Abstract
Acinetobacter baumannii emerges as one of the most worrisome pathogens causing a majority of hospital-acquired infections worldwide. However, the knowledge of its virulence factors remains obscure, in particular the bacterial lipooligosaccharides (LOS). Lipid A of the bacterial LOS exists as an inseparable mixture of homologues, making it impossible to study the immunological activity of each component. We herein report the synthesis of A. baumannii lipid A(s) (1−4) and corresponding monophosphate derivatives (1′, 3′ and 4′). The synthetic scheme features a short and stereoselective preparation of β-hydroxy acids and a convergent assembly of lipid A species with diverse structures. Subsequent immunological studies indicate that A. baumannii lipid A (2) having a [4 + 2]-acylation pattern displays the highest stimulatory potency. Further computational studies suggest that 2 and E. coli lipid A (5) sharing the same acylation pattern adopt an inverted binding mode in the TLR4/MD-2 receptor complex.

Similar content being viewed by others
Introduction
Acinetobacter baumannii (A. baumannii) is an opportunistic gram-negative pathogen causing a wide range of nosocomial infections1,2,3, including ventilator-associated pneumonia4, meningitis5, endocarditis6, catheter-related urinary tract infections7, and blood catheter tip-associated bacteremia8. With the propensity to withstand desiccation and rapid development of antibiotic resistance, the pathogen is able to spread rapidly throughout health care environments9,10. Despite the clinical significance, there still exists a knowledge gap on the infection biology of A. baumannii; only few virulence factors have been characterized so far, such as the efflux pumps for removal of antibiotic, unique transport systems for uptake of iron-(III) ions, and lipooligosaccharides (LOS) for modulation of the host immune responses11,12,13,14,15.
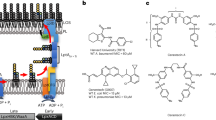
LOS is the structural component in the bacterial outer membrane, which is composed of a core oligosaccharide in conjugation with a glycolipid moiety, termed lipid A. Isolated LOS from wild-type A. baumannii exhibits potent stimulatory capacity for induction of the inflammatory signalling in human immune cells, though the lipid A component exists as a heterogenous mixture that vary in the number of fatty acyl chains16. Three lipid A species have been characterized in wild-type A. baumannii, including hepta-, hexa-, and penta-acylated lipid A(s) (1-3), (designated as acyl7, acyl6, and acyl5 lipid A hereafter) (Fig. 1)17. In particular mutants, symmetric derivative of acyl6 lipid A (4) and phosphoethanolamine derivative of acyl7 lipid A have also been identified10,17. Acyl7 1 is the most abundant species isolated from wild-type A. baumannii followed by is acyl6 2, and acyl5 3 is just barely detectable, according to MALDI-TOF mass spectrometric analysis. A unique feature of these lipid A species is the predominant 12 C acyl chains, which are two-carbon shorter than those found in Escherichia coli (E. coli) acyl6 lipid A (5), lipid IVA (6), and Salmonella serotype Typhimurium (S. Typhi) acyl7 lipid A (7). Furthermore, a hydroxylated secondary acyl chain is present at distal glucosamine (Glc2N) unit of 1-3, which is uncommon in enterobacterial lipid A(s)17,18,19.
A A. baumannii acyl7 lipid A (1) and monoP derivative 1ʹ. B A. baumannii acyl6 lipid A (2). C A. baumannii acyl5 lipid A (3) and monoP derivative 3ʹ. D Symmetric derivative of A. baumannii acyl6 lipid A (4) and monoP derivative (4′). E E. coli acyl6 lipid A (5) and lipid IVA (6). F. S. Typhi acyl7 lipid A (7).
Lipid A(s) belong to a class of pathogen-associated molecular patterns recognised by immune cells via the receptor complex of toll-like receptor 4 and myeloid differentiation factor 2 (TLR4/MD-2)20,21. The binding of lipid A to TLR4/MD-2 leads to dimerization and activation of the cytosolic Toll–interleukin-1 (IL-1) receptor (TIR) domain of the TLR4. Activated TIR domain recruits an adapter protein termed myeloid differentiation primary response protein 88 (MyD88) or Toll/IL-1R domain-containing adaptor-inducing IFN-β (TRIF), and then induces the activation of specific downstream signalling pathways22,23. In general, activation of the MyD88-dependent pathway mounts the primary innate immune responses24; while activation of the TRIF-dependent pathway is responsible for the induction of the adaptive immune responses25,26,27.
The key to induction of the immune response is the binding of lipid A to the TLR4/MD-2 receptor complex. A slight modification in the chemical structure of the lipid A is able to change the immunological responses by orders of magnitude, which is exemplified by E. coli lipid A (5) and its precursor lipid IVA (6)28,29,30,31. The former is a prototypical agonist to stimulate human immune cells, while the latter shows no agonistic activity. An acyl6 glucosaminide D-seryl conjugate has been shown to induce the activation of the TRIF signalling pathway selectively, but such selectivity was not observed for the L-seryl counterpart32.
As the lipid A component of A. baumannii LOS is heterogenous, it is imperative to acquire each of the lipid A species and elucidate their immunological properties. In the present study, we devised a versatile synthetic route to procure A. baumannii lipid A(s) (1−4) and corresponding monophosphate (monoP) derivatives (1′, 3′ and 4′) for investigating their structure–immune-activity relationship (SAR). In our study, A. baumannii lipid A(s) (1), (2), and symmetrically acylated derivative (4) were agonistic with stimulatory potencies varied with the number of acyl chains and their acylation patterns. In particular, acyl6 lipid A (2) was the most agonistic to induce the NF-κB activation in human monocytes. By contrast, acyl5 lipid A (3) showed no sign of stimulatory activity. Further computational docking and modelling analyses were applied to configure the binding mode of lipid A in the human TLR4/MD-2 receptor complex. The analyses suggest that the highest potency of 2 is attributable to the [4 + 2] acylation pattern and the dominant occupancy of the 12 C fatty acyl chains. Both features, coincidentally, are accompanied by an inverted binding mode of the di(Glc2N)2 scaffolds in the MD-2 pocket, which is different from the retained binding mode of other agonistic lipid A(s) 1, 4, and 7 with dissimilar acylation patterns.
Results and Discussion
Synthesis of A. baumannii lipid A(s)
In the past decades, dozens of synthetic routes have been explored for lipid A(s) and related analogues, which are broadly divided into two general strategies. One involves the prior preparation of lipidated Glc2N donor and acceptor followed by the convergent coupling33,34,35,36,37,38,39, while the other engages in iterative deprotection and acylation with an orthogonally protected di(Glc2N)2 scaffold40,41,42,43,44,45,46,47,48. Regardless the availability of these routes, the chemical synthesis of hyper-acylated lipid A(s) with six or more acyl chains remains as a daunting challenge. Major issues arise from (i) the tedious preparation of a variety of 3-hydroxyfatty and 3-acyloxyfatty acids, (ii) the propensity of the 3-acyloxyacyl chains to elimination, (iii) facile migration of the distal phosphate (PO4) group, (iv) the presence of a highly labile anomeric PO4 group36,38,39,40,44. Consequently, it is necessary to employ a short yet stereoselective scheme for the preparation of 3-hydroxyl and 3-acyloxyfatty acid building blocks and a reliable strategy to assemble the lipid A(s) with diverse structures.
