Abstract
Recent advances in spatial biology can reveal how tissue organization changes in disease; however, interpreting these datasets in a generalized, scalable way remains challenging. Existing computational approaches rely on pairwise comparisons or unsupervised clustering, which can lack statistical rigor and miss rare, clinically relevant cellular niches. Here we present QUICHE—an automated and scalable statistical framework designed to discover cellular niches differentially enriched in populations, histological structures or acellular regions. Using in silico models and spatial proteomic imaging of human tissues, we show that QUICHE can accurately detect low-prevalence, condition-specific niches, outperforming the next best algorithm threefold. To investigate how tumor structure influences recurrence risk in triple-negative breast cancer, we applied QUICHE to a multicenter spatial proteomics cohort of 314 primary tumor resections. We discovered niches consistently enriched in tumor border and extracellular-matrix-remodeling regions, including those associated with recurrence-free survival. These findings were validated in two independent cohorts, suggesting that antitumor responses are driven by coordinated engagement between innate and adaptive immune cells, rather than any single population. QUICHE is provided as an open-source Python package (https://github.com/jranek/quiche).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
High-resolution MIBI-TOF imaging data and cell segmentation masks for both the Spain and the Stanford TNBC cohorts are publicly available in the BioStudies repository (https://doi.org/10.6019/S-BIAD1507)78. Preprocessed anndata objects for all three TNBC cohorts are publicly available from Zenodo (https://doi.org/10.5281/zenodo.14290163)79. Publicly available imaging mass cytometry and clinical response data from the NeoTRIP study were accessed from a Zenodo repository (https://doi.org/10.5281/zenodo.7990870)71. Source data are provided with this paper.
Code availability
QUICHE is implemented as an open-source Python package and is publicly available from GitHub (https://github.com/jranek/quiche). Source code for preprocessing, benchmarking and figure generation is also publicly available from GitHub (https://github.com/angelolab/publications/tree/main/2024-Ranek_etal_QUICHE).
References
Keren, L. et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 5, eaax5851 (2019).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14, 8353 (2023).
Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 361, eaar7042 (2018).
Carstens, J. L. et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat. Commun. 8, 15095 (2017).
McCaffrey, E. F. et al. The immunoregulatory landscape of human tuberculosis granulomas. Nat. Immunol. 23, 318–329 (2022).
Vijayaragavan, K. et al. Single-cell spatial proteomic imaging for human neuropathology. Acta Neuropathol. Commun. 10, 158 (2022).
Risom, T. et al. Transition to invasive breast cancer is associated with progressive changes in the structure and composition of tumor stroma. Cell 185, 299–310 (2022).
Wang, X. Q. et al. Spatial predictors of immunotherapy response in triple-negative breast cancer. Nature 621, 868–876 (2023).
Jackson, H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Hirata, E. & Sahai, E. Tumor microenvironment and differential responses to therapy. Cold Spring Harb. Perspect. Med. 7, a026781 (2017).
Ibrahim, E. M., Al-Foheidi, M. E., Al-Mansour, M. M. & Kazkaz, G. A. The prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancer: a meta-analysis. Breast Cancer Res. Treat. 148, 467–476 (2014).
Edwards, J. et al. CD103+ tumor-resident CD8+ T cells are associated with improved survival in immunotherapy-naïve melanoma patients and expand significantly during anti-PD-1 treatment. Clin. Cancer Res. 24, 3036–3045 (2018).
Naito, Y. et al. CD8+ T cells infiltrated within cancer cell nests as a prognostic factor in human colorectal cancer. Cancer Res. 58, 3491–3494 (1998).
Fan, J. L., Nazaret, A. & Azizi, E. A thousand and one tumors: the promise of AI for cancer biology. Nat. Methods 21, 1403–1406 (2024).
Schapiro, D. et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 14, 873–876 (2017).
Ali, M. et al. GraphCompass: spatial metrics for differential analyses of cell organization across conditions. Bioinformatics 40, i548–i557 (2024).
Varrone, M., Tavernari, D., Santamaria-Martínez, A., Walsh, L. A. & Ciriello, G. CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity. Nat. Genet. 56, 74–84 (2024).
Yuan, Z. et al. SOTIP is a versatile method for microenvironment modeling with spatial omics data. Nat. Commun. 13, 7330 (2022).
