was highly enriched. Interestingly, chemokines associated with acute inflammation and monocytes recruitment, including Ccl2, Ccl12 and Ccl7, were also strongly upregulated. This may suggest LAMs in MI heart potentially participate in inflammatory response by contributing to monocyte recruitment. Cardiac HFpEF LAMs were defined by Ptgs2 expression. It has been shown that macrophage expression of Ptgs2 mediates proinflammatory responses by driving immune cell infiltration to inflammatory sites71,72. It is also involved in the resolution phase of inflammation and contributes to tissue homeostasis. This observation supports the idea that LAMs in HFpEF are indeed a regulator of chronic disease. Furthermore, adrenal LAMs exclusively express Hes1, which is an anti-inflammatory gene73, suggesting adrenal LAMs may utilize Hes1 to dampen inflammation by limiting immune cell infiltration (Fig. 4B, C). Lastly, adipose LAMs showed high expression of Folr2 (Fig. 4B, C), which in most tissues is restricted to a subpopulation of tissue resident macrophages8. In summary, differential analysis of LAMs across tissues has identified tissue-specific expression patterns that are likely associated with specialized functions in these pathologic responses.
A Venn diagram shown overlapping and unique gene count between tissues in LAM. B Top 5 differentially expressed genes that represent each tissue. C Top representative feature for each tissue. D Quantification of Folr2+ macrophages in tissue (n = 3 for each group). E Histogram showing Folr2 expression in different tissues. Error bars represent standard error of the mean.
To experimentally validate our observations regarding tissue specificity of LAMs, we utilized LDLR-/- mice fed an HFD for 16 weeks. This diet induces systemic elevation of cholesterol, therefore promoting atherosclerosis, metabolic dysregulation, and disease progression. Flow cytometry analysis was used to quantify target LAM molecules. Since only a limited number of LAM genes have commercially available antibodies for flow cytometry, we were restricted to the targets we could interrogate. Here, we aimed to measure Folr2 enrichment in the aorta, as well as the adrenal gland, liver, and WAT, as antibodies targeting Folr2 are widely available. We defined macrophages in tissues as sharing expression of F4/80 and CD11b. Total macrophages were then examined for significant Folr2 expression, where positivity in WAT macrophages was about 40% of WAT macrophages were classified positive for Folr2 (Fig. 4D, E, Supplemental Fig. 4). Consistent with transcriptional data, aortic macrophage Folr2 expression ranked second after WAT, while adrenal gland and liver macrophages showed negligible Folr2 expression (Fig. 4D, E). In conclusion, our computational and experimental approaches suggest LAM indeed possess unique transcriptional signatures despite their overlapping core LAM transcriptional profile.
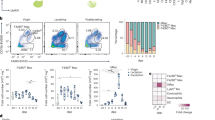
To validate the conserved LAM features in mice, we expanded the flow cytometry experiment that utilized the LDLR-/- mice. We targeted surface proteins selectively enriched in murine LAMs. In addition to canonical LAM features like CD36, Trem2, and CD9, we also investigated Adam8, Clec1b, Clec7a, and CD63 which were also identified as conserved markers across tissues (Supplemental Fig. 5). We first distinguished Trem2-high (Trem2 + ) and Trem2-low (Trem2-) macrophages (Supplemental Fig. 6), and then compared the expression of LAM-associated features between these populations. It is worth noting that Trem2 was dramatically expanded in all tissues of interest, where close to 90% of macrophages in WAT and liver and 50% in adrenal gland and heart, were classified as Trem2 + . In the adrenal glands, Trem2+ macrophages exhibited significantly higher expression of CD63, CD36, and Clec1b compared to Trem2- macrophages, while Clec7a and Adam8 showed no significant differences in protein abundance (Fig. 5A). A similar trend was observed in the heart, where CD63, CD36, and Clec7a were highly expressed in Trem2+ macrophages but not in Trem2- macrophages (Fig. 5B). In the liver, Trem2+ macrophages exhibited elevated CD63, CD36, Clec1b, and Clec7b expression (Fig. 5C), whereas in WAT, Trem2+ macrophages did not show enhanced Clec1B (Fig. 5D). In summary, Trem2+ macrophages represent LAMs across multiple tissues, upregulating core LAM proteins across several tissues that included Trem2, CD36, CD63, Clec1b, and Clec7b protein levels compared to non-LAM macrophages, which is consistent with transcript analysis.