Disconnection of the N-acyl chain at distal Glc2N moiety and the anomeric PO4 group at proximal Glc2N of the targets 1−4, 1′, 3′, and 4′ revealed the tri-, tetra-, and penta-lipidated disaccharide building blocks, and fatty acids 12b46 and 1419 (See Supplementary Fig. s1). The multi-lipidated disaccharides were derived from glycosylation coupling of Glc2N donors and acceptors 8–11. The acyl chains of 8–11 stemmed from fatty acids 12a19, 12b46, 13a49, and/or 13b50, and their Glc2N skeleton was built on thioglucosaminoside 1551. For Glc2N donors 8 and 9, the amino groups were masked with the trichloroethoxycarbonyl (Troc) protection, which controlled the β-stereoselectivity in glycosidic bond formation. Fatty acids 12a/b, 13a/b, and 14 were prepared by syn Aldol addition of (S)-4-benzyl-N-(bromoacetyl)-oxazolidinone 16 with appropriate alkanal, and/or lauric acid52,53,54,55.
At the onset, we prepared the fatty acid building blocks 12−14. Such fatty acids have been synthesized by a number of synthetic routes, but most of which suffer from some defects that limit the practicability46,49,50,56,57,58,59,60,61,62,63. We were inspired by the use of Evans asymmetric Aldol addition in chemical synthesis of several natural products52,64,65,66. In particular, the N-acetyl-(R)-4-benzylthiazolidine-2-thione has been used in preparation of (R)-3-hydroxyacyl components of solonamides via the non-Evans syn addition protocol, though the stereoselectivity was modest64. On the other hand, there remains no report on preparation of 3-hydroxy- and 3-acyloxyfatty acids such as 12-14 from N-bromoacetyloxazolidinone 16. Further, we reasoned that the jointed forces of the chiral auxiliary 16 and Evans syn addition conditions may improve the diastereoselectivity of the reaction65. Thus, 16 was enolized in the presence of TiCl4, diisopropylethylamine (DIEA), and N-methyl pyrrolidinone (NMP) (Fig. 2A). Subsequent addition of the titanium enolate intermediate to decanal (or dodecanal) afforded the expected Evans syn-addition product 17a (or 17b from dodecanal). From TLC examination and column chromatography purification, no anti-addition product was detected or isolated for characterization, suggesting an excellent anti/syn stereoselecitivity. This is presumably attributed to a non-chelated Zimmerman-Traxler transition state that proposed by Sunoj et al. (See inset)67.
Enantiomeric purity of of 17a and 17b was confirmed after debromination, which afforded N-(acyl)-oxazolidinone derivatives (3′R,4S)-18a and (3′R,4S)-18b, respectively. Meanwhile, the enantiomer of 18b, i.e., (3′S,4R)-ent-18b, was prepared from (4 R)-4-benzyl-N-(α-bromoacetyl)-oxazolidinone ent-16 (See inset of Fig. 2A). The enantiomeric purity of (3′S,4R)-ent-18b and (3′R,4S)-18b was examined with the chiral HPLC. In the HPLC chromatograms, no trace of undesired enantiomer was detected from 18b or ent-18b, hence confirming the enatiomeric purity of 18b prepared by present protocol (See Supplementary Fig. s2). The R-configuration of the 3-hydroxyl groups at 18a and 18b was confirmed by the negative optical rotation [α] of the hydrolysed products 19a and 19b, which agree with the literatures68,69,70.
To complete the preparation of (R)-3-benzyloxyfatty acids 12a and 12b, the 3-hydroxyl group at 18a and 18b was alkylated with benzyltrichloroacetimidate (BTCA) in acid-catalyzed conditions. This was followed by reductive hydrolysis of the oxazolidinone afforded the 3-benzyloxyfatty acids 12a and 12b. Preparation of the 3-acyloxyfatty acids 13a, 13b, and 14 started with advanced intermediates 19a and 19b, the carboxylic acid function of which was esterified with trichloroethanol to give trichloroethyl (TCE) esters 20a and 20b, respectively. Subsequent coupling of 20a and 20b with the lauric acid and 3-benzyloxylauric acid 12a followed by reductive cleavage of the TCE ester group, gave building blocks 13a, 13b, and 14 from 16 over six linear steps.
With the fatty acid building blocks in hands, the di- and tri-lipidated Glc2N acceptors 10 and 11 were prepared through standard protecting group manipulations and coupling procedues (Fig. 2B and Supplementary Fig. s3). The preparation of Glc2N donor 8 commenced with the coupling of thioglucosaminoside 15 with fatty acid 12a in the presence of dicyclohexylcarbodiimide (DCC) and 4-(N,N-dimethylamino)pyridine (DMAP) to provide 3-O-fatty acyl intermediate 21 (Fig. 3A). Subsequent reductive cleavage of the 4,6-O-benzylidene group followed by hydrolysis of the anomeric thioacetal obtained hemiacetal 23, which was converted to Glc2N imidate donor 8 by treatment with trichloroacetonitrile (Cl3CCN) and Cs2CO3.
Initial preparation of Glc2N donor 9 was analogous to that of 8 involving (i) the Steglich coupling of 15 with 3-acyloxyfatty acid 13a, (ii) reductive cleavage of the benzylidene acetal then (iii) followed by phosphorylation, thioacetal hydrolysis, and trichloroacetimidate formation (See Supplementary Fig. s4)42. Unexpectedly, elimination of the 3-acyloxyacyl moiety occurred in the reductive cleavage reaction despite the application of various reaction conditions34,38,70. To address the issue, 15 was alkylated by a known reductive etherification procedure to give 2-naphthylmethyl (Nap) protected thioglycoside 24 (Fig. 3B)71. Reductive cleavage of the 4,6-O-benzylidene acetal at 24 afforded thioglycoside 25 with a C4 hydroxyl for phosphorylation72,73,74,75. In the presence of dibenzyldiisopropyl-phosphoramidite [iPr2NP(OBn)2] and tetrazole73, 25 was converted to a phosphite intermediate, which via oxidation with dry trimethylamine N-oxide (TMAO) furnished phosphodiester 2676,77. Subsequent Nap deprotection and acylation with 3-acyloxyfatty acid 13a furnished 4-O-phosphoryl-3-O-acyloxyacyl intermediate 28. In the acylation, type of coupling reagent, reaction temperature, and amount of DMAP catalyst were optimized to prevent the elimination of the acyloxy group39,44. Final conversion of 28 to the designated Glc2N imidate donor 9 followed the same procedures as described in Fig. 3A.
