Abstract
Immune checkpoint blockade using anti-programmed cell death protein 1/programmed cell death 1 ligand 1 antibody effectively targets the tumor-T cell interaction in cancer treatment, yet the overall response rate of less than 30% necessitates the identification of additional immune checkpoints modulating T cell function. Here, we identified the tumor cell-expressed paired immunoglobulin-like type 2 receptor alpha (PILRα) as an immune suppressor targeting T cells using high-throughput screening. PILRα inhibits T cell activation, proliferation and effector function by targeting CD99, a T cell surface antigen, suppressing ZAP70/NFAT/IL-2/JAK/STAT signaling. A cluster of O-glycosylated serine and threonine residues within the stalk region is critical for PILRα–CD99 interactions. Blocking these interactions with a stalk-targeting anti-PILRα antibody enhances T cell antitumor immunity and suppresses tumor growth. When combined with programmed cell death protein 1 antibody, anti-PILRα antibody shows synergistic tumor suppression. Notably, PILRα is highly expressed in several human cancers and predicts poor prognosis. These findings unveil PILRα as an immune checkpoint with therapeutic potential for clinical cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) under accession no. GSE254013. The RNA-seq data used in this study are available in the GEO under accession no. GSE174617. All mass spectrometry raw files have been deposited in ProteomeXchange via the PRIDE database under accession no. PXD048903. The source data for Figs. 1–8 and Extended Data Figs. 1–10 have been provided as source data files. The remaining data are available in the article, its supplementary information, as source data files or from the corresponding author W.L. (w2liu@xmu.edu.cn) upon reasonable request. Source data are provided with this paper.
References
Dong, H. et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8, 793–800 (2002).
Chen, L. & Han, X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J. Clin. Invest. 125, 3384–3391 (2015).
Taube, J. M. et al. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci. Transl. Med. 4, 127ra137 (2012).
Sznol, M. & Chen, L. Antagonist antibodies to PD-1 and B7-H1 (PD-L1) in the treatment of advanced human cancer—response. Clin. Cancer Res. 19, 5542 (2013).
Emens, L. A. et al. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: a phase 1 study. JAMA Oncol. 5, 74–82 (2019).
Armand, P. et al. Pembrolizumab in relapsed or refractory primary mediastinal large B-cell lymphoma. J. Clin. Oncol. 37, 3291–3299 (2019).
Ansell, S. M. et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 372, 311–319 (2015).
Wang, J. et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 25, 656–666 (2019).
Gajewski, T. F., Schreiber, H. & Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013).
Johnson, L. A. & June, C. H. Driving gene-engineered T cell immunotherapy of cancer. Cell Res. 27, 38–58 (2017).
Mahoney, K. M., Rennert, P. D. & Freeman, G. J. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 14, 561–584 (2015).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Blankenstein, T., Coulie, P. G., Gilboa, E. & Jaffee, E. M. The determinants of tumour immunogenicity. Nat. Rev. Cancer 12, 307–313 (2012).
Dong, M. B. et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 178, 1189–1204 (2019).
Kim, T. K., Vandsemb, E. N., Herbst, R. S. & Chen, L. Adaptive immune resistance at the tumour site: mechanisms and therapeutic opportunities. Nat. Rev. Drug Discov. 21, 529–540 (2022).
Xia, L. et al. Targeting triple-negative breast cancer with combination therapy of EGFR CAR T cells and CDK7 inhibition. Cancer Immunol. Res. 9, 707–722 (2021).
Xia, L. et al. BRD4 inhibition boosts the therapeutic effects of epidermal growth factor receptor-targeted chimeric antigen receptor T cells in glioblastoma. Mol. Ther. 29, 3011–3026 (2021).
Lu, Q. et al. PILRα and PILRβ have a siglec fold and provide the basis of binding to sialic acid. Proc. Natl Acad. Sci. USA 111, 8221–8226 (2014).
Mousseau, D. D., Banville, D., L’Abbé, D., Bouchard, P. & Shen, S. H. PILRα, a novel immunoreceptor tyrosine-based inhibitory motif-bearing protein, recruits SHP-1 upon tyrosine phosphorylation and is paired with the truncated counterpart PILRβ. J. Biol. Chem. 275, 4467–4474 (2000).
