Abstract
Climate warming is one factor increasing the severity of harmful algal blooms (HABs). Innovative exposure models are needed to understand how HABs affect brain health. Here, we examined HAB exposure on the brain transcriptome of dolphins found stranded in Florida’s Indian River Lagoon. We report the neurotoxin 2,4-diaminobutyric acid (2,4-DAB) is 2900 times more concentrated in dolphin brains during bloom seasons compared to non-bloom seasons. The same dolphins show 536 differentially expressed genes whose enrichment reveal impairment in GABAergic synapses, basement membrane alteration, and Alzheimer’s disease (AD) risk factors that increase with each subsequent season. Dolphins also display concurrent AD-like neuropathological changes and elevated AD gene expression with 2,4-DAB exposure. Our study demonstrates disproportionate seasonal exposure to 2,4-DAB increases AD signatures in the brain transcriptome. As our climate warms, HABs will continue to intensify. Understanding the impact of HAB exposures will help to identify populations at risk for neurological illnesses.

Similar content being viewed by others
Main
Climate warming and nutrient pollution have been associated with the frequent occurrence of harmful algal blooms (HABs) in the Indian River Lagoon (IRL), a 251-km estuary with brackish water occupying nearly half of Florida’s east coast1. Super blooms, like the ones experienced in the IRL over the last 15 years have been shown to deplete oxygen, kill fish and reduce seagrass2. It is estimated that HABs can cause a loss in the billions of dollars by impacting tourism, recreational fishing, property damage, and job loss3. The last decade has recorded some of the warmest surface temperatures in modern times in Florida and surrounding states4. Rising marine and fresh water temperatures will cause HABs to occur earlier in season and to occur for a longer duration, thus putting more stress on ecosystems5. Therefore, organisms will be more impacted during bloom seasons, which typically occur during warm weather5.
Toxins produced during HABs can bio-magnify in the aquatic food web, posing a significant health risk to marine life, terrestrial mammals, and humans6. Acute effects of these toxins are well defined; however, long-term and chronic exposures are an area of intense research. Several studies have shown neurotoxins originating from HABs to be concentrated in brain tissues of individuals diagnosed with neurodegenerative diseases, suggesting these toxins may be correlated with disease etiology7,8. Thus, there is an urgent need for innovative experimental models to help guide our understanding of how exposures to HAB toxins impact brain health across the lifespan.
Dolphins are long-lived marine mammals, a well-regarded sentinel species for toxin exposure, and can provide us with a range of relative exposures to HAB toxins and other pollutants in our environment9. Dolphins are also considered a natural model of Alzheimer’s disease (AD), because with age they develop the two main neuropathological changes associated with the disease: amyloid beta (Aβ) plaques and tau neurofibrillary tangles (NFTs)10,11,12,13. The co-occurrence of AD neuropathological changes and the natural accumulation of algal toxins observed in dolphins allows a unique opportunity to study the impact of these two converging events on the brain. In the IRL, dolphins have increased mortality rates during summer months when HABs are most common, suggesting seasonality may also play a critical role in increasing the susceptibility to algal toxin neurotoxicity14,15. Therefore, studies investigating the interaction between season and toxin exposure are needed to predict risk of neurological diseases, especially since the brain transcriptome shows the highest impact of gene seasonality16. The analysis of the dolphin skin transcriptome has provided insight on gene seasonality for persistent organic pollutants17. However, the interaction of gene seasonality and HAB toxin effects on the brain are unknown.
Here, we investigate the role of seasonal variation in HAB toxin exposure on the brain transcriptome of dolphins found stranded in the IRL, a water body located less than 200 miles from Miami-Dade County, Florida, USA. A county which had the highest prevalence of individuals living with AD in 202418. Opportunistic samples were collected over nearly a decade, starting in 2010, a year before the substantial change in the IRL ecologic system occurred caused by long-lasting phytoplankton blooms15. Archived brain tissues collected at necropsy was used to measure HAB neurotoxin exposure, AD related dementia gene expression, whole transcriptome analysis, and AD histopathology (Supplementary Fig. 1).
Results
Indian River Lagoon dolphins
Twenty common bottlenose dolphins (Tursiops truncatus truncatus) found stranded in the IRL from 2010 to 2019 were examined in this study. Adult (n = 15) and juvenile (n = 5) were sampled over a 155 km stretch of the IRL including: two from the Banana River, six from the northern Indian River, five from the Mosquito Lagoon, six from the Northern Central Indian River, and one dolphin from the Halifax River (Tomoka River tributary). Our cohort consisted of a one-to-one ratio of females to males with a median body length of 243.5 cm, with a minimum of 212 cm and a maximum of 261 cm. Causes of death included acute trauma, emaciation and inanition, viral infection and pneumonia, which were present in both the control and experimental groups. For our analysis dolphins were classified into two seasonal groups based on their date of stranding: non-bloom (n = 11, December–May) and bloom (n = 9, June–November) (Supplementary Tables 1 and 2).
Bloom season and exposure to 2,4-diaminobutyric acid
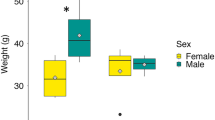
All dolphin cerebral cortex samples were positive for 2,4-diaminobutyric acid (2,4-DAB) or DABA, a neurotoxin produced by cyanobacteria, diatoms and dinoflagellates and a structural isomer of the neurodegenerative agent β-N-methylamino-L-alanine (BMAA)19. Brain concentrations of 2,4-DAB were observed from the limit of detection (LOD = 0.01 ng/g) to 89 ng/g using trichloroacetic acid (TCA) extraction. 2,4-DAB detection was confirmed in the cerebellum of nine dolphins where those archived samples were available (Fig. 1a, b, Supplementary Table 2). A second extraction method using hydrochloric acid (HCl) yielded four positive dolphins with a larger concentration range (not detected to 1042 ng/g). Dolphins from the North Indian River and Banana River had the highest frequency of positive testing. The highest concentrations of 2,4-DAB were detected in June and August (Fig. 1b; Supplementary Tables 2–3). The median brain concentration of 2,4-DAB in dolphins from bloom seasons was 29.0 ng/g (95% CI: 22.0–36.0) (p = 0.0037, Mann–Whitney U), which was approximately 2900-fold more concentrated than those in the non-bloom season (Fig. 1c). BMAA was only detected in one dolphin from the non-bloom season and the structural isomer N-(2-aminoethyl) glycine (AEG) was not detected (Supplementary Table 3).