With glycosyl donors and acceptors 8−11 in hands, the stage was set for assembly of the targets. Herein, we discussed only the syntheses of lipid A(s) 2, 3, and monoP derivative 3′ (Fig. 4) and the synthesis of 1, 4, and monoP derivatives 1′ and 4′ were given in the supplementary information due to the similarity of the synthetic routes (See Supplementary Fig. s5). Thus, Glc2N acceptor 10 was coupled with donors 8 and 9 to afford the tri- and tetra-lipidated disaccharides 30 and 31, respectively, in good to excellent yields (Fig. 4A). Next, the amino protecting group of 30 and 31 was removed for DCC-mediated coupling with (R)-3-((R)-3-hydroxyacyl)oxyfatty acid 14 to obtain the desired penta-lipidated and hexa-lipidated disaccharides 32 (derived from 30 and 14) and 33 (from 31 and 14). It should be noted that different carbodiimide reagents were used during the reaction course, such as EDC, DCC, and DIC. The reagent of choice depends on the cost effectiveness and ability to resist the elimination reaction. For example, DIC and catalytic DMAP were employed for the coupling of 4-O-phosphoryl 27 and 3-acyloxyfatty acid 13a, as the same acylation conditions have been found practical in previous synthesis of lipid A analogues39,44.
After the introduction of the fatty acyl chains, the anomeric allyl protecting group was cleaved to release the hydroxyl for phosphorylation. Thus, treatment of lipidated disaccharides 32 and 33 with OsO4 and NaIO4 deprotected the allyl group and obtained the hemiacetals 34 and 35, respectively (Fig. 4B)78. 34 and 35 were next subjected to phosphorylation, producing the fully protected lipid A(s) 36 and 37. In subsequent hydrogenolysis of the benzyl ether protections, a MeOH-THF cosolvent mixture and Pearlman’s reagent were employed (Conditions A)79. Under standard H2 (balloon) conditions, the hydrogenolysis proceeded gradually to give the desired lipid A(s) 2 and 3 with no cleavage of the anomeric PO4 group observed. However, when acetic acid (AcOH) was present in the above hydrogenolysis conditions, the anomeric PO4 was cleaved completely (Conditions B). Such a result was different from the previous synthesis of lipid A(s); in which the PO4 group remained viable in the presence of AcOH19,45. The inconsistency may be attributed to different solvent systems of the reactions. Nevertheless, the hydrogenolysis in the presence of the acetic acid, i.e., conditions B was still useful in converting the advanced intermediate 36 to monoP derivative 3′. In the end, the acidic protons of PO4 groups at 2, 3, and 3ʹ were exchanged with triethylammonium ions (Et3NH+) via treatment with the Et3NH+-bound resins.
The synthesis of 1, 4, and their monoP derivative 1′, and 4′ commenced the preparation of pentalipidated disaccharide 38 by glycosylation coupling of Glc2N donor 9 and acceptor 11 (Fig. 4C). Subsequent manipulations followed the aforementioned procedures used in the syntheses of 2, 3, and 3′ (See Supplementary Fig. s5).
Immunological properties
With the defined lipid A species (1-4) and monoP derivatives (1′, 3′, and 4′) in hand, we investigated their immunological properties and the related SAR. In the beginning, human THP1-Dual™ cells (InvivoGen)80 were treated with each of 1-3, 3′ and 5 [E. coli lipid A, isolated from E. coli F583 (Rd mutant), Molecular Depot] at different concentrations (i.e., 10−4 to 1 μg mL−1). After incubation, the level of NF-κB activation was measured with the secreted embryonic alkaline phosphatase activity81. Among the lipid A species, A. baumannii acyl7 1, acyl6 2, and E. coli lipid A 5 induced NF-κB activation in a dose-dependent fashion and both of 1 and 2 were more potent than 5 (Fig. 5).
On the other hand, neither of the A. baumannii acyl5 3 nor its monoP derivative 3′ displayed NF-κB activation at concentrations up to 1 μg mL−1. Furthermore, the treatment with 1 or 2 also stimulated the production of downstream pro-inflammatory cytokines such as TNF-α and IL-6 (Fig. 6)82,83. Although the trends for production levels of the cytokines are similar, the level of production is significantly lower for IL-6 than for TNF-α. The lower level of IL-6 production may be to unoptimized incubation time and/or undifferentiated THP1 cells used in the present assays84. Despite the difference, both results indicate that A. baumannii lipid A(s) 1 and 2, in particular the latter, are highly potent agonists, and that a minimum of six acyl chains appears to be critical for activation of the THP1 cells.
The THP-1 cells used are known to express a panel of TLRs, including TLR4 and TLR285. Previous studies of LPS(s) with atypical acyl chains showed that both TLR4 and TLR2 are involved to induce inflammatory responses86,87. To validate which of the TLR(s) is responsible for the NF-κB activation, we examined the stimulatory property with human embryonic kidney (HEK) cells that had been specifically transfected with either human TLR2 or TLR4 gene alongside with the human MD-2 and CD14 genes, i.e., HEK-Blue™ hTLR4 or hTLR2 cell lines (InvivoGen), respectively. In addition to E. coli lipid A 5, 6 (lipid IVA, a synthetic product from Biosynth Ltd) and 7 (S. Typhi lipid A, a synthetic product from Molecular Depot) were examined.
As shown in Fig. 7, lipid A(s) 1, 2, 4 and 5 with at six or seven acyl chains induced dose-dependently the NF-κB activation in HEK-Blue™ hTLR4 cells to different extents. S. Typhi acyl7 lipid A (7) and monoP derivatives 1′ and 4′ could induce the activation at a higher concentration, i.e., 100 ng mL−1 or above. On the other hand, no trace of NF-κB activation was induced by hypo-acylated 3, monoP derivative 3′ and lipid IVA (6) that have four or five acyl chains, even at 1 μg mL−1. In agreement with the conclusion drawn previously, six acyl chains are required for the lipid A species to exhibit immune-stimulatory property, though such a conclusion is based on those lipid A species with 12 C and/or 14 C acyl chains as the predominant lipid components86,88. Further, the impact of the number of PO4 groups on the immuno-stimulatory activity was manifested by comparing the potencies of 1 and its monoP derivative 1′, as well as those of 4 and 4′. Removal of the anomeric PO4 at 1 or 4 (i.e., 1′ or 4′) greatly reduced the activity, which is consistent with the previous dephosphorylation studies31,89.