Wang, J., Shiratori, I., Uehori, J., Ikawa, M. & Arase, H. Neutrophil infiltration during inflammation is regulated by PILRα via modulation of integrin activation. Nat. Immunol. 14, 34–40 (2013).
Sun, Y. et al. PILRα negatively regulates mouse inflammatory arthritis. J. Immunol. 193, 860–870 (2014).
Satoh, T. et al. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell 132, 935–944 (2008).
Arii, J. et al. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor α. J. Virol. 83, 4520–4527 (2009).
Rathore, N. et al. Paired immunoglobulin-like type 2 receptor alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLoS Genet. 14, e1007427 (2018).
Dong, H., Zhu, G., Tamada, K. & Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 5, 1365–1369 (1999).
Tabata, S. et al. Biophysical characterization of O-glycosylated CD99 recognition by paired Ig-like type 2 receptors. J. Biol. Chem. 283, 8893–8901 (2008).
Moody, A. M. et al. Developmentally regulated glycosylation of the CD8αβ coreceptor stalk modulates ligand binding. Cell 107, 501–512 (2001).
Wilson, M. D., Cheung, J., Martindale, D. W., Scherer, S. W. & Koop, B. F. Comparative analysis of the paired immunoglobulin-like receptor (PILR) locus in six mammalian genomes: duplication, conversion, and the birth of new genes. Physiol. Genomics 27, 201–218 (2006).
Fournier, N. et al. FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells. J. Immunol. 165, 1197–1209 (2000).
Fredriksson, R., Lagerström, M. C., Höglund, P. J. & Schiöth, H. B. Novel human G protein-coupled receptors with long N-terminals containing GPS domains and Ser/Thr-rich regions. FEBS Lett. 531, 407–414 (2002).
Zheng, L. et al. The CD8α–PILRα interaction maintains CD8+ T cell quiescence. Science 376, 996–1001 (2022).
Wingett, D., Forcier, K. & Nielson, C. P. A role for CD99 in T cell activation. Cell. Immunol. 193, 17–23 (1999).
Oh, K. I. et al. CD99 activates T cells via a costimulatory function that promotes raft association of TCR complex and tyrosine phosphorylation of TCR ζ. Exp. Mol. Med. 39, 176–184 (2007).
van der Merwe, P. A. & Davis, S. J. Molecular interactions mediating T cell antigen recognition. Annu. Rev. Immunol. 21, 659–684 (2003).
Gaud, G., Lesourne, R. & Love, P. E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018).
Alberts, B. et al. Helper T Cells and Lymphocyte Activation. In Molecular Biology of the Cell, 4th edn. (Garland Science, 2002).
Rochman, Y., Spolski, R. & Leonard, W. J. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol. 9, 480–490 (2009).
Chan, A. C., Iwashima, M., Turck, C. W. & Weiss, A. ZAP-70: a 70 kd protein-tyrosine kinase that associates with the TCR ζ chain. Cell 71, 649–662 (1992).
Waclavicek, M. et al. CD99 engagement on human peripheral blood T cells results in TCR/CD3-dependent cellular activation and allows for Th1-restricted cytokine production. J. Immunol. 161, 4671–4678 (1998).
Basu, S. et al. Small molecule inhibitors of NFkB reverse iron overload and hepcidin deregulation in a zebrafish model for hereditary hemochromatosis type 3. ACS Chem. Biol. 13, 2143–2152 (2018).
Aramburu, J. et al. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285, 2129–2133 (1999).
Zhang, T. et al. Discovery of potent and selective covalent inhibitors of JNK. Chem. Biol. 19, 140–154 (2012).
Uchihashi, S. et al. Metabolism of the c-Fos/activator protein-1 inhibitor T-5224 by multiple human UDP-glucuronosyltransferase isoforms. Drug Metab. Dispos. 39, 803–813 (2011).
Sallusto, F., Geginat, J. & Lanzavecchia, A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu. Rev. Immunol. 22, 745–763 (2004).
Reiser, J. & Banerjee, A. Effector, memory, and dysfunctional CD8+ T cell fates in the antitumor immune response. J. Immunol. Res. 2016, 8941260 (2016).