a UHPLC-MS/MS chromatography of the presence of 2,4-diaminobutyric acid (2,4-DAB) in the cerebral cortex of an Indian River Lagoon dolphin. Structural isomers β-N-methylamino-L-alanine (BMAA) and N-(2-aminoethyl) glycine (AEG) were also evaluated. b 2,4-DAB was detected in all dolphin brain samples using TCA extraction, with those from the North Indian River and Banana River having the highest median brain concentration. BMAA was detected in one dolphin from the Mosquito Lagoon using an HCL extraction method. AEG was not detected with either method. c Dolphins stranded in the bloom seasons had a median concentration of 29 ng/g (95% CI: 22.0–36.0) concentration of 2,4-DAB in their brains (Mann–Whitney U; n = 11, 9 dolphins, effect size d = 1.07). Error bars indicate the minimum and maximum values. For the limit of detection (LOD) value (0.01 ng/g) were used for calculation. NB non-bloom. a was designed using ChemDraw and b was created in BioRender. Davis, D. (2025) https://BioRender.com/p96wek4.
Bloom season and the dolphin brain transcriptome
Whole transcriptome analysis was performed on cerebral cortex RNAs with a median RIN of 9.1 (95% CI: 8.9–9.5) comparing bloom and non-bloom seasons (Supplementary Table 4). A principal component analysis using all differentially expressed (DE) gene transcripts using a false discovery rate (FDR) ≤0.05, stranding seasons, and 2,4-DAB brain concentration showed three distinct clusters. Dolphins stranded during non-bloom seasons were tightly clustered. Whereas, dolphins sampled during bloom seasons had two clusters with larger variances in PC1 and PC2 (Fig. 2a). Stranded dolphins in the bloom vs non-bloom seasons had 536 DE protein encoding genes, which included 420 up- and 116 down-regulated genes using a FDR cut-off of ≤0.05. The top three upregulated (VIT, MT-ND1 & MT-ND5) and most significant genes (ACAP1, ZNF235 & ARHGEF19) are involved in mental, metabolic, and neuronal diseases. The top three downregulated genes (CCR2, PIK3CG & ANKRD22) and most significant genes (NCF2, RBM3, & ETNK1) were associated with blood, immune, and neuronal diseases (Fig. 2b). A table of top 20 upregulated and downregulated genes can be found in the supplementary material (Supplementary Tables 5 and 6). Hierarchical cluster analysis of the 536 DE genes and dolphins show grouping of dolphins by season (Fig. 2c). Similarity matrix analysis of the 536 DE genes shows the relationship of each dolphin in non-bloom and bloom seasons. Again, brain transcriptomes of stranded dolphins in the bloom seasons were more positively correlated with those stranded in the same season (Fig. 2d). Dolphins in the bloom seasons also showed increased TARBDP expression, a marker of neurodegenerative disease, which has been shown to be increased with BMAA isomer exposure20,21. TARBDP expression was confirmed by qPCR measurements p = 0.0244; Spearman r = 0.5490) (Fig. 2e).
a Principal component analysis comparing 536 differentially expressed (DE) genes, stranding season, and 2,4-DAB brain concentration. b Volcano plot of the 536 DE genes comparing bloom vs. non-bloom (NB) seasons. Dolphins beached in the bloom seasons had 420 upregulated genes (green) and 116 downregulated genes (orange) (FDR ≤ 0.05). Genes with the highest fold change and most significant p-values are highlighted in blue. c, d Heatmaps displaying hierarchical cluster and similarity matrix analysis of the relationship of all 536 DE genes. e Spearman r correlation analysis of normalized expression of TARDBP from qPCR and RNA Seq assays (n = 17 dolphins for all analyses a–e).
Altered synaptic transmission and blood brain barrier
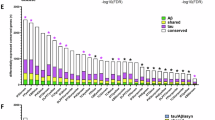
Gene Ontology (GO) database search of the 116 downregulated dolphin gene transcripts against the Homo sapiens catalog showed enrichment in the following top GO terms: glutamate decarboxylase activity (>100-fold; GO:0004351), the neuronal cell body (3.8-fold; GO:0043025), and cellular processes (1.5-fold; GO:0009987). GO database search of the 420 upregulated gene transcripts were associated with the following top GO terms: laminin-3 complex (48.7-fold; GO:0005608), positive regulation of integrin-mediated signaling pathway (23.3-fold; GO:2001046), and actin binding (2.5-fold; GO:0003779) (Fig. 3a, Supplementary Tables 7–9).
a Gene Ontology (GO) enrichment analysis of molecular, cellular, and biological functions for upregulated (green; n = 420) and downregulated (orange; n = 116) dolphin genes using a human (Homo sapiens) database search. Glutamate decarboxylase activity (>100-Fold; GO:0004351) and laminin-3 complex (48.7-fold GO:0005608) were the top enriched GO terms (labeled with star). b Comparative protein-protein interaction networks of GO terms, GO:0004351 and GO:0005608, for IRL dolphins and Alzheimer’s disease (AD) patients. Relative gene expression from the frontal pole of AD versus normal controls was obtained from the Alzheimer DataLens bulk transcriptomic database. Differential expression of bloom vs non-bloom (NB) season dolphins is depicted. c Reactome pathway enrichment for GO:0004351 and GO:0005608 for dolphin (Tursiops truncatus). d Heatmap and cluster analysis of dolphin genes related to the GABAergic synapse. b was created in BioRender. Davis, D. (2025) https://BioRender.com/ovsibtb. c was generated with STRING.
Comparative assessment of predictive protein-protein interaction networks (PPINs) functional enrichment analysis of human and dolphin were performed on the top most enriched GO terms for up- and downregulated transcripts: glutamate decarboxylase activity and laminin-3 complex (Fig. 3a). We show that PPINs for both species are highly similar, except for an interaction between ALDH5A1 and SLC32A1. Individual gene expression levels for each GO term were also compared between stranded dolphins in the bloom season and bulk transcriptomic data from AD patients. Stranded dolphins in the bloom season displayed a similar decrease in expression of glutamate decarboxylase activity and increased expression of the laminin-3 complex as seen in AD patients (Fig. 3b and Supplementary Fig. 2). Reactome pathway enrichment of these two protein networks for both species were most significant for GABA synthesis, release, release cycle, reuptake and degradation well as laminin interactions across both species (Fig. 3c and Supplementary Table 10). A heat map of GABA synapse related genes highlighting differential expression for non-bloom and bloom seasons (Fig. 3d).