To confirm the agonistic lipid A(s) to be TLR4 targeting, THP1-Dual™ cells were treated with 5 μg mL−1 of anti-hTLR4 antibody (InvivoGen, San Diego) and mouse IgG1 isotype control antibody (the negatiuve control; InvivoGen, San Diego) separately for 2 h. The antibody-pretreated THP-1 cells were then subjected to activation with each of the E. coli lipid A 5, A. baumannii acyl7 1, acyl6 2 at different concentrations for 20 h. Comparison with the treatment with the control antibody, the treatment with the anti-hTLR4 antibody completely abolished the NF-κB activation (See Supplementary Fig. s6). Further support is the significant difference in extent of NF-κB activation between the THP1-cells pretreated with the anti-hTLR4 and control antibodies.
To clarify whether or not the TLR2 receptor was involved in the NF-κB activation, HEK-Blue™ hTLR2 cells were separately subjected to activation with lipid A(s) 1, 2, 3, 5, and monoP derivative 3′ alongside with tripalmitoyl-CysSerLys4 (Pam3CSK4, a positive control) (See Supplementary Fig. s7). With the exception of for the Pam3CSK4, no NF-κB activation was observed for 1, 2, 3, 5, and 3′, suggesting that the observed NF-κB activation in Fig. 7 was solely due to the binding of 1, 2, and 5 to the TLR4 receptor.
Several structural features of lipid A molecules have been related to the agonistic potency, such as the number, chain length, and distribution pattern of the acyl chains, as well as the number of the PO4 groups90. In particular, the chain number and chain length have long been regarded as the dominant factors40,41,44. To delve deeper, we cross-compared the structures of 1-7 with their stimulatory activities in HEK-Blue™ hTLR4 cells.
Both of the hepta-acylated A. baumannii lipid A (1) and S. Typhi lipid A (7) sharing the same distribution pattern and number of the acyl chains. However, 1 exhibited much higher potency in the induction of the NF-κB activation than 7. There are two possible structural factors accounting for the lower potency of 7. The first should relate to the longer average chain length of the acyl chains in 7 and the second is the presence of is a 16 C secondary acyl chain at C2 position of its proximal Glc2N moiety (versus a 12 C secondary acyl chain in 1).
Next, we compared three hexa-acylated lipid A(s), including A. baumannii acyl6 2, acyl6 symmetric derivative 4, and E. coli acyl6 5. The chain length of acyl chains of 2 and 4 are almost identical but different only in the distribution pattern of acyl chains, i.e., [4 + 2] in 2 versus [3 + 3] in 4. Meanwhile, 2 and 5 have the same [4 + 2]-distribution pattern, but the chain lengths are different. With the identical chain length, [4 + 2]-acylated 2 exhibited a significantly higher potency than [3 + 3]-acylated 4. Moreover, 2 (in which four out of the six acyl chains are of 12 C and the remaining two are 14 C acyl chains) is more potent than 5 (in which five out of the six acyl chains are of 14 C and the other one is 12 C acyl chain). Apparently, both the 12C-chain length and [4 + 2]-distribution pattern are crucial for full expression of the TLR4-mediated stimulation. Note, the impact of the distribution pattern of acyl chains has been discussed only in a few publications19,41.
Furthermore, the hypoacylated lipid IVA (6) has been shown to compete with E. coli LPS for TLR4 binding. It is of interest to investigate if the hypoacylated A. baumannii lipid A (3) with shorter acyl chains also displayed similar competitive capability. Thus, the HEK-Blue™ hTLR4 cells were first pre-treated with each of 3, 3′, and 6, followed by stimulation with E. coli LPS. After incubation for 20 h, bis(phosphorylated) 3 and 6 showed inhibition on NF-κB activation to different extents; whereas, under the same conditions, no sign of inhibition was observed for monoP derivative 3′ (See Supplementary Fig. s8)91.
Computational docking and modelling analysis
To gain insight into the binding interactions of the synthesized lipid A molecules with TLR4/MD2 receptor complex, we conducted the computational docking and modelling analysis in accordance with the crystal structure of human TLR4/MD-2/E. coli LPS tetrameric complex (PDB: 3FXI)22. In this structure, the primary MD-2 displays a hydrophobic pocket for the binding with the lipid A moiety of the LPS. The binding of lipid A is critical to the dimerization of two TLR4/MD-2/LPS complexes, resulting in a tetrameric structure. In the following molecular models, we marked the secondary TLR4 and their amino acid residues in the tetramer complex with an asterisk (e.g., TLR4*) to distinguish them from those of the primary TLR4. The molecular models of bisphosphorylated lipid A(s), including 1, 2, 4, 5, and 7, were analyzed by AutoDock Vina 1.5.6. Several possible binding modes were generated from each lipid A binding with the TLR4/MD-2 complex; the optimized mode was determined based on two criteria: (i) the calculated lowest binding energy, and (ii) the distance between a PO4 group of the lipid A and cationic R264 of the primary TLR4. The distance should be no greater than or close to 3.5 Å according to the previous structural analysis22.
We first compared the binding mode of the lipid A component of E. coli LPS (which was extracted from the reported complex structure of human TLR4/MD-2/E.coli LPS) with that of A. baumannii acyl6 2 (Fig. 8A and 8B)22. Both were suggested to adopt an inverted binding mode in the TLR4/MD-2 receptor complex, in which the majority of the acyl chains are buried inside a hydrophobic pocket of the MD-2. The bis(phosphorylated) disaccharide scaffold is located at the opening rim of the pocket with the proximal and distal Glc2N units facing the secondary TLR4* and primary TLR4, respectively. In the literature, the binding of lipid A induces a conformation change of the MD-2 pocket, such as to shift the N-acyl chain of the proximal Glc2N to MD-2 surface22,85,92. Together, the proximal PO4 group and the surface-exposed acyl chain create the ionic and hydrophobic interfaces for interactions with the cationic and hydrophobic residues of the second TLR4. By contrast, non-agonistic lipid IVA (6) adopts a retained binding mode with MD-2 (PDB: 2E59, shown in Fig. 8C) where the positions of the PO4 groups and Glc2N moieties are reversed with respect to those mentioned in the inverted binding mode.
Since the PO4 groups of an agonistic lipid A play a critical role in inducing the TLR4/MD-2 dimerization via electrostatic interactions, it is of interest to probe if there are any connections between the binding mode, ionic distance, and stimulatory potency. To this end, we examined the optimized molecular models of 1, 2, 4, 5 and 7 to compare the ionic distances between their PO4 group and several cationic residues at TLR4* and TLR4 (Fig. 9). A. baumannii [4 + 2]-acylated 2 and 5 both bind to the MD-2 pocket with an inverted binding mode (Fig. 9B and 9D), similar to that of the E. coli LPS in the complex structure of TLR4/MD-2/E.coli LPS (Fig. 9A). In contrast, a retained binding mode was displayed by [4 + 3]-acylated lipid A(s) 1, 7, and [3 + 3]-acylated lipid A 4 (Fig. 9C, 9E, and 9F). The results suggest that the [4 + 2] acylation pattern togther with 12 C fatty acyl chains favour the inverted binding mode in the MD-2 pocket, which correlates with the highest potency of 2 in aforementioned immunological studies. Furthermore, to validate the binding mode predicted by the AutoDock Vina 1.5.6, we applied AlphaFold 3.0 to predict the binding modes of lipid A(s) 2 and 4 with TLR4/MD-2 receptor complex93. AlphaFold 3.0 has been shown to accurately predict the protein-ligand biomolecular interactions94,95. To our delight, the binding modes of 2 and 4 with TLR4/MD2 complex predicted by AlphaFold 3.0 were also in agreement with those predicted by AutoDock Vina 1.5.6.