LAMs signatures are conserved in humans
Despite the number of studies reporting LAMs in mouse, only a few studies have directly investigated gene programming for LAM between species67,69,74. To investigate this idea, we integrated monocytes and macrophages from four human studies, including samples from obese WAT, liver cirrhosis, atherosclerotic plaque, and dilated cardiomyopathy (DCM) biopsy (Fig. 6A, Table 2). MoMACs were first isolated according to the author’s annotations. Next, we used CD14 expression to validate the annotation and purify the MoMAC population, followed by integration of individual datasets and downstream analysis. Clustering of the integrated dataset resulted in seven MoMAC clusters (Fig. 6B). Although the overall composition of clusters was similar between tissues, adipose tissue showed a lower proportion of cluster 2 and enrichment of cluster 1 and 5 (Fig. 6C). Interestingly, the heart exhibited a unique cluster 6 which was absent in the other tissues (Fig. 6C–E). Differential analysis was performed to generate transcriptional features for each cluster. Overlapping features between 0 and 6 also included C1QA, PLTP, DAB2, TXNIP, LYVE1, and F13A1 (Fig. 6F). SEPP1 was unique to cluster 0, and ATP6V0C and NME2 were unique to cluster 6. Cluster 1 was distinguished by SPP1, FABP4, APOC1, TREM2, and MMP9, likely the LAM population. Cluster 2 exhibited mostly ribosomal genes, suggesting this might be stressed macrophages. Cluster 3 showed monocyte features like S100A8, S100A9 and S100A12. Interestingly, we observed immunoglobulin-associated gene expression, such as IGHM, IGLC2 and IGLC3 in cluster 4. CCL5 and CD69 were also highly expressed in this cluster, which exhibited a B cell signature. We suspect that this might be macrophages contaminated by B cells. Cluster 5 showed unique expression of ITLN1, KRT18, SLPI, and TM4SF1, which would likely represent an activated epithelial cell population that was contaminating the datasets. Overall, human MoMACs exhibited high heterogeneity as cell clusters differentially expressed unique features. However, consistent with recent studies comparing tissue-resident macrophages between mouse and human, several key overlapping signatures were conserved.
Next, we took a similar approach to our mouse study to generate LAM features in human cells. We first calculated DEGs of cluster 1 for each tissue dataset, as cluster 1 co-expresses multiple lipid associated features, such as TREM2 and LIPA. We aimed to find overlapping genes across all tissues and named it LAM score. A total of 15 genes were generated, including FABP4, FABP5, APOC1, CSTB, SPP1, LIPA, TREM2, CTSD, CTSB, CD9, APOE, LGALS3, ACP5, GPNMB, and PLD3 (Fig. 7A–C). Interestingly, most of these genes overlapped with mouse LAM signatures. To address whether LAM programming was conserved between human and mice, we overlaid the 14 mouse LAM consensus genes on the human LAM DEG background. Mouse LAM features were strikingly consistent with those of human (Fig. 7D). To assess the overall conservation of transcriptional landscapes between mice and humans, we overlaid statistically significant DEGs from human LAM subclusters onto those of mouse LAMs (Supplemental Fig. 7). This analysis revealed a strong transcriptional correlation between mouse and human subpopulations. Specifically, mouse LAM cluster 0 corresponded closely to human LAM cluster 2, while mouse LAM clusters 1–3 aligned with human LAM clusters 1, 0, and 3, respectively. Overall, our findings highlight both conserved LAM genes and distinct LAM subcluster gene signatures between mouse and human LAMs, suggesting that core LAM programming is conserved between human and mouse.