Toxin exposure and temporal adaptation in gene expression
The IRL dolphin brain transcriptome was evaluated with respect to 2,4-DAB tissue concentration. We observed 63 gene transcripts that showed expression which were positively (44 genes) or negatively (19 genes) correlated with 2,4-DAB brain concentration (Fig. 4a and Supplementary Table 11). Of these 63 genes, 31 genes (49%) were identified in the following PPINs: positive regulation of cell migration, galactose metabolism, clathrin binding and RNA binding. One network did not have a significant enrichment. The differential effect of 2,4-DAB concentration on CCL27 and CCR2 gene expression, a chemokine and chemokine receptor respectfully, were the most significant (Supplementary Fig. 3).
a Correlation analysis for 2,4-DAB brain tissue concentration (n = 17 dolphins) vs. differential expressed (DE) genes. b Correlation analysis of year of stranding for non-bloom (n = 8 dolphins) and bloom (n = 9 dolphins) seasons vs. DE genes. c Venn diagram analysis of genes correlated with seasons (19 and 35 transcripts) and those correlated with 2,4-DAB brain tissue concentration (63 transcripts) identifies 15 overlapping DE gene transcripts significantly correlated with season, year, and 2,4-DAB brain tissue concentrations. d Correlation plots showing the impact of sequential bloom seasons on genes associated with Alzheimer’s disease (AD) susceptibility increased in bloom seasons. The gene encoding for cold-inducible RNA-binding protein (CIRBP), a stress response protein was increased across the entire dolphin cohort. e AD risk factor gene, apolipoprotein E (APOE; Cohen’s d = 1.07) along with early onset marker (NRG3; Cohen’s d = −1.53), death receptor 3 gene (TNFRSF25; Cohen’s d = 1.43) and mitochondrial dysfunction (MT-ND1; Cohen’s d = 0.59) were regulated by bloom seasons (n = 8 vs n = 9 dolphins per season). Error bars on boxplots indicate the minimum and maximum values. a & b were created in BioRender. Davis, D. (2025) https://BioRender.com/26bppmd and ChemDraw.
We also evaluated dolphin brain transcriptome expression by year of stranding. We observed 19 genes (non-bloom season) and 35 genes (bloom season) whose expression correlated with stranding year (Fig. 4b). We found 15 of these transcripts overlapped with the gene transcripts that were correlated with 2,4-DAB tissue concentration (Fig. 4c, Supplementary Tables 12 and 13). These overlapping genes observed are linked to neurological disorders such as AD, amyloidosis, diabetic neuropathy; sensorineural hearing loss, etc. Genes related to the development of AD, such as of advanced glycosylation end product receptor (AGER) gene, ARHGEF19 were observed to increase with each sequential bloom season Fig. 4d. These data suggest both 2,4-DAB toxin exposure, bloom season, and time may interact to regulate these transcripts.
Bloom season and Alzheimer’s disease risk factors
We observed increased transcription of the apolipoprotein E (APOE) gene, a risk factor and best predictive biomarker of AD up to 6.5-fold in some dolphins (Fig. 4e)22. APOE gene expression was also positively correlated with the expression of the AGER gene, which has shown to also be correlated with 2,4-DAB tissue concentration (Fig. 4d and Supplementary Fig. 4). Studies have shown increased in AGE receptor activity is associated with APOE e4 in humans22. Neuregulin 3 (NRG3) a promoter of excitatory synapse formation and crucial player in the development and function of the nervous system, was decreased by 36% (p = 0.0025, Mann–Whitney U). We also observed a 3-fold increase in the tumor necrosis factor receptor superfamily member 25 (TNFRSF25) or death receptor 3, responsible for apoptosis signaling and AD neurodegeneration (p = 0.0003, Mann U). Lastly, as mentioned above, we observed an increased in mitochondrially encoded NADH:Ubiquinone (MT-ND1 or ND1) during bloom seasons (Fig. 4e, also see Fig. 2b). MT-ND1 has been shown to be a blood biomarker for early stage AD23.
Dolphins and Alzheimer’s disease-like neuropathology
Genes encoding for the hallmark neuropathological changes observed in AD, such as the amyloid-β precursor protein (APP), microtubule-associated protein tau (MAPT), and TAR DNA binding protein (TARDBP), were found to be elevated with increased brain concentrations of 2,4-DAB and bloom season (Fig. 5a, e, i). Genes involved in familial forms of AD (PSEN1 and PSEN2), and frontotemporal dementia (GRN and C9orf72) expression was unchanged (Supplementary Fig. 5). The increased expression of APP, MAPT, and TARDBP genes was further supported by the presence of AD-like neuropathological changes in the dolphin brain, such as intracellular Aβ deposits, Aβ plaques (Fig. 5b–d), phosphorylated tau intracellular staining of pre-NFTs, tau neuropil threads (Fig. 5f–h), and phosphorylated TDP-43 intracytoplasmic inclusions (Fig. 5j–l)21,24. We observed varying degrees of AD-like neuropathological changes in all dolphins regardless of season. Thus, our data suggests 2,4-DAB exposure and bloom season may interact to exacerbate AD pathology.
a Expression of the amyloid precursor protein gene (APP) was increased with 2,4-DAB exposure. APP is cleaved to form the toxic peptide amyloid-β (Aβ). b Aβ deposits can be shown throughout the dolphin cortical layers. c Intracellular Aβ deposits and d Aβ plaques are the earliest event in the progression of AD neuropathological changes. e The microtubule associated protein tau (MAPT) gene was also increased with 2,4-DAB exposure. Abnormal tau proteins are hyperphosphorylated to form neurofibrillary tangles. f Superficial cortical layers 2 and 3 in our IRL dolphins were positive for Tau pThr217, a clinical biomarker for detection of early stage AD. g Differential expression of Tau pThr217 can be observed in neurons and h long neuritic threads. i Transactive response DNA binding protein 43 gene TARDBP encodes for the TDP-43 protein was increased with 2,4-DAB exposure. Phosphorylated TDP-43 intracytoplasmic inclusions have been shown in more than 50% of AD patients with a more progressive disease course. j, k TDP-43 inclusions are observed in neurons and glia cells in the cortex and in cells around the perivascular space (l). LOD: limit of detection; POS+: quantifiable levels of toxin. Sample size for all bar graphs NB LOD n = 5, NB POS+ n = 3, and Bloom POS+ n = 7, unpaired t-test (a, ND LOD vs. NB POS+) and Kruskal–Wallis (e, i). Error bars on bar graphs indicate the minimum and maximum values. Digital scan magnification = b, f (×10) j (×20) c, d, g, h, k, l (×40). Scale bar = b, f (500 μm); j (100 μm); c, d, g, h, k, l (50 μm).