A Binding modes of lipid A moiety of E. coli LPS. B Binding mode of A. baumannii acyl6 lipid A (2). C Binding mode of A. baumannii acyl7 lipid A (1). D Binding mode of E. coli acyl6 lipid A (5). E Binding mode of A. baumannii symmetric acyl6 lipid A derivative (4). F Binding mode of S. Typhi acyl7 lipid A (7). Cationic residues of TLR4, MD-2, and TLR4* are shown in blue, wheat, and green, respectively. The distances between the PO4 groups and the residues are depicted by black dotted lines.
In addition to the binding mode, we examined the distance between the PO4 group of synthetic lipid A (or E. coli LPS) and specific cationic amino acid residue at the TLR4 receptors (See supplementary Table s1). For E. coli LPS, A. baumannii acyl6 2 and A. baumannii acyl7 1, the ionic distances between the K388 of the TLR4* and the PO4 group are 7.3, 8.2, and 9.0 Å, respectively (Table s1, entries 1−3). The distances are shorter than those observed from E. coli acyl6 5 (12.3 Å), A. baumannii acyl6 symmetric derivative 4 (14.2 Å), and S. Typhi acyl7 7 (15.2 Å) (Table s1, entries 4−6). Apparently, the shorter is the distance, the stronger is the stimulatory potency.
It should be mentioned that the computational docking methods including AutoDock Vina 1.5.6. and AlphaFold 3.0 rely on static conformation extracted from the crystal structure of TLR4/MD-2/E. coli LPS complexes; which however do not account for conformational changes of the protein receptors upon interacting with lipid A. Although the predicted binding modes of 1-7 in Fig. 8 provide insight to relationships between the structure, TLR4-binding, and stimulatory activity, the accuracy is still limited. Further validation with the molecular dynamic (MD) simulation would be done in future endeavour96,97.
Conclusions
To understand how the lipid A component of A. baumannii LOS functions in the pathogenesis, herein we report for the first total synthesis of A. baumannii lipid A(s) (1−4) and preparation of monophosphate derivatives 1ʹ, 3ʹ and 4ʹ for immunological studies. The results indicate that A. bau acyl7 lipid A (1), acyl6 lipid A (2) are highly agonistic, while hypoacylated acyl5 lipid A (3) shows no immune-stimulatory activity. Supported by further SAR analysis, both the chain length and asymmetric [4 + 2]-acylation pattern are essential for full expression of the agonistic potency, which is likely associated with the inverted binding mode in the TLR4/MD-2 receptor complex. In addition, we identified the correlation of the stimulatory potency with the ionic distance between the PO4 group of lipid A and K388 residue in TLR*. The acquired SAR should be valuable to chemists for designing artificial immune-modulating agents.
Experimental
Experimental procedures for syntheses of A. baumannii lipid A(s) 1-4, monophosphoryl derivatives 1′, 3′, and 4′, corresponding spectroscopic data, and NMR spectra are available in the supplementary information and supplementary data 1.
NF-kB activation, TNF-α, and IL-6 in THP1-Dual™ cells
THP1-Dual™ cells, purchased from InvivoGen (San Diego), were cultured in RPMI 1640 (Gibco, Hampton) and supplemented with 10% heat-inactivated foetal bovine serum (Gibco, Hampton), 100 μgmL−1 Normocin™ (InvivoGen, San Diego), 100 U mL−1 penicillin and 100 μg mL−1 Strep (Biological Industries, Kibbutz Belt-Haemek). Synthesized or purchased lipid A compounds were first dissolved in DMSO and diluted to appropriate concentrations (as shown in the figures). To determine the degree of NF-κB activation, THP1-Dual™ cells were seeded at the seedling density of 1 × 105 cells, treated with each lipid A, and incubated for 20 h at 37 oC in the presence of 5% CO2. The culture supernatant (20 μL) was collected and added to Quanti-Blue™ solution (180 μL), then incubated at 37 oC for 1 h. The level of secreted embryonic alkaline phosphatase was quantified with a spectrophotometer at 620 nm. Meanwhile, the culture supernatant was also used to determine the concentrations of the induced cytokines TNF-α and IL-6 by using commercially available ELISA kits (Duo Set Kit, RandD Systems, Minneapolis) according to manufacturer’s protocol.
NF-kB activation in HEK-Blue™ hTLR4 and hTLR2 cells
HEK-Blue™ hTLR2 and HEK-Blue™ hTLR4, purchased from InvivoGen (San Diego), were cultured in DMEM (Gibco, Hampton) and supplemented with 4.5 g L−1 D-glucose, L-glutamine, 10% heat-inactivated foetal bovine serum (Gibco, Hampton), 100 μg mL−1 Normocin™ (Invivogen, San Diego), 100 U mL−1 penicillin and 100 μg mL−1 Strep (Biological Industries, Kibbutz Belt-Haemek). To determine the degree of NF-κB activation, the remaining procedure was the same as the aforementioned.
Docking and MD simulation of A. baumannii lipid A(s) (1-4), E. coli lipid A (5), lipid IVa (6), and S. typhi lipid A (7). Predicted binding modes to human TLR4
Computational studies of lipid A(s) were performed by using Auto Duck Vina 1.5.6. Molecular models of agonistic lipid A(s) (1, 2, 4, 5, and 7) were calculated based on the complex structures of hTLR4/hMD-2/E. coli LPS (PDB: 3FXI). The complex structure of hMD-2/lipid IVa (PDB: 2E56) was utilized to calculate the binding mode of non-agonistic lipid A (6). Kollman charges and polar hydrogen atoms, ligands were docked individually to the binding site of MD-2 with a grid box, performed with AutoDockTools 1.5.7. The grid size setting was at 90 × 66 × 88 (x, y, and z) points, and the grid centers were designated at x, y, and z dimensions of −11.164, −17.738, and −31.462, respectively, with a grid spacing of 0.375 Å.
Data availability
Synthetic procedures, characterization data, and source data for the biological assays and computational studies supporting the findings of the research have been included in the main text, supplementary information (SI), supplementary data 1 & 2, or will be available from the corresponding authors upon request.
References
Peleg, A. Y., Seifert, H. & Paterson, D. L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 (2008).