Despite the myriad of conserved features, a few genes were not shared across human and mouse LAMs. Human LAMs specifically featured APOC1, APOE, ACP5, FABP4, and CD9 (Fig. 7A), while mouse exhibit high expressions of Cd63, Cd68, Lpl, and Plin2 (Fig. 2A), in their core shared LAM genesets. Rather surprisingly, mouse LAMs features did not include Cd9, as this gene has been well-documented in previous studies involved in the context of atherosclerosis, liver, and WAT LAMs18,43,69. Absence of Cd9 led us to speculate that some of the tissue may exhibit low expression of the gene. To test this, we examined Cd9 expression in each tissue. Interestingly, adrenal, aortic, liver, and WAT LAMs showed profound enrichment of Cd9, but it was faintly detected in either MI or HEpEF cardiac LAMs (Supplemental Fig. 8A). Additionally, we performed experimental validation of CD9 protein in adrenal gland, aorta, liver, and WAT. As introduced previously (Fig. 4D), LDLR-/- mice fed HFD were utilized to disturb lipid metabolism in mice. Consistent with the scRNAseq data, CD9 protein was observed in the four tissues after physiological challenge with HFD (Supplemental Fig. 8B–E), suggesting CD9 can indeed be reliable for identifying LAMs in mouse tissues, with the exception of the heart. In conclusion, we observed both conserved and distinct gene signatures of LAMs between conditions and species, underscoring the importance of understanding the tissue-specific LAM signatures in translational studies.
Discussion
This study provides a comprehensive analysis of LAM heterogeneity in mice and humans across tissues in response to sterile inflammation, highlighting both conserved and tissue-specific transcriptional alterations. Using an integration approach of scRNAseq data from multiple metabolic disease models, we documented various macrophage subsets that arose during disease. One subset was found to be LAMs. These findings expand our understanding of how macrophages adapt to metabolic stress and serve as a resource for future studies investigating the function of LAMs in different disease models.
Our analysis revealed that LAMs share common transcriptional features in response to chronic and sterile inflammation. The conservation of LAM transcriptional signatures across different tissues and conditions highlights the role of these cells in maintaining lipid homeostasis and inflammation regulation. The shared set of features across human and mouse suggests that the fundamental mechanisms governing LAM generation and functions are evolutionarily preserved. Interestingly, all LAM populations show consistent expression of genes such as Trem2, Lpl, and Fabp5, across organs, suggesting that LAMs may serve as a critical component in the body’s response to systemic metabolic challenges, regardless of the tissue context. Notably, given the ubiquitous expression of Trem2 by LAMs across tissues and species, therapeutic interventions of Trem2 or other LAM-specific gene targets could represent a promising strategy for modulating dysregulated metabolism in human.
In fact, immunotherapeutic approaches agonizing and antagonizing Trem2 are underway in the clinic75,76,77,78. According to a recent study targeting Trem2 in the context of atherosclerosis41,79, Trem2 agonist treatment stabilizes atherosclerotic plaque by promoting Trem2-expressing foamy macrophage survival and reducing cell apoptosis. Interestingly, this treatment led to increased collagen deposition in the atherosclerotic niche, which also likely contributes to plaque stability. This study not only emphasizes the importance of LAMs in disease progression but also highlights that therapeutic strategies targeting LAMs are viable to regulate disease outcomes. In the context of breast cancer, Trem2-expressing LAMs have been demonstrated to mediate immune suppression by limiting T cell effector function and proliferative capacity20,80,81. Interestingly, anti-Trem2 treatment can indeed enhance anti-PD-1 efficacy and promote immune infiltration in the cancer microenvironment, thereby enhancing T cell functions and limiting tumor growth82. Together, Trem2 has been shown to possess therapeutic potential. Targeting Trem2+ LAMs could serve as a promising approach against various diseases where LAMs are found.
Despite the conservation of LAM features, our study also highlighted tissue specificity in LAM gene expression, reflecting tissue-specific function of LAMs. For instance, aortic LAMs exhibited high expression of Abca1, which is associated with cholesterol efflux. Heart-resident LAMs in both myocardial infarction and HFpEF conditions upregulated inflammatory cytokines such as Ccl2 and Il1b, suggesting LAMs may be vital in promoting inflammatory response in the context of the heart. This phenomenon indicates that while LAMs share a core set of functions, their specific roles may be modulated by the local tissue environment, contributing to the unique pathophysiological outcomes observed in different metabolic diseases. In several of these contexts, it may be feasible to specifically target a subset of LAMs based on their tissue-restricted gene signatures to avoid systemic LAM modulation.