Giles, J. R., Globig, A.-M., Kaech, S. M. & Wherry, E. J. CD8+ T cells in the cancer-immunity cycle. Immunity 56, 2231–2253 (2023).
Deng, M. et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018).
Wu, G. et al. LILRB3 supports acute myeloid leukemia development and regulates T-cell antitumor immune responses through the TRAF2–cFLIP–NF-κB signaling axis. Nat. Cancer 2, 1170–1184 (2021).
Shields, R. L. et al. High resolution mapping of the binding site on human IgG1 for FcγRI, FcγRII, FcγRIII, and FcRn and design of IgG1 variants with improved binding to the FcγR. J. Biol. Chem. 276, 6591–6604 (2001).
Xia, L. et al. EGFR-targeted CAR-T cells are potent and specific in suppressing triple-negative breast cancer both in vitro and in vivo. Clin. Transl. Immunol. 9, e01135 (2020).
Omuro, A. et al. Nivolumab with or without ipilimumab in patients with recurrent glioblastoma: results from exploratory phase I cohorts of CheckMate 143. Neuro Oncol. 20, 674–686 (2018).
Reardon, D. A. et al. Effect of nivolumab vs bevacizumab in patients with recurrent glioblastoma: the CheckMate 143 phase 3 randomized clinical trial. JAMA Oncol. 6, 1003–1010 (2020).
Yang, T., Kong, Z. & Ma, W. PD-1/PD-L1 immune checkpoint inhibitors in glioblastoma: clinical studies, challenges and potential. Hum. Vaccin. Immunother. 17, 546–553 (2021).
Mei, Y. et al. Siglec-9 acts as an immune-checkpoint molecule on macrophages in glioblastoma, restricting T-cell priming and immunotherapy response. Nat. Cancer 4, 1273–1291 (2023).
Croft, M. Co-stimulatory members of the TNFR family: keys to effective T-cell immunity? Nat. Rev. Immunol. 3, 609–620 (2003).
Li, C.-W. et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell 33, 187–201 (2018).
Buettner, M. J., Shah, S. R., Saeui, C. T., Ariss, R. & Yarema, K. J. Improving immunotherapy through glycodesign. Front. Immunol. 9, 2485 (2018).
Posey, A. D. Jr. et al. Engineered CAR T cells targeting the cancer-associated Tn-glycoform of the membrane mucin MUC1 control adenocarcinoma. Immunity 44, 1444–1454 (2016).
Lu, X. Structure and functions of T-cell immunoglobulin-domain and mucin- domain protein 3 in cancer. Curr. Med. Chem. 29, 1851–1865 (2022).
Sun, Y. et al. Blockade of the CD93 pathway normalizes tumor vasculature to facilitate drug delivery and immunotherapy. Sci. Transl. Med. 13, eabc8922 (2021).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (no. 82472769), Guangdong Basic and Applied Basic Research Foundation (no. 2024A1515030245) and the Scientific Research Foundation of State Key Laboratory of Vaccines for Infectious Diseases, Xiang An Biomedicine Laboratory (no. 2023XAKJ0102059) to L.X. This work was also supported by the National Natural Science Foundation of China (nos. 82125028, U22A20320, 31871319 and 91953114), the National Key Research and Development Program of China (nos. 2020YFA0803600 and 2020YFA0112300), the Shenzhen Science and Technology Program (no. JCYJ20230807091159001) and the Fundamental Research Funds for the Central University (nos. 20720190145 and 20720220003) to W.L. We thank K. Mao at Xiamen University for providing the OT-I mice. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
W.L. and L.X. conceived and designed the study, and wrote the manuscript with input from W.-X.L. and N.-S.X. J.-Y.L. performed the main experiments. C.Y. performed the Ab screening. H.-W.L. and Ya-hong Hu performed the protein–protein interaction experiment. G.-S.H. performed the bioinformatics analysis. Yao-hui He performed the mass spectrometry analysis. Y.-Y.C. performed the CAR T preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Glycosylation and functional analysis of PILRα.