Finally, shellfish poisoning (SP) is a syndrome which can cause dolphin stranding25. Neurotoxins associated with SP, such as okadaic acid and domoic acid, have been associated with memory impairment and have been used to induced AD-like pathology in experimental animal models26,27. To determine the extant SP may have contributed to transcriptomic changes in our cohort, a curated dataset of 8092 genes associated with SP was compared to our 536 DE dolphin genes. A third dataset of 29,056 curated genes associated with AD was also included to compare the similarity of all three datasets. We found 98% of the dolphin genes overlap with AD and roughly 42% of the genes overlapped with both AD and SP. However, we did not observe any of our dolphin genes overlapping exclusively with SP. Furthermore, transcription of genes encoding for proteins involved in SP illness (PPPC1A and GCLM) were unchanged in our dolphin cohort (Supplementary Fig. 6). Although, it is difficult to rule out the contributions of other HAB toxins in our cohort, our data suggests that the toxins responsible for SP do not play a major role in the observed transcriptomic changes reported here. However, further studies are needed to understand the complex interactions of HAB neurotoxins on the brain across the life span.
Discussion
In 2024, surface temperatures on Earth were the warmest recorded in modern times28. As global temperatures rise, HABs will continue to intensify, increasing the risk of exposure to algal toxins5. For the last 25 years, Florida’s IRL has experienced severe HABs that have been paralleled by a state-wide 3.3 °C average annual increase in temperature29. Here, we show that dolphins stranded during bloom seasons in the IRL had disproportionate exposures to the neurotoxin 2,4-DAB, increased expression of Alzheimer’s disease (AD) related genes, and transcriptomic changes reflecting altered GABAergic synapse and basement membrane alteration. Albeit the sample size of this study was limited due to the challenge of obtaining beached dolphins with the highest quality brain RNAs. With this stringent inclusion criteria, we analyzed dolphins with a median RNA integrity number (RIN) of 9.1. Nevertheless, the toxin associated increases in AD related gene expression in the IRL bottlenose dolphins (Tursiops truncatus) and concurrent AD-like neuropathological changes were similar to those observed in other cetaceans (Delphinus delphis, Phocoena phocoena) that had been exposed to cyanotoxins and found stranded in brackish water systems10,21.
Originally described as a neurolarthyrogen, 2,4-DAB has been shown to provoke hyperirritability, tremors, and convulsions within hours of exposure30. The toxin has also been classified as an excitatory amino acid due to its ability to stimulate membrane depolarization in a concentration dependent manner31. In addition, 2,4-DAB functions as a non-linear, non-competitive inhibitor of the enzyme gamma-aminobutyric acid (GABA) transaminase, which can caused elevated levels of GABA in the synapse and neurotoxicity32. Exposure to 2,4-DAB is suggested to be more toxic than its structural isomer BMAA, which has been linked to AD and has been co-detected in autopsied brains from AD patients as well as dolphins with AD-like changes7,10,33. Further studies are needed to elucidate the role 2,4-DAB plays in Alzheimer’s disease and related dementias and its potential synergistic effects with other HAB neurotoxins.
Dolphins stranded during bloom seasons had decreased expression of glutamate decarboxylase (GAD), an enzyme responsible for the conversion of the neurotransmitter glutamate to GABA34. Clinically, reduced function of GAD activity has been implicated in dyskinesia, mental, psychiatric, neurodegenerative, and neurodevelopmental disorders35. In AD, expression of both GAD1 and GAD2 genes are decreased, suggesting 2,4-DAB exposure could further exacerbate this pathological change36. Transcriptomic signatures of laminin-3 complex or S-laminin, also termed laminin-121 was also observed to be associated with 2,4-DAB exposure in the bloom season. Laminins are part of the basement membrane and interact with integrins to regulate many biological functions. Laminin-121 is located in the synapse, consists of in α1β2γ1 chain, supports neurite outgrowth, synaptogenesis, the blood brain barrier (BBB) and topologically complex assemblies of cells37,38. IRL dolphins had upregulation of the LAMA1, LAMA3 and LAMB2 genes that encode for laminin-121 and -321 isomers found in brain, liver, and muscle fibers. In high concentrations, the β2 chain is toxic to neurons and has been observed in Aβ plaques of AD patients39. The upregulation of laminin-3 complex could suggest a response to BBB injury and neuroinflammation in our dolphin cohort40.
Lastly, it has been reported that at least one half of stranded bottlenose dolphins have severe to profound hearing loss41. Many factors can contribute to hearing loss in dolphins, including age, birth defects, and noise pollution. Even acute loss of hearing is associated with behavioral effects42. In this study, we did not have data available regarding the hearing status in our dolphin cohort. However, we show several gene transcripts (MY1OF, STRC, SYNE4) correlated with both 2,4-DAB exposure, bloom season and year of stranding. In human, hearing loss is a risk factor for AD and can accelerate AD related neuropathology43. Given the effects of 2,4-DAB on the brain transcriptome, future studies are needed to understand the role of this molecule in the etiology of neurodegenerative diseases.
Methods
Study site
Florida’s IRL is defined as a shallow (<1 m), 250 km long microtidal estuarine system, which extends along five counties on the east coast of Florida from Ponce de Leon to the Jupiter Inlets and consists of three main interconnected sub-basins (Indian River, Banana River, and Mosquito Lagoon). In this study, the sample sites extended from Sebastian Inlet north to the through Halifax River and included the following sub-basins (Mosquito Lagoon, Banana River, Northern Indian River, North-Central Indian River, and Halifax River and tributaries (Tomoka River). The IRL has been subjected to several large-scale ecosystem changes in the last decade, including diminishing seagrass coverage associated with nutrient accumulation and declining water quality44,45. Likewise, multiple HABs have been documented within the IRL for decades.