In conclusion, our integrative analysis of mouse and human scRNAseq data offers a valuable reference for the study of LAMs, potentially serving as a foundation for future research. We emphasize the significant heterogeneity of LAMs in both mouse and human models, with LAMs displaying conserved transcriptional features across different tissues. Despite these conserved characteristics, LAMs also exhibit tissue-specific transcriptional profiles, which we have validated through experimental approaches. Importantly, the strong overlap in LAM features between mouse and human underscores the utility of mouse models in translational studies, particularly for testing therapeutics aimed at targeting LAMs in models of human diseases. Overall, our findings highlight the potential for leveraging conserved or tissue-specific LAM signatures in both species to develop therapeutic strategies that can address a range of LAM-related conditions across different tissues and diseases.
Methods
Animal maintenance
LDLR-/- mice were purchased from the Jackson Laboratory (Jax 002207). Animals, including both males and females, were housed in the University of Minnesota (UMN) Research Animal Resources facility under 12 h dark and light cycle and specific pathogen-free conditions. Mice had unrestricted access to food and water. HFD used in this study was purchased from Envigo Teklad (TD.88137, adjusted calories diet, 42% fat). Mice started on HFD at 7 weeks of age and stayed on HFD for 16 weeks. Animals were sacrificed by CO2 asphyxiation. We have complied with all relevant ethical regulations for animal use. All experiments performed in this study were approved and performed in accordance with the UMN Institutional Animal care and use committee.
scRNAseq analysis
Preprocessing
Single cell RNA sequencing (scRNAseq) datasets were downloaded from gene expression omnibus (GEO) database (Table 1, Table 2)30,41,83,84,85,86,87,88,89,90,91. Data was first loaded and transformed to Seurat object using Read10X and CreateSeuratObject functions from the Seurat v4.2.1 package. We skipped preprocessing procedures on datasets that were annotated and processed by the original authors. Otherwise, datasets were filtered on cell mitochondrial content, where cells with over 25% mitochondrial transcription were removed. Normalization and scaling were performed using the NormalizeData and ScaleData functions. Doublets were detected and removed using the DoubletFinder v2.0.3 package.
Data integration
All datasets were first merged as one meta dataset using the merge function. FindVariableFeatures, and RunPCA functions were subsequently performed. A total of 33,947 cells were generated in the mouse metadata, and human data consisted of 11,767 cells. To correct batch effects among datasets, RunHarmony function from the Harmony v0.1.0 package was applied to the merged object. Next, cells were projected to a UMAP embedding using the RunUMAP function from Seurat, with the “reduction” command set to “harmony.” Determination of significant dimensions was facilitated by an elbow plot generated using the Seurat ElbowPlot. FindNeighbors and FindClusters functions were used to calculate clusters. For LAM reclustering, the LAM population was first identified according to the transcriptional profile of the cluster. Then, the cluster corresponding to the LAMs was extracted. A total of 5941 LAMs were selected. The data integration process described above was repeated on the isolated LAM cluster.
LAM feature identification
FindAllMarkers function was first used to generate DEGs for each cluster. The cluster featuring lipid-associated genes was annotated as the LAM cluster. Next, FindAllMarkers was performed on each dataset between the potential LAM cluster and all other clusters to generate LAM-representing features for each tissue and condition. Next, we found the overlapping features across all tissues. In mouse, 14 features were identified. In human, 15 features were identified. Merging of all LAM features was achieved using the addModuleScore function and named “LAM score”.
Trajectory
Pseudotime trajectory analysis was supported by the Monocle v2.18.0 and Monocle3 v0.2.3.3 packages. First, as.cell_data_set function from the SeuratWrapper v0.3.0 package was used to transform Seurat object to Monocle 3 cell_data_set. Root of the pseudotime trajectory was set to classical monocytes identified by transcriptional features, including Ly6c2. Next, cluster_cells, lean_graph, and order_cells were subsequently performed.
KEGG pathway analysis
FindMarkers function was used to generate the gene background for LAM cluster, with logfc.threshold and min.pct argument set to 0. Functions from the FGSEA v 1.18.0 package were used to performed pathway analysis. KEGG pathway database was imported from the msigdbr v7.2.1 package using the msigdbr function.