a, The identity and similarity of IgV domain among inhibitory or stimulatory molecules on T cell activity are shown. The identity refers to the percentage of identical residues between the IgV domain of PILRα and those of other inhibitory or stimulatory molecules, while the similarity refers to the protein sequence similarity between the IgV domain of PILRα and those of other inhibitory or stimulatory molecules. b, Primary T cells labeled with CFSE in αCD3-coated plates (1 μg ml−1) were treated with or without PILRα (ECD)-hIg (20 μg ml−1) for 72 h, followed by flow cytometry to measure T cell divisions (n = 5 biologically independent experiments). The discrete peaks represent individual generations of cells indicated by numbers, where generation 0 represents undivided cells. c,d,e,i,k, Quantification of CD25+, CD44+, and CD69+ staining in Fig. 2e (c), Fig. 2f (d and e), Fig. 2l (i), and Fig. 2p (k) is shown (n = 5 biologically independent experiments). f, PILRα (ECD)-hIg, PILRβ (ECD)-hIg, PILRα-iso1-hIg, and PILRα-iso2-hIg proteins were subjected to IB analysis using an anti-IgG1 Fc antibody. The IB image is representative of three individual experiments (n = 3 biologically independent experiments). g, PILRα (ECD)-hIg and PILRα (IgV)-hIg proteins were subjected to IB analysis using an anti-IgG1 Fc antibody. The IB image is representative of three individual experiments (n = 3 biologically independent experiments). h, The O-glycosylation sites in PILRα (ECD) predicted using NetOGlyc 3.1 algorithm (www.cbs.dtu.dk/services/NetOGlyc/). j, The O-glycosylation sites in PILRα (ECD) identified by LC-MS/MS analysis. Data are presented as mean ± s.e.m. c,d,e,i,k, P values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test.
Extended Data Fig. 2 Anti-PILRα monoclonal antibody C21 specifically targets the stalk region of PILRα.
a,j, CD25+ quantification in Fig. 3d (a) and 3n (j) (n = 5 biologically independent experiments). b, Expression kinetics of CD99, PD-1, and TIM-3 in T cells with or without αCD3-stimulation for 48 h (n = 3 biologically independent experiments). c, Representative flow cytometry of CD99-Fc (10 μg ml−1) binding to WT or PILRAOE-MDA-MB-231 cells using Dylight 650-conjugated anti-human IgG Fc antibody (n = 3 biologically independent experiments). d, ELISA of hybridoma supernatants binding to PILRα-coated plates (OD450/OD630) (n = 3 biologically independent experiments). e,f, PILRα-expressing U87 cells incubated with hybridoma supernatants were stained with Alexa Fluor 488-conjugated anti-mouse IgG (H + L) antibody for high-content imaging (e), and immunofluorescence (IF) quantification (f) (n = 3 biologically independent experiments). g, Primary T cells were co-cultured with aAPC-293T cells expressing PILRα, with or without hybridoma supernatants for 72 h, followed by ELISA for IFNγ (n = 3 biologically independent experiments). Red clones were further purified. h,i, αCD3-stimulated (1 μg ml−1) NFAT-Lucia reporter Jurkat T cells were treated with or without PILRα (ECD)-hIg (20 μg ml−1) and anti-PILRα antibodies (50 μg ml−1), for luciferase activity (h) (n = 6 biologically independent experiments) and IFNγ detection (i) (n = 3 biologically independent experiments). k, PILRα (ECD)-hIg (2 μg ml−1) was pre-incubated with His-CD8-coated plates, followed by blocking ELISA using isotype, C21, or C126 (n = 3 biologically independent experiments). l, ELISA of PILRα (IgV)-hIg binding to C126-coated plate (n = 3 biologically independent experiments). m,n, αCD3-stimuated (1 μg ml−1) T cells were treated with or without PILRα (ECD)-hIg (20 μg ml−1) and isotype, C21, or C126 (50 μg ml−1) for 72 h, followed by flow cytometry for CD25 expression (m), and ELISA for IFNγ, granzyme B, and IL-2 (n) (n = 5 biologically independent experiments). Data are presented as mean ± s.e.m. a, P values were calculated using a two-way ANOVA with Tukey’s multiple comparisons test. j,n, P values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test.