Dolphins
Common bottlenose dolphins (Tursiops truncatus truncatus) that stranded within the study area were examined under a Stranding Agreement between Hubbs-SeaWorld Research Institute (HSWRI) and National Oceanic and Atmospheric Administration (NOAA) Fisheries. Dolphin carcasses were examined by established protocols46. Biological sex was determined by external examination of the genital region and internal examination of the reproductive organs. Total body length was measured as a straight-line length from the tip of the rostrum to the fluke notch and was used to estimate age class: adult male (>246 cm), adult female (>231 cm), juvenile male (161–245 cm), juvenile female (161–230 cm)47. To best eliminate influencing factors of demographics, nutritional status, and agonal states on toxin uptake and transcriptomic data, we selected dolphins with the highest possible quality postmortem brain specimens and ensured that control and experimental groups had equal representation in age class, biological sex, decomposition level, and cause of death. For our analysis dolphins were classified into two seasonal groups based on their date of stranding: non-bloom (December–May; n = 11) and bloom (June–November; n = 9) (Supplementary Table 1). The brain was extracted, cerebral cortex and cerebellum excised (~150 g), placed in a Whirl-Pak® bag (Wisconsin USA), and frozen at −80 °C until further analysis. A permit to obtain, store, and analyze dolphin brain samples was obtained from the National Marine Fisheries Service (NMFS) by the University of Miami (file: 1514-08b). The University of Miami Institutional Animal Care and Use Committee (IACUC) reviewed and authorized this study before receiving brain specimens. The handling of dolphin brain biospecimens satisfies the requirements of the Marine Mammal Protection Act pursuant to 50 CFR 216.22.
UHPLC-MS/MS
Triple quadrupole tandem mass spectrometry (UHPLC-MS/MS) was used to determine the presence of HAB toxins in IRL dolphin brain48. Dolphin brain samples (100 mg, wet weight) were hydrolyzed in HCl (6.0 M) [200 mg/ml] at 110 °C for 18 h. The pellet was hydrolyzed in 6.0 M HCl (333 mg/ml) at 110 °C for 16 h. Both the supernatant and hydrolyzed protein fractions were filtered through a centrifuge/ filtered (0.2 µm, ultrafree-MC GV Sigma–Aldrich) at 14,171 × g for 3 min and diluted (1/10 supernatant; 1/50 HCl) before derivatization with aminoquinolyl-N-hydroxysuccinimidyl carbamate (AQC, WAT052880, Waters Corp, Milford, MA) using 10 µl sample + 70 µl borate buffer mixed with internal standard (β-N-methyl-D3-amino-DL-alanine-15N2, Sigma Aldrich Custom Synthesis, 2011) and 20 µl AQC. The final concentration of internal standard in the derivatized sample was 0.055 ng/g. Each sample was injected three times into the mass spectrometer (3 µl injection volume) and the concentration of each isomer was calculated based on a standard curve with R2 = 0.99. The limit of detection (LOD) for all three isomers was 0.01 ng/g and lower limit of quantification (LLOQ) was calculated to be 0.037 ng/g for each isomer based on the US Food and Drug Administration methods for Instrument validation (2018-10926 (83 FR 23690). Analysis was conducted on a TSQ Quantiva triple quadrupole mass spectrometer attached to a Thermo Vanquish Ultra High-Pressure Liquid Chromatography Autosampler equipped with a Vanquish pump and heated column compartment. Separation of two BMAA isomers: N-[2-aminoethyl] glycine (AEG), 2,4-diaminobutyric acid (2,4-DAB)) from BMAA was achieved with gradient elution using 20 mM ammonium acetate, pH 5.0 (Fisher Scientific LiChropur™ ≥ 99%) (A) and 100% methanol (Chromasolv; Honeywell Burdick & Jackson, Muskegon, MI) (B) as follows: flow rate of 0.5 ml/min, initial conditions 10% B, 1.0 min 10% B, 4.8 mins 40% B (curve 5), 5.0 min 90% B (curve 5), 6.8 min 90% B, 6.81 min 10% B (curve 5), and 8 min 10% B. Separation was performed using a Thermo Hypersil Gold C-18 column (PN 25002–102130) 100 × 2.1 mm, particle size 1.9 µm heated to 65 °C. Samples were analyzed in positive ion, single reaction monitoring mode using heated electrospray ionization using previously published ion transitions. Mass spectrometer ion source properties were as follows: 3500 V-positive ion, 50 Arb Sheath gas, 10 Arb Aux gas, Sweep gas 0.1 Arb, vaporizer temperature 400 °C, and ion transfer tube temperature 350 °C. Validation curves and parameters were performed passing all criteria exceeding minimum requirements for a single-laboratory validation48. All samples were run and normalized with an internal BMAA standard. System blanks (AQC derivatized blanks, internal standards, and deionized water) were injected between sample injections.
RNA extraction and cDNA synthesis
Total RNA was extracted from approximately 100 mg of frozen dolphin cerebral cortex using RNeasy Lipid Tissue Mini Kit (Qiagen Inc., USA) following the manufacturer’s instructions. DNase I on-column treatment (Qiagen Inc., USA) was applied to samples to eliminate any genomic DNA contamination. RNA concentration was measured for each sample using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA). To determine the quality of RNA, Agilent 2100 Bioanalyzer (Agilent Technologies Inc., USA) was used to obtain an RNA integrity number (RIN) (Supplementary Table 4). cDNA library was generated from 5 μg of total RNA for each sample using the High-Capacity Reverse Transcription Kit (Thermo Fisher Scientific, USA).
Quantitative PCR
Seven genes that are known to be commonly involved in the pathogenesis of Alzheimer’s disease (AD) and several related dementias (APP, C9orf72, GRN, MAPT, PSEN1, PSEN2, and TARDBP) were selected for analysis. Gene expression was measured using custom dolphin AD PCR assays with a TaqMan Universal PCR Master Mix on a QuantStudio® 6 Flex Real-Time PCR System (Thermo Fisher Scientific, USA). Custom TaqMan assays were designed by the Thermo Fisher Bioinformatics Team based on the Tursiops truncatus genome turTur149 (Supplementary Table 14). Gene expression levels were normalized to 40S ribosomal protein S9 gene (RPS9) one of the most stable genes in cetacean species50. All samples were measured in triplicate and no template control (NTC) was performed for each assay. For each sample, 100 ng cDNA was amplified and run at the following conditions: 2 min at 50 °C, 10 min at 95 °C, 40 cycles: 15 s at 95 °C, and 1 min at 60 °C. Experimental set-up and data analysis were performed using QuantStudio® 6 Flex Real-Time PCR System Software v1.0 (Thermo Fisher Scientific, USA). All real-time PCR data files were imported into ExpressionSuite Software v1.0.4 (Applied Biosystems, USA) to analyze relative expression across all plates using comparative CT (ΔΔCT) method21. After the raw data was normalized to the internal control, the fold change of genes was calculated using the groups with the lowest detectable level of 2,4-DAB toxin as controls and the results were grouped by season.