Flow cytometry
Freshly harvested adrenal gland, liver, aorta, and white adipose were finely minced. Adrenal glands were enzymatically dissociated using Liberase DH (Roche 5401054001, 2.5 mg/mL in double:200-distilled water) at 10% dilution for 35 min. Liver, aorta, and white adipose were processed in 1.5 mg/mL collagenase A (COLLARO 10103578001) for 35 min. Dissociation procedure was performed at 37 °C in orbital shaker at 800 revolutions per minute. Dissociated single cell suspensions were next filtered through 100 μm nylon mesh (McMaster Carr) to remove debris. Cell solutions next underwent centrifuging at 1450 revolutions per minute for 5 min. For antibody staining, all cells were stained at 1:200 diluted flow antibodies (Table 3) in FACS buffer at 4 °C for 25 min. Cells were washed twice in FACS buffer post-staining, and data were collected using the BD LSRFortessa instrument maintained at the flow cytometry core facility at the UMN.
Statistics
Flow cytometry graphs were made in Flowjo software. Graphpad Prism was used to generate bar graphs and perform statistical analysis, where comparisons of groups of two were performed using a students t test, and groups of more than two were performed by ANOVA. Statistical significance was determined for P < 0.05, with exact p values being presented in the figures. ScRNAseq data differential expression was based on the non-parametric Wilcoxon rank sum text, supported by the Seurat package. P value adjustment shown in this study used the Bonferroni correction method.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
References
Park, M. D., Silvin, A., Ginhoux, F. & Merad, M. Macrophages in health and disease. Cell 185, 4259–4279 (2022).
Gautier, E. L. et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 (2012).
Wynn, T. A. & Vannella, K. M. Macrophages in tissue repair, regeneration, and fibrosis. Immunity 44, 450–462 (2016).
Gautier, E. L. et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J. Exp. Med. 211, 1525–1531 (2014).
Kohyama, M. et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457, 318–321 (2009).
Haldar, M. et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell 156, 1223–1234 (2014).
Ninni, A. et al. MACanalyzeR scRNAseq analysis tool reveals PPARγHIGH/GDF15HIGH lipid-associated macrophages facilitate thermogenic expansion in BAT. Nat. Commun. 16, 5063 (2025).
Dick, S. A. et al. Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Sci. Immunol. 7, eabf7777 (2022).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Li, R.-Y. et al. TREM2 in the pathogenesis of AD: a lipid metabolism regulator and potential metabolic therapeutic target. Mol. Neurodegener. 17, 40 (2022).
Xu, R. et al. Lipid-associated macrophages between aggravation and alleviation of metabolic diseases. Trends Endocrinol. Metab. 31, 981–995 (2024).
Silvin, A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465 (2022).
Marelli, G. et al. Lipid-loaded macrophages as new therapeutic target in cancer. J. Immunother. Cancer 10, e004584 (2022).
Ren, Y., Wang, M., Yuan, H., Wang, Z. & Yu, L. A novel insight into cancer therapy: lipid metabolism in tumor-associated macrophages. Int. Immunopharmacol. 135, 112319 (2024).
Vonderlin, J., Chavakis, T., Sieweke, M. & Tacke, F. The multifaceted roles of macrophages in NAFLD pathogenesis. Cell. Mol. Gastroenterol. Hepatol. 15, 1311–1324 (2023).
Alabdulaali, B. et al. Macrophages and the development and progression of non-alcoholic fatty liver disease. Front. Immunol. 14, 1195699 (2023).
Dib, L. et al. Lipid-associated macrophages transition to an inflammatory state in human atherosclerosis increasing the risk of cerebrovascular complications. Nat. Cardiovasc. Res. 2, 656–672 (2023).
Kim, K., Park, S., Park, J. & Choi, J.-H. Characteristics of plaque lipid-associated macrophages and their possible roles in the pathogenesis of atherosclerosis. Curr. Opin. Lipidol. 33, 283 (2022).
Lavine, K. Identification of inflammatory lipid-associated macrophages in human carotid atherosclerosis. Nat. Cardiovasc. Res. 2, 604–605 (2023).
Liu, Z. et al. Lipid-associated macrophages in the tumor-adipose microenvironment facilitate breast cancer progression. Oncoimmunology 11, 2085432 (2022).