Extended Data Fig. 3 PILRα inhibits CD99-induced signaling in T cells.
a,c, The intensity of the protein bands in Fig. 4d (a) and Fig. 4i (c) was quantified using Image J. The phosphorylated protein levels, normalized to the total protein levels, are shown (n = 3 biologically independent experiments). b, Quantification of CD25+ staining in Fig. 4h (n = 5 biologically independent experiments). Data are presented as mean ± s.e.m. a, P values were calculated using an unpaired two-tailed Student’s t-test. b,c, P values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test.
Extended Data Fig. 4 Flow cytometry analysis of spleen and tumor tissues from PILRα-engineered cell xenografts.
a, Representative flow cytometry of U87 and MDA-MB-231 cells stained with C21 antibody (n = 3 biologically independent experiments). b,c, Representative flow cytometry plots depicting the percentage of CD25+, IFNγ+, granzyme B+, and IL-2+ cells within CD8+ T cells in spleen or tumor tissues, as summarized in Fig. 5m (b) and Fig. 5s (c) (n = 5 independent mice).
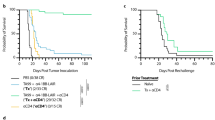
Extended Data Fig. 5 PILRα suppresses antitumor T cell immunity in the MC38 allograft model.
a, Control MC38 cells (control) and MC38 cells with Pilra knockdown (PilraKD) or Pilra overexpression (PilraOE) were subjected to IB analysis. The IB image is representative of three individual experiments (n = 3 biologically independent experiments). b-d, C57BL/6 mice were inoculated subcutaneously with control, PilraKD, or PilraOE MC38 cells (1.0 × 106 cells per mouse). Tumor growth curve (b) (n = 7 independent mice), tumor weight (c) (n = 7 independent mice), and mice survival (d) (n = 5 independent mice) are shown. e, Spleen and tumor tissues from mice as described in (b) were subjected to flow cytometry analysis. The percentage of CD3+ T cells, CD3+CD8+ T cells, CD3+CD4+ T cells, CD19+ B cells, NK1.1+ NK cells, CD11b+ myeloid cells, and CD11b+F4/80+ macrophages in CD45+ leukocytes were counted (n = 5 independent mice). The box plots display the minima and maxima as the whiskers, the median as the central line, the bounds of the box as the first and third quartiles. f, Spleen and tumor tissues from mice as described in (b) were subjected to flow cytometry analysis, and the percentage of CD25+, IFNγ+, granzyme B+, and IL-2+ cells within CD8+ T cells is shown (n = 3 independent mice). Data are presented as mean ± s.e.m. b, P values were calculated using a two-way ANOVA with Geisser-Greenhouse’s correlation. c, left panel of e,f, P values were calculated using a one-way ANOVA with Holm-Šídák’s multiple comparisons test. Right panel of e,f, P values were determined using an unpaired two-tailed Student’s t-test. d, P values were determined using a log-rank (Mantel-Cox) test for survival, which was independently repeated twice with similar results.
Extended Data Fig. 6 PILRα suppresses antigen-specific T cell responses in vitro and in vivo.
a,c,e OT-I T cells were co-cultured with or without control, PilraKD, or PilraOE MC38-OVA cells for 48 h, followed by flow cytometry for CD25 (a), IFNγ (c), and IL-2 (e) expression in OT-I cells (n = 3 biologically independent experiments). b,d,f, Quantification of CD25+ (b), IFNγ+ (d), and IL-2+ staining (f) in OT-I T cells from panels (a), (c), and (e), respectively (n = 3 biologically independent experiments). g,h, C57BL/6 mice were inoculated subcutaneously with control, PilraKD, or PilraOE MC38-OVA cells (5.0 × 106 cells per mouse, n = 3). Purified OT-I T cells were adoptively transferred intravenously into mice on day 7 (5.0 × 106 cells per mouse, n = 3). Representative flow cytometry plots depicting the percentage of TCR Vα2+ CD8+ OT-I T cells in spleen (g) and tumor (h) on day 12 are shown (n = 3 independent mice). i, The percentage of TCR Vα2+ CD8+ OT-I T cells in the spleen (g) and tumor (h) (n = 3 independent mice). j,k, Representative flow cytometry plots depicting the percentage of CD44+, CD25+, IFNγ+, and IL-2+ cells within CD8+ OT-I T cells in the spleen (j) and tumor (k) (n = 3 independent mice). l, The percentage of CD44+, CD25+, IFNγ+, and IL-2+ staining in CD8+ OT-I T cells in the spleen (j) and tumor (k) (n = 3 independent mice). Data are presented as mean ± s.e.m. b,d,f, left panel of i,l, P values were calculated using a one-way ANOVA with Holm-Šídák’s multiple comparisons test. Right panel of i,l, P values were calculated using an unpaired two-tailed Student’s t-test.