Transcriptomics
Total dolphin RNAs extracted from the cerebral cortex were submitted for RNA sequencing at the John P. Hussman Institute for Human Genomics Center for Genome Technology at the University of Miami Miller School of Medicine (Miami, Florida, USA). RNAs (n = 17, median RIN of 9.1 (CI: 8.9–9.5) were prepared with the Nugen Universal Plus mRNA-Seq (M01442 v2) using 600 ng via Qubit and 14 cycles PCR. Libraries were sequenced on Illumina Novaseq 6000 and ~30 M reads were generated per sample. Primary RNAseq bioinformatics raw FASTQs were processed through a bioinformatics pipeline including adapter trimming by TrimGalore (v0.6.1) and Cutadapt (v2.3) with Python (v3.6.11) alignment with the STAR aligner 2.5.2a algorithm and gene counts or reads quantified against the Ensembl v101gene release database for turTru1 using the Gene Counts function in STAR. All protein encoding genes that had a raw read count were analyzed. Differential gene expression of gene counts for each dolphin were transformed and normalized with R software v4.2.1. We analyzed the differentially expressed genes for enriched pathways with GO Ontology for Biological Processes, Cellular Processes and Molecular Process using PANTHER Overrepresentation Test (Released 20240807) for GO Ontology database DOI: 10.5281/zenodo.14083199 Released 2024-11-03. Dolphin genes were compared against Homo sapiens (all genes in database). Statistical analyses used FISHER test with FDR correction. To identify potential protein-protein interaction networks (PPINs) between differentially expressed genes, we utilized the STRINGdb database accessed at https://string-db.org/with default parameters using medium confidence interaction (0.4). For comparative expression of GO terms in Alzheimer’s disease patients and dolphins, bulk transcriptomic data values from the frontal pole of AD vs non-cognitively impairment (NCI) patients were used from the Alzheimer’s DataLENS database (https://alzdatalens.partners.org/). For comparison of dolphin genes with curated genes for SP (MESH ID: D057096) and AD (MESH ID: D000544), curated gene and toxin interaction databases were obtained from the Comparative Toxicogenomic Database (CTD) (https://ctdbase.org/).
Immunohistochemistry
Archived formalin-fixed paraffin-embedded blocks from IRL dolphin necropsies (n = 7) were used to prepared brain sections on a Leica microtome (Leica Biosystems, Deer Park, IL, USA). Brain sections (7 μm) were mounted to positive charge slides dehydrated, deparaffinized and placed into 3% hydrogen peroxide for 10 min. Antigen retrieval was performed using heated 10 mM citric acid (pH 6.0) for 30 min or formic acid (Sigma-Aldrich, St. Louis, MO, USA) for 5 min. Following antigen retrieval, 5% goat serum (Vector Laboratories, Newark, CA, USA) was applied to tissues sections for 20 min to block nonspecific antibody binding. The following primary antibodies were incubated on tissue sections overnight at 4 °C: mouse monoclonal β-Amyloid 1–16 (6E10; 1.25 μg/mL; BioLegend, San Diego, CA, USA), rabbit polyclonal Phospho-Tau (Thr217; 2 μg/mL; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA); mouse monoclonal phospho- TDP-43 (Ser409/410; 1.0 μg/mL; Cosmo Bio, Carlsbad, CA, USA) in phosphate-buffered saline (PBS). The next day, goat-anti-rabbit IgG or horse-anti-mouse IgG biotinylated secondary antibodies were applied to brain sections (15 μg/mL; Vector Laboratories, Newark, CA, USA) for 60 min before adding the avidin–biotin complex for an additional 60 min (1:200, ABC; Vector Laboratories, Newark, CA, USA). To visualize the reaction, 3,3′-diaminobenzidine or DAB (Millipore Sigma, Burlington, VT, USA) was added to brain sections for 10 min. Postmortem brain tissue sections from AD patients were used as positive pathology controls. A 40× digital scan of each slide was generated using an EasyScan Pro 6 (Motic, Schertz, TX, USA) and exported to ObjectiveViewTM (Objective Pathology, Halton Hills, ON, Canada) and FIJI ImageJ VER2.00-rc-69/1.52p (National Institute of Health, USA) for analysis.
Statistical analyses
Statistical analyses and graphing of data sets were performed using Prism 9 (GraphPad Version 9.5.1 (528). Normality of datasets were determined using D’Agostino & Pearson test and the Shapiro–Wilk test. Single comparisons used Mann–Whitney U test and multiple comparisons Kruskal–Wallis with Dunn’s multiple comparisons test. Spearman R correlation was used to determine linear relationships of data sets. All statistical analyses performed were two-sided. Data is presented as the median and 95% confidence interval. Effect size (Cohen’s d) was calculated for single comparisons using mean values and standard deviations of the population. Hierarchical cluster and similarity matrix analysis of differentially expressed genes was performed using Morpheus software https://software.broadinstitute.org/morpheus. Venn diagrams were generated using Venny 2.1 software (https://bioinfogp.cnb.csic.es/tools/venny/index.html).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data presented here that support the findings of this study are available within the manuscript and the Supplementary Information. The numerical source data used to generate heatmaps and graphs are available for download in the figshare data repository (https://doi.org/10.6084/m9.figshare.29618978)51. RNA sequencing of brain tissue from estuarine dolphins can also be accessed by at NCBI Gene Expression Omnibus under accession number GSE303874 (https://www.ncbi.nlm.nih.gov/geo/).
Code availability
The code used in this study is publicly available and can be found on a GitHub repository at https://github.com/FelixKrueger/TrimGalore and also on Zenoob https://doi.org/10.5281/zenodo.7598955.
Change history
22 October 2025
In this article in the title lower case characters are used for “estuarine dolphins”, and university name for affiliation 3 were incorrectly given as “University of Miami School of Miami" but shoud have been “University of Miami School of Medicine “. The original article has been corrected.