Xu, Y., Schrank, P. R. & Williams, J. W. Macrophage fate mapping. Curr. Protoc. 2, e456 (2022).
Dolfi, B. et al. Unravelling the sex-specific diversity and functions of adrenal gland macrophages. Cell Rep. 39, 110949 (2022).
Hill, D. A. et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl. Acad. Sci. USA115, E5096–E5105 (2018).
Kado, T., Nishimura, A. & Tobe, K. History and future perspectives of adipose tissue macrophage biology. Front. Pharmacol. 15, 1373182 (2024).
Vogel, A., Brunner, J. S., Hajto, A., Sharif, O. & Schabbauer, G. Lipid scavenging macrophages and inflammation. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1867, 159066 (2022).
Loving, B. A. et al. Lipoprotein lipase regulates microglial lipid droplet accumulation. Cells 10, 198 (2021).
Nugent, A. A. et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 105, 837–854 (2020).
Do, T. H. et al. TREM2 macrophages induced by human lipids drive inflammation in acne lesions. Sci. Immunol. 7, eabo2787 (2022).
Ye, H. et al. Triggering receptor expressed on myeloid Cells-2 (TREM2) inhibits steroidogenesis in adrenocortical cell by macrophage-derived exosomes in lipopolysaccharide-induced septic shock. Mol. Cell. Endocrinol. 525, 111178 (2021).
Xu, Y. et al. Adrenal gland macrophages regulate glucocorticoid production through Trem2 and TGF-β. JCI Insight 9, e174746 (2024).
Remmerie, A., Martens, L. & Scott, C. L. Macrophage subsets in obesity, aligning the liver and adipose tissue. Front. Endocrinol. 11, 259 (2020).
Li, X. et al. Adipose tissue macrophages as potential targets for obesity and metabolic diseases. Front. Immunol. 14, 1153915 (2023).
Dahik, V. D., Frisdal, E. & Le Goff, W. Rewiring of lipid metabolism in adipose tissue macrophages in obesity: impact on insulin resistance and type 2 diabetes. Int. J. Mol. Sci. 21, 5505 (2020).
Kawai, T., Autieri, M. V. & Scalia, R. Adipose tissue inflammation and metabolic dysfunction in obesity. Am. J. Physiol. Cell Physiol. 320, C375–C391 (2021).
Chakarov, S., Blériot, C. & Ginhoux, F. Role of adipose tissue macrophages in obesity-related disorders. J. Exp. Med. 219, e20211948 (2022).
Park, S.-J., Garcia Diaz, J., Um, E. & Hahn, Y. S. Major roles of kupffer cells and macrophages in NAFLD development. Front. Endocrinol. 14, 1150118 (2023).
Kazankov, K. et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 16, 145–159 (2019).
Moore, K. J., Sheedy, F. J. & Fisher, E. A. Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol. 13, 709–721 (2013).
Bobryshev, Y. V., Ivanova, E. A., Chistiakov, D. A., Nikiforov, N. G. & Orekhov, A. N. Macrophages and their role in atherosclerosis: pathophysiology and transcriptome analysis. BioMed. Res. Int. 2016, 9582430 (2016).
Patterson, M. T. et al. Trem2 promotes foamy macrophage lipid uptake and survival in atherosclerosis. Nat. Cardiovasc. Res. 2, 1015–1031 (2023).
Patterson, M. T. et al. Trem2 agonist reprograms foamy macrophages to promote atherosclerotic plaque stability—brief report. Arterioscler. Thromb. Vasc. Biol. 44, 1646–1657 (2024).
Patterson, M. T. & Williams, J. W. Metabolic regulation of macrophage proliferation and function in atherosclerosis. Curr. Opin. Lipidol. 32, 293–300 (2021).
Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a TREM2-dependent manner. Cell 178, 686–698 (2019).
Sun, J. et al. The role of peroxisome proliferator-activated receptors in the tumor microenvironment, tumor cell metabolism, and anticancer therapy. Front. Pharmacol. 18, 1184794 (2023).
Gaffar, S. & Aathirah, A. S. Fatty-acid-binding proteins: from lipid transporters to disease biomarkers. Biomolecules 13, 1753 (2023).