Extended Data Fig. 7 Anti-PILRα monoclonal antibody C21 potently inhibits tumor growth and is relatively safe in humanized mouse xenograft models.
a,b, Tumor tissues from humanized MDA-MB-231 xenograft models described in Fig. 6a were collected on day 20 for flow cytometry analysis. CD8+ (CD45+CD3+CD8+), CD8+ TEM (CD45+CD3+CD8+CD62L-CD45RO+), and CD8+ TEFF (CD45+CD3+CD8+CD62L-CD45RO-) TILs gated by anti-CD45 antibody conjugated with APC (APC-CD45), anti-CD3 antibody conjugated with PerCP/Cy5.5 (PerCP/Cy5.5-CD3), anti-CD8 antibody conjugated with PE/Cy7 (PE/Cy7-CD8), anti-CD62L antibody conjugated with Alexa Fluor 700 (AF700-CD62L), and anti-CD45RO conjugated with FITC (FITC-CD45RO) were further analyzed by staining of anti-Granzyme B antibody conjugated with BV421 (BV421-Granzyme B) and anti-IFNγ antibody conjugated with PE (PE-IFNγ), or anti-CD25 antibody conjugated with BV421 (BV421-CD25). Representative flow cytometry plots depicting the percentage of IFNγ+ and granzyme B+ (a) and CD25+ (b) cells are shown (n = 5 independent mice). c, The percentage of CD25+, IFNγ+, and granzyme B+ cells within CD8+ (CD45+CD3+CD8+), CD8+ TEM (CD45+CD3+CD8+CD62L-CD45RO+), and CD8+ TEFF (CD45+CD3+CD8+CD62L-CD45RO-) subsets as described in (a) and (b) (n = 5 independent mice). d,g, Body weight of mice from the treatment as described in Fig. 6a (d) and Fig. 6h (g) (n = 5 independent mice). e,h, Representative gross morphology of major organs including heart, liver, spleen, lung, and kidney collected from mice in Fig. 6b (e) and Fig. 6i (h) (n = 5 independent mice). f,i, Representative H&E staining images of tissue sections from organs in panel e (f) and h (i) (n = 5 independent mice). Data are presented as mean ± s.e.m. c, P values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test. f,i, Scale bar, 100 μm.
Extended Data Fig. 8 Anti-PILRα monoclonal antibody C21 enhances EGFR CAR T cell efficacy in a MDA-MB-231 xenograft model.
a, NCG mice were subcutaneously (s.c.) inoculated with MDA-MB-231 cells (5.0 × 106 cells per mouse, n = 5). After 15 days, EGFR CAR T cells (5.0 × 106 cells per mouse) were administered intravenously (i.v.), followed by intraperitoneally (i.p.) injection of either C21 or C21mut antibodies (Abs) (10 mg kg−1, n = 5) 1 h post CAR T infusion. A second dose of either CAR T or antibody was administered one week later. b-d, Tumor growth curve (b), tumor weight (c), and tumors at day 27 (d) are shown (n = 5 independent mice). e,f Tumors at day 20 were collected for flow cytometry analysis. CD8+ (CD45+CD3+CD8+), CD8+ TEM (CD45+CD3+CD8+CD62L-CD45RO+), and CD8+ TEFF (CD45+CD3+CD8+CD62L-CD45RO-) TILs gated by anti-CD45 antibody conjugated with APC (APC-CD45), anti-CD3 antibody conjugated with PerCP/Cy5.5 (PerCP/Cy5.5-CD3), anti-CD8 antibody conjugated with PE/Cy7 (PE/Cy7-CD8), anti-CD62L antibody conjugated with Alexa Fluor 700 (AF700-CD62L), and anti-human CD45RO-FITC (FITC-CD45RO) were further analyzed by staining of anti-CD25 antibody conjugated with BV421 (BV421-CD25), or anti-Granzyme B antibody conjugated with BV421 (BV421-Granzyme B), and anti-IFNγ antibody conjugated with PE (PE-IFNγ). Representative flow cytometry plots depicting the percentage of CD25+ (e) and IFNγ+/granzyme B+ (f) cells are shown (n = 5 independent mice). g, The percentage of CD25+, IFNγ+, and granzyme B+ cells within CD8+ (CD45+CD3+CD8+), CD8+ TEM (CD45+CD3+CD8+CD62L-CD45RO+), and CD8+ TEFF (CD45+CD3+CD8+CD62L-CD45RO-) subsets as described in (e) and (f) (n = 5 independent mice). Data are presented as mean ± s.e.m. b, P values were calculated using a two-way ANOVA with Geisser-Greenhouse’s correlation. c, P values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test. g, P values were calculated using a one-way ANOVA with Holm-Šídák’s multiple comparisons test.