References
Sneed, J. M. et al. Bloom dynamics and chemical defenses of benthic cyanobacteria in the Indian River Lagoon, Florida. Harmful Algae 69, 75–82 (2017).
Lapointe, B. E., Herren, L. W., Brewton, R. A. & Alderman, P. K. Nutrient over-enrichment and light limitation of seagrass communities in the Indian River Lagoon, an urbanized subtropical estuary. Sci. Total Environ. 699, 134068 (2020).
Alvarez, S., Brown, C. E., Garcia Diaz, M., O’Leary, H. & Solis, D. Non-linear impacts of harmful algae blooms on the coastal tourism economy. J. Environ. Manag. 351, 119811 (2024).
GISTEMP. GISS Surface Temperature Analysis (GISTEMP), version 4. NASA Goddard Institute for Space Studies. Dataset accessed 20YY-MM-DD, https://data.giss.nasa.gov/gistemp/ (2024).
Griffith, A. W. & Gobler, C. J. Harmful algal blooms: a climate change co-stressor in marine and freshwater ecosystems. Harmful Algae 91, 101590 (2020).
Brand, L. E., Pablo, J., Compton, A., Hammerschlag, N. & Mash, D. C. Cyanobacterial blooms and the occurrence of the neurotoxin beta-N-methylamino-L-alanine (BMAA) in South Florida Aquatic Food Webs. Harmful Algae 9, 620–635 (2010).
Garamszegi, S. P. et al. Detection of beta-N-methylamino-l-alanine in postmortem olfactory bulbs of Alzheimer’s disease patients using UHPLC-MS/MS: an autopsy case-series study. Toxicol. Rep. 10, 87–96 (2023).
Murch, S. J., Cox, P. A. & Banack, S. A. A mechanism for slow release of biomagnified cyanobacterial neurotoxins and neurodegenerative disease in Guam. Proc. Natl. Acad. Sci. USA 101, 12228–12231 (2004).
Hazen, E. L. et al. Marine top predators as climate and ecosystem sentinels. Front. Ecol. Environ. 17, 565–574 (2019).
Davis, D. A. et al. Cyanobacterial neurotoxin BMAA and brain pathology in stranded dolphins. PLoS ONE 14, e0213346 (2019).
Di Guardo, G. Do dolphins get Alzheimer’s disease? Vet. Rec. 185, 762 (2019).
Gunn-Moore, D., Kaidanovich-Beilin, O., Gallego Iradi, M. C., Gunn-Moore, F. & Lovestone, S. Alzheimer’s disease in humans and other animals: a consequence of postreproductive life span and longevity rather than aging. Alzheimers Dement. 14, 195–204 (2018).
Vacher, M. C. et al. Alzheimer’s disease-like neuropathology in three species of oceanic dolphin. Eur. J. Neurosci. 57, 1161–1179 (2023).
Durden, W. N. et al. Morbidity and mortality patterns of Indian River Lagoon common bottlenose dolphins (Tursiops Truncatus Truncatus) 2002-2020. J. Wildl. Dis. 59, 616–628 (2023).
Stolen, M. K. et al. An unusual mortality event for bottlenose dolphins links to altered diets resulting from ecological changes. Front. Mar. Sci. 12, https://doi.org/10.3389/fmars.2025.1531742 (2025).
Wucher, V., Sodaei, R., Amador, R., Irimia, M. & Guigo, R. Day-night and seasonal variation of human gene expression across tissues. PLoS Biol. 21, e3001986 (2023).
Van Dolah, F. M. et al. Seasonal variation in the skin transcriptome of common bottlenose dolphins (Tursiops truncatus) from the northern Gulf of Mexico. PLoS ONE 10, e0130934 (2015).
Better, M.A. 2024 Alzheimer’s disease facts and figures. Alzheimers Dement 20, 3708–3821 (2024).
Violi, J. P., Facey, J. A., Mitrovic, S. M., Colville, A. & Rodgers, K. J. Production of beta-methylamino-L-alanine (BMAA) and Its Isomers by Freshwater Diatoms. Toxins 11, https://doi.org/10.3390/toxins11090512 (2019).
Jo, M. et al. The role of TDP-43 propagation in neurodegenerative diseases: integrating insights from clinical and experimental studies. Exp. Mol. Med. 52, 1652–1662 (2020).
Garamszegi, S. P., Brzostowicki, D. J., Coyne, T. M., Vontell, R. T. & Davis, D. A. TDP-43 and Alzheimer’s disease pathology in the brain of a harbor porpoise exposed to the cyanobacterial toxin BMAA. Toxins 16, https://doi.org/10.3390/toxins16010042 (2024).
Deo, P., Dhillon, V. S., Chua, A., Thomas, P. & Fenech, M. APOE epsilon4 carriers have a greater propensity to glycation and sRAGE which is further influenced by RAGE G82S polymorphism. J. Gerontol. A Biol. Sci. Med. Sci. 75, 1899–1905 (2020).
Lunnon, K. et al. Mitochondrial genes are altered in blood early in Alzheimer’s disease. Neurobiol. Aging 53, 36–47 (2017).
LaFerla, F. M., Green, K. N. & Oddo, S. Intracellular amyloid-beta in Alzheimer’s disease. Nat. Rev. Neurosci. 8, 499–509 (2007).
Fire, S. E. et al. Co-occurrence of multiple classes of harmful algal toxins in bottlenose dolphins (Tursiops truncatus) stranding during an unusual mortality event in Texas, USA. Harmful Algae 10, 330–336 (2011).
Kaushal, A., Wani, W. Y., Bal, A., Gill, K. D. & Kaur, J. Okadaic acid and hypoxia induced dementia model of Alzheimer’s type in rats. Neurotox. Res. 35, 621–634 (2019).
Grattan, L. M. et al. Repeated dietary exposure to low levels of domoic acid and problems with everyday memory: research to public health outreach. Toxins 10, https://doi.org/10.3390/toxins10030103 (2018).
NOAA. (ed. NATIONAL CENTERS FOR ENVIRONMENTAL INFORMATION AND GLOBAL PRECIPITATION CLIMATOLOGY PROJECT (UNIVERSITY OF MARYLAND)) (https://www.climate.gov/, (2024).
Information, N. C. f. E. Florida statewide averaged mean monthly and annual temperature data, https://climatecenter.fsu.edu/products-services/data/statewide-averages/temperature (2024).