Chen, Y., Zhang, J., Cui, W. & Silverstein, R. L. CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J. Exp. Med. 219, e20211314 (2022).
Yvan-Charvet, L. et al. Increased inflammatory gene expression in ABC transporter deficient macrophages: free cholesterol accumulation, increased signaling via Toll-like receptors and neutrophil infiltration of atherosclerotic lesions. Circulation 118, 1837–1847 (2008).
Kotlyarov, S. & Kotlyarova, A. Clinical significance of lipid transport function of ABC transporters in the innate immune system. Membranes 12, 1083 (2022).
Murray, P. J. & Wynn, T. A. Protective and pathogenic functions of macrophage subsets. Nat. Rev. Immunol. 11, 723–737 (2011).
Chen, S. et al. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 8, 1–35 (2023).
Jiang, M. et al. Dysregulated expression of miR-146a contributes to age-related dysfunction of macrophages. Aging Cell 11, 29–40 (2012).
Gao, J. et al. Tumoral IL-10-activated SHP2 in macrophages promotes mammary carcinoma progression. Fundam. Res. 9, 1957–1969 (2024).
Fredrickson, G. et al. TREM2 macrophages mediate the beneficial effects of bariatric surgery against MASH. Hepatology 81, 1776–1791 (2025).
Florance, I. & Ramasubbu, S. Current understanding on the role of lipids in macrophages and associated diseases. Int. J. Mol. Sci. 24, 589 (2022).
Batista-Gonzalez, A. et al. New insights on the role of lipid metabolism in the metabolic reprogramming of macrophages. Front. Immunol. 10, 2993 (2020).
Coats, B. R. et al. Metabolically activated adipose tissue macrophages perform detrimental and beneficial functions during diet-induced obesity. Cell Rep. 20, 3149–3161 (2017).
Lumeng, C. N. & Saltiel, A. R. Inflammatory links between obesity and metabolic disease. J. Clin. Invest. 121, 2111–2117 (2011).
Chistiakov, D. A., Bobryshev, Y. V. & Orekhov, A. N. Macrophage-mediated cholesterol handling in atherosclerosis. J. Cell. Mol. Med. 20, 17–28 (2016).
Zigmond, E. et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Colombo, A. et al. Loss of NPC1 enhances phagocytic uptake and impairs lipid trafficking in microglia. Nat. Commun. 12, 1158 (2021).
Götzl, J. K. et al. Early lysosomal maturation deficits in microglia triggers enhanced lysosomal activity in other brain cells of progranulin knockout mice. Mol. Neurodegener. 13, 48 (2018).
Khantakova, D., Brioschi, S. & Molgora, M. Exploring the impact of TREM2 in tumor-associated macrophages. Vaccines 10, 943 (2022).
Nickl, B., Qadri, F. & Bader, M. Anti-inflammatory role of GPNMB in adipose tissue of mice. Sci. Rep. 11, 19614 (2021).
Saade, M., Araujo de Souza, G., Scavone, C. & Kinoshita, P. F. The role of GPNMB in inflammation. Front. Immunol. 12, 674739 (2021).
Rajendran, S. et al. Single-cell RNA sequencing reveals immunosuppressive myeloid cell diversity during malignant progression in a murine model of glioma. Cell Rep. 42, 112197 (2023).
Williams, J. W. et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat. Immunol. 21, 1194–1204 (2020).
T’Jonck, W., Guilliams, M. & Bonnardel, J. Niche signals and transcription factors involved in tissue-resident macrophage development. Cell. Immunol. 330, 43–53 (2018).
Stansbury, C. M. et al. A lipid-associated macrophage lineage rewires the spatial landscape of adipose tissue in early obesity. JCI Insight 8, e171701 (2023).
McDowell, S. A. C. et al. Obesity alters monocyte developmental trajectories to enhance metastasis. J. Exp. Med. 220, e20220509 (2023).
Martín-Vázquez, E., Cobo-Vuilleumier, N., López-Noriega, L., Lorenzo, P. I. & Gauthier, B. R. The PTGS2/COX2-PGE2 signaling cascade in inflammation: pro or anti? A case study with type 1 diabetes mellitus. Int. J. Biol. Sci. 19, 4157–4165 (2023).
Cox, D. G. et al. Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br. J. Cancer 91, 339–343 (2004).