Extended Data Fig. 9 Combination therapy with anti-PILRα antibody C21 and anti-PD-1 antibody is relatively safe in a humanized U87 xenograft model.
a, Representative gross morphology of major organs including heart, liver, spleen, lungs, and kidneys collected from the mice in Fig. 7b are shown (n = 5 independent mice). b, Representative H&E staining images of tissue sections from organs in panel a (n = 5 independent mice). Scale bar, 100 μm.
Extended Data Fig. 10 PILRα expression correlates with CD99, immune checkpoints, and patient survival across multiple cancer types.
a, Positive correlation between PILRA and CD99 mRNA expression in tumor tissues form patients with TNBC (n = 1100), GBM (n = 516), HNSC (n = 522), KIRP (n = 290), THCA (n = 509), and STAD (n = 415), as analyzed by TIMER. Spearman’s correlation coefficient and P-value are given (two-sided t-test). False discovery rate-adjusted p < 0.05. Regression line with standard error interval is shown. b, Kaplan–Meier analysis of overall survival stratified by median baseline of CD99 levels in TNBC (low = 82, high = 85), KIRC (low = 350, high = 180), KIRP (low = 209, high = 78), and STAD patients (low = 152, high = 219). Kaplan–Meier Plotter curves are typically analyzed using the log-rank (Mantel-Cox) test to compare survival distributions between groups. c,d, Positive correlation between the expression of PILRA mRNA and mRNA encoding immune checkpoints PD-1 (PDCD1), CTLA-4 (CTLA4), and TIM-3 (HAVCR2) in TNBC (n = 191) (c) and GBM (n = 153) (d) patients, as determined by TIMER analysis. Spearman’s correlation coefficient and P-value are given (two-sided t-test). False discovery rate-adjusted p < 0.05. Regression line with standard error interval is shown.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2.
Supplementary Tables 1–5
Supplementary Table 1: List of genes induced by EGFR CAR T treatment in U87 GBM cells (n = 600). Supplementary Table 2: List of genes from sheet 1 that encode membrane proteins (n = 148). Supplementary Table 3: List of genes from sheet 2 that are overexpressed in GBM tumors compared to normal samples based on the TCGA database (n = 63). Supplementary Table 4: List of genes from sheet 2 that are overexpressed in GBM tumors compared to normal samples based on the TCGA database and are predictive of poor prognosis (n = 32). Supplementary Table 5: List of antibodies, chemicals, protein, mouse and cell lines, key commercial kits, instrument, software, algorithm and tools.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed immunoblots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed immunoblots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed immunoblots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed immunoblots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed immunoblots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed immunoblots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xia, L., Liu, Jy., Yu, C. et al. PILRα on tumor cells interacts with the T cell surface protein CD99 to suppress antitumor immunity. Nat Cancer 6, 1184–1201 (2025). https://doi.org/10.1038/s43018-025-00958-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s43018-025-00958-7
This article is cited by
-
CD99 is a potential diagnostic and immunological biomarker in pan-cancer
Discover Oncology (2025)