Russo, A. et al. Multidrug-resistant Acinetobacter baumannii infections in COVID-19 patients hospitalized in intensive care unit. Infection 20, 83–92 (2022).
Rangel, K., Chagas, T. P. G. & De-Simone, S. G. Acinetobacter baumannii infections in times of COVID-19 pandemic. Pathogens 10, 1006 (2021). article.
Garnacho-Montero, J. et al. Acinetobacter baumannii ventilator-associated pneumonia: epidemiological and clinical findings. Intensive Care Med. 31, 649–655 (2005).
Krol, V., Hamid, N. S. & Cunha, B. A. Neurosurgically related nosocomial Acinetobacter baumannii meningitis: report of two cases and literature review. J. Hosp. Infect. 71, 176–180 (2009).
Laganà, P., Melcarne, L. & Delia, S. Acinetobacter baumannii and endocarditis, rare complication but important clinical relevance. Int. J. Cardiol. 187, 678–679 (2015).
Pour, N. K. et al. Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Microbiol. Immunol. 62, 328–338 (2011).
Beck-Saguė, C. M. et al. Epidemic bacteremia due to Acinetobacter baumannii in five intensive care units. Am. J. Epidemiol. 132, 723–733 (1990).
Karalewitza, A. P.-A. & Miller, S. I. Multidrug-resistant Acinetobacter baumannii chloramphenicol resistance requires an inner membrane permease. Antimicrob. Agents Chemother. 62, e00513–e00518 (2018).
Boll, J. M. et al. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 6, e00478–15 (2015).
March, C. et al. Dissection of host cell signal transduction during Acinetobacter baumannii – triggered inflammatory response. PLoS One 5, e10033 (2010).
Aliramezani, A., Soleimani, M., Mazaheri, R., Fard, N. & Nojoomi, F. Virulence determinants and biofilm formation of Acinetobacter baumannii isolated from hospitalized patients. Germs 9, 148–153 (2019).
Bartholomew, T. L., Kidd, T. J., Pessoa, J. S. á, Álvarez, R. C. & Bengoechea, J. A. 2-Hydroxylation of Acinetobacter baumannii lipid A contributes to virulence. Infect. Immun. 87, e00066–19 (2019).
Griffiss, J. M. & Schneider, H. The chemistry and biology of lipooligosaccharides: The endotoxins of bacteria of the respiratory and genital mucosae in endotoxin in health and disease. Editor: Helmut Brade, 1st Ed., 1999, C. R. C.: Boca Raton.
Erridge, C., Bennett-Guerrero, E. & Poxton, I. R. Structure and function of lipopolysaccharides. Microbes Infect. 4, 837–838 (2002).
Erridge, C., Moncayo-Nieto, O. L., Morgan, R., Young, M. & Poxton, I. R. Acinetobacter baumannii lipopolysaccharides are potent stimulators of human monocyte activation via Toll-like receptor 4 signaling. J. Med. Microbiol. 56, 165–171 (2007).
Beceiro, A. et al. Phosphoethanolamine Modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55, 3370–3379 (2011).
Pelletier, M. R. et al. Unique structural modifications are present in the Lipopolysaccharide from Colistin-resistant strains of Acinetobacter baumannii. Antimicrob. Agents Chemother. 57, 4831–4840 (2013).
Shimoyama, A. et al. Lipopolysaccharide from gut-associated lymphoid-tissue-resident Alcaligenes faecalis: Complete structure determination and chemical synthesis of its lipid A. Angew. Chem. Int. Ed. 60, 10023–10031 (2021).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Di Lorenzo, F. et al. A journey from structure to function of bacterial lipopolysaccharides. Chem. Rev. 122, 15767–15821 (2022).
Park, B. et al. The structural basis of lipopolysaccharide recognition by the TLR4/MD-2 complex. Nature 458, 1191–1195 (2009).
Deguine, J. & Barton, G. M. MyD88: a central player in innate immune signaling. F1000Prime Rep. 6, 97 (2014).
Yamamoto, M. et al. Role of Adaptor TRIF in the MyD88-Independent Toll-Like Receptor Signaling Pathway. Science 301, 640–642 (2003).
Ullah, M. O., Sweet, M. J., Mansell, A., Kellie, S. & Kobe, B. TRIF-dependent TLR signaling, its functions in host defense and inflammation, and its potential as a therapeutic target. J. Leukoc. Biol. 100, 27–45 (2016).
Sica, A. & Mantovani, A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 122, 787–795 (2012).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signalling in inflammation. Sig. Transduct. Target Ther. 2, 17023 (2017).
van der Poll, T. & van Deventer, S. J. H. Cytokines and anti-cytokines in the pathogenesis of sepsis. Infect. Dis. Clin. North Am. 13, 413–426 (1999).
Saitoh, S.-I. et al. Lipid A antagonist, lipid IVa, is distinct from lipid A in interaction with Toll-like receptor 4 (TLR4)-MD-2 and ligand-induced TLR4 oligomerization. Int. Immunol. 16, 961–969 (2004).
Aybay, C. & Imir, T. Comparison of the effects of Salmonella minnesota Re595 lipopolysaccharide, lipid A and monophosphoryl lipid A on nitric oxide, TNF-alpha, and IL-6 induction from RAW 264.7 macrophages. FEMS Immunol. Med. Microbiol. 22, 263–273 (1998).
Johnson, D. A. et al. 3-O-Desacyl monophosphoryl lipid A derivatives: Synthesis and immunostimulant activities. J. Med. Chem. 42, 4640–4649 (1999).
Bowen, W. S. et al. Selective TRIF-dependent signaling by a synthetic Toll-like receptor 4 agonist. Sci. Signal. 5, ra13 (2012).
Imoto, M., Yoshimura, H., Sakaguchi, N., Kusumoto, S. & Shiba, T. Total synthesis of Escherichia coli Lipid A, the endotoxically active principle of cell-surface lipopolysaccharide. Bull. Chem. Soc. Jpn. 60, 2205–2214 (1987).
Liu, W., Oikawa, M., Fukase, K., Suda, Y. & Kusumoto, S. A Divergent synthesis of lipid A and its chemically stable unnatural Analogues. Bull. Chem. Soc. Jpn. 72, 1377–1385 (1999).
Christ, W. J. et al. Total synthesis of proposed structure of Rhodobacter sphaeroides lipid A resulting in the synthesis of new potent lipopolysaccharide antagonist. J. Am. Chem. Soc. 116, 3637–3638 (1994).
Zamyatina, A., Sekljic, H., Brade, H. & Kosma, P. Synthesis and purity assessment of tetra- and pentaacyl lipid A of Chlamydia containing (R)-3-hydroxyicosanoic acid. Tetrahedron 60, 12113–12137 (2004).
Kumada, H. et al. Biological properties of the native and synthetic lipid A of Porphyromonas gingivalis lipopolysaccharide. Oral. Microbiol. Immunol. 23, 60–69 (2008).