Shang, Y. et al. The transcriptional repressor Hes1 attenuates inflammation via regulating transcriptional elongation. Nat. Immunol. 17, 930–937 (2016).
Sciarretta, F. et al. Lipid-associated macrophages reshape BAT cell identity in obesity. Cell Rep. 43, 114447 (2024).
Deczkowska, A., Weiner, A. & Amit, I. The physiology, pathology, and potential therapeutic applications of the TREM2 signaling pathway. Cell 181, 1207–1217 (2020).
Etxeberria, A. et al. Neutral or detrimental effects of TREM2 agonist antibodies in preclinical models of alzheimer’s disease and multiple sclerosis. J. Neurosci. 44, 2347 (2024).
Schlepckow, K., Morenas-Rodríguez, E., Hong, S. & Haass, C. Stimulation of TREM2 with agonistic antibodies—an emerging therapeutic option for Alzheimer’s disease. Lancet Neurol. 22, 1048–1060 (2023).
Reifschneider, A. et al. Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J. 41, e109108 (2022).
Piollet, M. et al. TREM2 protects from atherosclerosis by limiting necrotic core formation. Nat. Cardiovasc. Res. 3, 269–282 (2024).
Timperi, E. et al. Lipid-associated macrophages are induced by cancer-associated fibroblasts and mediate immune suppression in breast cancer. Cancer Res. 82, 3291–3306 (2022).
Huggins, D. N. et al. Characterizing macrophage diversity in metastasis-bearing lungs reveals a lipid-associated macrophage subset. Cancer Res. 81, 5284–5295 (2021).
Molgora, M. et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell 182, 886–900 (2020).
Cottam, M. A., Caslin, H. L., Winn, N. C. & Hasty, A. H. Multiomics reveals persistence of obesity-associated immune cell phenotypes in adipose tissue during weight loss and weight regain in mice. Nat. Commun. 13, 2950 (2022).
Su, Q. et al. Single-cell RNA transcriptome landscape of hepatocytes and non-parenchymal cells in healthy and NAFLD mouse liver. iScience 24, 103233 (2021).
Kim, K. et al. Transcriptome analysis reveals nonfoamy rather than foamy plaque macrophages are proinflammatory in atherosclerotic murine models. Circ. Res. 123, 1127–1142 (2018).
Jin, K. et al. Single-cell RNA sequencing reveals the temporal diversity and dynamics of cardiac immunity after myocardial infarction. Small Methods 6, e2100752 (2022).
Li, G. et al. Single-cell transcriptomic profiling of heart reveals ANGPTL4 linking fibroblasts and angiogenesis in heart failure with preserved ejection fraction. J. Adv. Res. 68, 215–230 (2024).
Massier, L. et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 14, 1438 (2023).
Koenig, A. L. et al. Single-cell transcriptomics reveals cell-type-specific diversification in human heart failure. Nat. Cardiovasc. Res. 1, 263–280 (2022).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518 (2019).
Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat. Med. 25, 1280–1289 (2019).
Acknowledgements
This study was supported by the National Institutes of Health (NIH) R01 AI165553 (JWW) and NIH R01 HL166843 (JWW). HH was supported by American Heart Association predoctoral fellowship 25PRE1361476, and MC was supported by NIH T32 HL166142. Thanks to the UMN Flow Cytometry Resource (UFCR) and UMN Minnesota Supercomputing Institute (MSI) for support and assistance in data acquisition and analysis.
Author information
Authors and Affiliations
Contributions
Y.X., S.I., X.S.R. and J.W.W. conceived and designed the project. Y.X., H.H., M.C. and F.B. conducted the experiments. Y.X. conducted the flow cytometry and RNA sequencing data analysis. Y.X. and J.W.W. wrote, and all authors participated in editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Jian-Da Lin and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, Y., Hillman, H., Chang, M. et al. Identification of conserved and tissue-restricted transcriptional profiles for lipid associated macrophages. Commun Biol 8, 953 (2025). https://doi.org/10.1038/s42003-025-08387-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08387-z
This article is cited by
-
Mechanisms of Innate Immune Modulation by High-Fat Diet: Implications for Obesity and Asthma
Current Allergy and Asthma Reports (2025)