O’Neal, R. M., Chen, C. H., Reynolds, C. S., Meghal, S. K. & Koeppe, R. E. The ‘neurotoxicity’ of L-2,4-diaminobutyric acid. Biochem. J. 106, 699–706 (1968).
Spasic, S. et al. Extensive depolarization and lack of recovery of leech Retzius neurons caused by 2,4 diaminobutyric acid. Aquat. Toxicol. 199, 269–275 (2018).
Beart, P. M. & Bilal, K. l-2,4-diaminobutyric acid and the GABA system. Neurosci. Lett. 5, 193–198 (1977).
Martin, R. M., Bereman, M. S. & Marsden, K. C. The cyanotoxin 2,4-DAB reduces viability and causes behavioral and molecular dysfunctions associated with neurodegeneration in Larval Zebrafish. Neurotox. Res. 40, 347–364 (2022).
Erlander, M. G., Tillakaratne, N. J., Feldblum, S., Patel, N. & Tobin, A. J. Two genes encode distinct glutamate decarboxylases. Neuron 7, 91–100 (1991).
Monfort, J. C., Javoy-Agid, F., Hauw, J. J., Dubois, B. & Agid, Y. Brain glutamate decarboxylase in Parkinson’s disease with particular reference to a premortem severity index. Brain 108, 301–313 (1985).
Hill, M. A. & Gammie, S. C. Alzheimer’s disease large-scale gene expression portrait identifies exercise as the top theoretical treatment. Sci. Rep. 12, 17189 (2022).
Hall, Z. W. Laminin beta 2 (S-laminin): a new player at the synapse. Science 269, 362–363 (1995).
Sasaki, T. et al. Laminin-121-recombinant expression and interactions with integrins. Matrix Biol. 29, 484–493 (2010).
Murtomaki, S. et al. Laminin and its neurite outgrowth-promoting domain in the brain in Alzheimer’s disease and Down’s syndrome patients. J. Neurosci. Res. 32, 261–273 (1992).
Zapata-Acevedo, J. F. et al. Laminin as a biomarker of blood-brain barrier disruption under neuroinflammation: a systematic review. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms23126788 (2022).
Mann, D. et al. Hearing loss in stranded odontocete dolphins and whales. PLoS ONE 5, e13824 (2010).
Mooney, T. A., Nachtigall, P. E. & Vlachos, S. Sonar-induced temporary hearing loss in dolphins. Biol. Lett. 5, 565–567 (2009).
Lin, F. R. et al. Hearing loss and incident dementia. Arch. Neurol. 68, 214–220 (2011).
Sigua, G. C. & Tweedale, W. A. Watershed scale assessment of nitrogen and phosphorus loadings in the Indian River Lagoon basin, Florida. J. Environ. Manag. 67, 363–372 (2003).
Morris, L. J. et al. Seagrass in a Changing Estuary, the Indian River Lagoon, Florida, United States. Front. Mar. Sci. 8, https://doi.org/10.3389/fmars.2021.789818 (2022).
Geraci, J. R. & Lounsbury, V. J. Marine Mammals Ashore: A Field Guide for Strandings (National Aquarium in Baltimore, 1993).
The Committee on Marine Mammals American Society of, M & Norris, K. S. Standardized methods for measuring and recording data on the smaller cetaceans. J. Mammal. 42, 471–476 (1961).
Banack, S. A. Second laboratory validation of beta-N-methylamino-L-alanine, N-(2aminoethyl)glycine, and 2,4-diaminobuytric acid by ultra-performance liquid chromatography and tandem mass spectrometry. Neurotox. Res. 39, 107–116 (2021).
Leduc, R. G., Perrin, W. F. & Dizon, A. E. PHYLOGENETIC RELATIONSHIPS AMONG THE DELPHINID CETACEANS BASED ON FULL CYTOCHROME B SEQUENCES. Mar. Mamm. Sci. 15, https://doi.org/10.1111/j.1748-7692.1999.tb00833.x (2006).
Chen, I. H. et al. Selection of suitable reference genes for normalization of quantitative RT-PCR in peripheral blood samples of bottlenose dolphins (Tursiops truncatus). Sci. Rep. 5, 15425 (2015).
Davis, D. A. et al. Alzheimer’s disease signatures in the brain transcriptome of Estuarine Dolphins (Datasets). figshare. Datasets, https://doi.org/10.6084/m9.figshare.29618978 (2025).
Acknowledgements
We thank Teresa Jablonski of Hubbs SeaWorld Research Institute’s Marine Mammal Response Team for assistance with dolphin brain specimen collection. We thank Anthony Griswold and William Hulme of the John P. Hussman Institute for Human Genomics (HIHG) Genomic Center for Genome Technology for performing the RNA sequencing and differential gene expression analysis. We are very grateful for the helpful discussions of dolphin histology with Thomas C. Coyne of the District 8 Medical Examiner Office of Florida. We would also like to thank Joseph D. Ortiz of Kent State University for his helpful discussions. We greatly appreciate University of Miami Brain Endowment Bank staff members Wizner Canate and Nathalie Augustine-Adras for managing the storage and temperature monitoring of dolphin brain biospecimens. We are very grateful for the financial support of the Herbert W. Hoover Foundation. This work was also funded in part by the John H. Prescott Marine Mammal Rescue, Assistance Grant, SeaWorld Busch Gardens Conservation Fund, Discover Florida Ocean’s License Plate, Brevard County Tourism and Development Council.
Author information
Authors and Affiliations
Contributions
Conception and design: W.D., M.S., S.G. and D.D. Dolphin biospecimen collection: W.D. and M.S. Dolphin biospecimen dissection: D.D. Toxin testing: S.B. and P.C. qPCR analysis: S.G. Immunohistochemistry: D.B. and R.V. Data analysis: W.D., M.S., S.G., S.B. and D.D. Initial drafting of the manuscript: W.D., M.S., S.G. and D.D. Critical review and editing of the manuscript: W.D., M.S., S.G., S.B., D.B., R.V., L.B., P.C. and D.D.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Ibrahim Javed and Benjamin Bessieres. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Noke Durden, W., Stolen, M.K., Garamszegi, S.P. et al. Alzheimer’s disease signatures in the brain transcriptome of estuarine dolphins. Commun Biol 8, 1400 (2025). https://doi.org/10.1038/s42003-025-08796-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08796-0