Tang, S.-C., Wang, Q.-L. & Guo, Z.-W. Synthesis of a monophosphoryl derivative of Escherichia coli lipid A and its efficient coupling to a tumor-associated carbohydrate antigen. Chem. Eur. J. 16, 1319–1325 (2010).
Hollaus, R. et al. Chemical synthesis of Burkholderia lipid A modified with glycosyl phosphodiester-linked 4-amino-4-deoxy-β-L-arabinose and its Immunomodulatory potential. Chem. Eur. J. 21, 4102–4114 (2015).
Zhang, Y.-H., Gaekwad, J., Wolfert, M. A. & Boons, G.-J. Modulation of innate immune responses with synthetic lipid A derivatives. J. Am. Chem. Soc. 129, 5200–5216 (2007).
Zhang, Y.-H., Gaekwad, J., Wolfert, M. A. & Boons, G.-J. Innate immune responses of synthetic lipid A derivatives of Neisseria meningitidis. Chem. -Eur. J. 14, 558–569 (2008).
Fujimoto, Y., Shimoyama, A., Suda, Y. & Fukase, K. Synthesis and immunomodulatory activities of Helicobacter pylori lipophilic terminus of lipopolysaccharide including lipid A. Carbohydr. Res. 356, 37–43 (2012).
Fujimoto, Y. et al. Innate immunomodulation by lipophilic termini of lipopolysaccharide; synthesis of lipid A from Porphyromonas gingivalis and other bacteria and their immunomodulative responses. Mol. BioSyst. 9, 987–996 (2013).
Adanitsch, F. et al. Development of αGlcN(1↔1)αMan-based lipid A mimetics as a novel class of potent Toll-like receptor 4 agonists. J. Med. Chem. 57, 8056–8071 (2014).
Yamaura, H. et al. Chemical synthesis of Acetobacter pasteurianus lipid A with a unique tetrasaccharide backbone and evaluation of its immunological functions. Angew. Chem. Int. Ed. 63, e202402922 (2024).
Fukase, K. et al. Divergent synthesis and biological activities of lipid A analogues of shorter acyl chains. Tetrahedron 54, 4033–4050 (1998).
Fukase, Y. et al. New efficient route for synthesis of lipid A by using affinity separation. Synlett 2001, 1693–1698 (2001).
Shimoyama, A. et al. Chemical synthesis of Helicobacter pylori lipopolysaccharide partial structures and their selective proinflammatory responses. Chem. Eur. J. 17, 14464–14474 (2011).
Gao, L.-Q. et al. Full synthesis and bioactivity evaluation of Tn-RC-529 derivative conjugates as self-adjuvanting cancer vaccines. Chin. Chem. Lett. 32, 3011–3014 (2021).
Oikawa, M. & Kusumoto, S. On a practical synthesis of β-hydroxy fatty acid derivatives. Tetrahedron.: Asymmetry 6, 961–966 (1995).
Lin, S.-C., Chao, C.-S., Chang, C.-C. & Mong, K.-K. T. Joined use of oxazolidinone and desymmetric amino protection: a new strategy for protection of glucosamine. Tetrahedron Lett. 51, 1910–1913 (2010).
Evans, D. A. & Weber, A. E. Synthesis of the cyclic hexapeptide Echinocandin D. New approaches to the asymmetric synthesis of β-hydroxy-α-amino acids. J. Am. Chem. Soc. 109, 7151–7157 (1987).
Evans, D. A., Bartroli, J. & Shih, T.-L. Enantioselective aldol condensations. 2. Erythro-selective chiral Aldol condensations via boron enolates. J. Am. Chem. Soc. 103, 2127–2129 (1981).
Crimmins, M. T., King, B. W. & Tabet, E. A. Asymmetric aldol additions with titanium enolates of N-acyl oxazolidinethiones: Dependence of selectivity on amine base and Lewis acid stoichiometry. J. Am. Chem. Soc. 119, 7883–7884 (1997).
Crimmins, M. T., King, B. W., Tabet, E. A. & Chaudhary, K. Asymmetric aldol additions: use of titanium tetrachloride and (-)-sparteine for the soft enolization of N-acyl oxazolidinones, oxazolidinethiones, and thiazolidinethiones. J. Org. Chem. 66, 894–902 (2001).
Keegan, D. S., Hagen, S. R. & Johnson, D. A. Efficient asymmetric synthesis of (R)-3-hydroxy- and alkanoyloxytetradecanoic acids and method for the determination of enantiomeric purity. Tetrahedron.: Asymmetry 7, 3559–3564 (1996).
Reddy, R. G. et al. Fellutamide B synthetic path intermediates with in vitro neuroactive function shows mood-elevating effect in stress-induced zebrafish model. ACS Omega 3, 10534–10544 (2018).
Bourboula, A., Limnios, D., Kokotou, M. G., Mountanea, O. G. & Kokotos, G. Enantioselective Organocatalysis-Based Synthesis of 3-Hydroxy Fatty Acids and Fatty γ-Lactones. Molecules 24, 2081 (2019).
Jadhav, P. K. Asymmetric synthesis of (3R)-alkanoyloxytetradecanoic acids-components of bacterial lipopolysaccharides. Tetrahedron Lett. 30, 4763–4766 (1989).
Tai, A. et al. A facile method for preparation of the optically pure 3-hydroxytetradecanonic acid by an application of asymmetrically modified Nickel catalyst. Chem. Lett. 9, 1125–1126 (1989).
Cameron, G., Nguyen, T., Ciula, M., Williams, S. J. & Godfrey, D. I. Glycolipids from the gut symbiont Bacteroides fragilis are agonists for natural killer T cells and induce their regulatory differentiation. Chem. Sci. 14, 7887–7896 (2023).
Bauer, J., Brandenburg, K., Zähringer, U. & Rademann, J. Chemical synthesis of a Glycolipid library by a solid-phase strategy allows elucidation of the structural specificity of immune-stimulation by Rhamnolipids. Chem. Eur. J. 12, 7116–7124 (2006).
Guaragna, A., De Nisco, M., Pedatella, S. & Palumbo, G. Studies towards lipid A: a synthetic strategy for the enantioselective preparation of 3-hydroxy fatty acids. Tetrahedron: Asymmetry 17, 2839–2841 (2006).
Kitir, B., Baldry, M., Ingmer, H. & Olsen, C. A. Total synthesis and structural validation of cyclodepsipeptides solonamide A and B. Tetrahedron 70, 7721–7732 (2014).
Sato, E., Tanabe, Y., Nakajima, N., Ohkubo, A. & Suenaga, K. Total synthesis of biselyngbyolide B. Org. Lett. 18, 2047–2049 (2016).
Ohtawa, M. et al. Total synthesis and absolute configuration of Simpotentin, a potentiator of amphotericin B activity. Org. Lett. 21, 5596–5599 (2019).
Sreenithya, A. & Sunoj, R. aghavanB. Non-innocent role of N‑methyl pyrrolidinone in thiazolidinethione-promoted asymmetric Aldol reactions. Org. Lett. 14, 5752–5755 (2012).
Nakahata, M., Imaida, M., Ozaki, H., Harada, T. & Tai, A. The preparation of optically pure 3-hydroxyalkanoic Acid. The enantioface-differentiating hydrogenation of the C=O double bond with modified raney nickel. Bull. Chem. Soc. Jpn. 55, 2186–2218 (1982).
Th. Rietschel, E., Gottert, H., Lüderitz, O. & Westph, O. Nature and linkages of the fatty acids present in the lipid-A component of Salrnonella lipopolysaccharides. Eur. J. Biochem 28, 166–173 (1972).
Verpalen, E. C. J. M., Brouwer, A. J. & Boons, G.-J. Synthesis of monophosphoryl lipid A using 2-naphtylmethyl ethers as permanent protecting groups. Carbohydr. Res. 498, 108152 (2020).
Chen, C.-W., Wang, C.-C., Li, X. R., Witek, H. & Mong, K.-K. T. Sub-stoichiometric reductive etherification of carbohydrate substrates and one-pot protecting group manipulation. Org. Biomol. Chem. 81, 3135–3141 (2020).
Owa, T. & Ouchi, S. The facile synthesis of 5′-nucleotides by the selective phosphorylation of a primary hydroxyl group of nucleosides with phosphoryl chloride. Bull. Chem. Soc. Jpn. 48, 2084–2090 (1975).
Perich, J. W. & Johns, R. B. A new, convenient and efficient general procedure for the conversion of alcohols into their dibenzyl phosphorotriesters using N,N-diethyl dibenzyl phosphoramidite. Tetrahedron Lett. 28, 101–102 (1987).
Watanabe, Y., Komoda, Y., Ebisuya, K. & Ozaki, S. An efficient phosphorylation method using a new phosphitylating agent, 2-diethylamino-1,3,2-benzodioxaphosphepane. Tetrahedron Lett. 31, 255–256 (1990).
Domon, K. et al. Catalytic chemoselective O‑phosphorylation of alcohols. ACS Cent. Sci. 6, 283–292 (2020).
Hayakawa, Y., Uchiyama, M. & Noyori, R. Non-aqueous oxidation of nucleoside phosphites to the phosphates. Tetrahedron Lett. 27, 4191–4194 (1986).
Soderquist, J. A. & Anderson, C. L. Crystalline anhydrous trimethylamine N-oxide. Tetrahedron Lett. 27, 3961–3962 (1986).
Kitov, P. I. & Bundle, D. R. Mild oxidative one-pot Allyl group cleavage. Org. Lett. 3, 2835–2838 (2001).
Pearlman, W. M. Noble metal hydroxides on carbon nonpyrophoric dry catalysts. Tetrahedron Lett. 8, 1663–1664 (1967).
Chanput, W., Mes, J. J. & Wichers, H. J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 23, 37–45 (2014).
Wang, M.-Y., Wang, X.-Y. & Ye, H.-F. Measurement of secreted embryonic alkaline phosphatase. Bio Protoc. 13, e4600 (2023).
Adachi, O. et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9, 143–150 (1998).
Ulevitch, R. J. Therapeutics targeting the innate immune system. Nat. Rev. Immunol. 4, 512–520 (2004).
Liu, X.-F. et al. LPS‑induced proinflammatory cytokine expression in human airway epithelial cells and macrophages via NF‑κB, STAT3 or AP‑1 activation. Mol. Med. Rep. 17, 5484–5491 (2018).
Heine, H. et al. Tailored modulation of cellular pro-inflammatory responses with disaccharide lipid A mimetics. Front. Immunol. 12, 631797 (2021).
Lorenzo, F. D. et al. Pairing Bacteroides vulgatus LPS structure with its immunomodulatory effects on human cellular models. ACS Cent. Sci. 6, 1602–1616 (2020).
Francisco, S. et al. Induction of TLR4/TLR2 interaction and heterodimer formation by low endotoxic atypical LPS. Front. Immunol. 12, 748303 (2022).
Herath, T. D. et al. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-κB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS One 8, e58496 (2013).
Bentala, H. et al. Removal of phosphate from lipid A as a strategy to detoxify lipopolysaccharide. Shock 18, 561–566 (2002).
Molinaro, A. et al. Chemistry of lipid A: At the heart of innate immunity. Chem. Eur. J. 21, 500–519 (2015).
Lai, H.-C. et al. Gut microbiota modulates COPD pathogenesis: role of anti-inflammatory Parabacteroides goldsteinii lipopolysaccharides. Gut 71, 309–321 (2022).
Maeshima, N, & R. C. Femandez R. C. Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front. Cell. Infect. Microbiol. 2013, 3, article 3.
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Hsieh, S.-J. et al. Characterization of anti-EBNA-1 antibodies and exploration of their molecular mimicry potential in an EBV-infected Sjögren’s syndrome patient. Biochem. Biophys. Res. Commun. 735, 150839 (2024).
Vo, D. N. K. et al. Broad-spectrum antiviral activity of Ganoderma microsporum immunomodulatory protein: Targeting glycoprotein gB to inhibit EBV and HSV-1 infections via viral fusion blockage. Int. J. Biol. Macromol. 307, 142179 (2025).
Forli, S. et al. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 11, 906–919 (2016).
Sarkar, A., Concilio, S., Sessa, L., Marrafino, F. & Piotto, S. Advancements and novel approaches in modified AutoDock Vina algorithms for enhanced molecular docking. Results Chem. 7, 101319 (2024).
Acknowledgements
We thank the National Science and Technology Council (NSTC) of Taiwan for financial support (Grant no. NSTC 112-2113-M-A49-012-MY2).
Author information
Authors and Affiliations
Contributions
XR Li is the main contributor in the synthesis of the lipid A(s) and related derivatives for immunological studies done mainly by Dr. JY Tan. BH Goh performed additional assays during the revision. CW Chen participated in the initial phase of the synthesis of lipid A, and JY Huang established the synthetic protocol for β-hydroxy fatty acids. Computational docking and modelling experiments were done by XR Li and HY Lin. KC Tsai validated the binding modes of lipid A(s) by AlphaFold 3.0 and gave advice in revision. KK Mong & CH Lin jointly supervised the project, and the primary corresponding author is KKMong.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Antonio Molinaro and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, XR., Tan, J.JY., Chen, CW. et al. Synthesis of Acinetobacter baumannii Lipid A(s) and derivatives and their structure-immunostimulatory activity relationships. Commun Chem 8, 234 (2025). https://doi.org/10.1038/s42004-025-01628-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-025-01628-6