Hu, J. et al. SpaGCN: integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Wu, Z. et al. Graph deep learning for the characterization of tumour microenvironments from spatial protein profiles in tissue specimens. Nat. Biomed. Eng. 6, 1435–1448 (2022).
Dong, K. & Zhang, S. Deciphering spatial domains from spatially resolved transcriptomics with an adaptive graph attention auto-encoder. Nat. Commun. 13, 1739 (2022).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Sorin, M. et al. Single-cell spatial landscapes of the lung tumour immune microenvironment. Nature 614, 548–554 (2023).
Karimi, E. et al. Single-cell spatial immune landscapes of primary and metastatic brain tumours. Nature 614, 555–563 (2023).
Salié, H. et al. Spatial single-cell profiling and neighbourhood analysis reveal the determinants of immune architecture connected to checkpoint inhibitor therapy outcome in hepatocellular carcinoma. Gut 74, 451–466 (2025).
Baskaran, V. A., Ranek, J., Shan, S., Stanley, N. & Oliva, J. B. Distribution-based sketching of single-cell samples. In Proc. 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (eds Bubak, M. et al.) (ACM, 2022).
Dann, E., Henderson, N. C., Teichmann, S. A., Morgan, M. D. & Marioni, J. C. Differential abundance testing on single-cell data using k-nearest neighbor graphs. Nat. Biotechnol. 40, 245–253 (2022).
Missarova, A., Dann, E., Rosen, L., Satija, R. & Marioni, J. Leveraging neighborhood representations of single-cell data to achieve sensitive DE testing with miloDE. Genome Biol. 25, 189 (2024).
Alice, K., Marioni, J. C. & Morgan, M. D. Milo2.0 unlocks population genetic analyses of cell state abundance using a count-based mixed model. Preprint at bioRxiv https://doi.org/10.1101/2023.11.08.566176 (2023).
Lun, A. T. L., Richard, A. C. & Marioni, J. C. Testing for differential abundance in mass cytometry data. Nat. Methods 14, 707–709 (2017).
Arthur, D. & Vassilvitskii, S. K-means++: the advantages of careful seeding. In Proc. 18th Annual ACM-SIAM Symposium on Discrete Algorithms (ed. Gabow, H.) 1027–1035 (Society for Industrial and Applied Mathematics, 2007).
Reiss, D. J. et al. High-plex imaging and cellular neighborhood spatial analysis reveals multiple immune escape and suppression patterns in diffuse large B-cell lymphoma. Leukemia 38, 1164–1168 (2024).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Liu, C. C. et al. Robust phenotyping of highly multiplexed tissue imaging data using pixel-level clustering. Nat. Commun. 14, 4618 (2023).
Kalluri, R. & Weinberg, R. A. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009).
Pallotta, M. T. et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 12, 870–878 (2011).
da Silva, I. P. et al. Reversal of NK-cell exhaustion in advanced melanoma by Tim-3 blockade. Cancer Immunol. Res. 2, 410–422 (2014).
Zhang, J. et al. Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells. Nat. Commun. 12, 5446 (2021).
Costa, A. et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 33, 463–479 (2018).
Greenwald, N. F. et al. Temporal and spatial composition of the tumor microenvironment predicts response to immune checkpoint inhibition. Preprint at bioRxiv https://doi.org/10.1101/2025.01.26.634557 (2025).
Keren, L. et al. A structured tumor–immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387 (2018).
de Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell 41, 374–403 (2023).
Hammerl, D. et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 12, 5668 (2021).
Ma, J. et al. Spatial immunophenotypes orchestrate prognosis in triple-negative breast cancer with miller-payne grade 4 following neoadjuvant chemotherapy. NPJ Breast Cancer 9, 57 (2023).
Winkler, J., Abisoye-Ogunniyan, A., Metcalf, K. J. & Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 11, 5120 (2020).
Song, K. et al. Collagen remodeling along cancer progression providing a novel opportunity for cancer diagnosis and treatment. Int. J. Mol. Sci. 23, 10509 (2022).
Conklin, M. W. et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011).
Provenzano, P. P. et al. Collagen reorganization at the tumor–stromal interface facilitates local invasion. BMC Med. 4, 38 (2006).
Teillaud, J.-L., Houel, A., Panouillot, M., Riffard, C. & Dieu-Nosjean, M.-C. Tertiary lymphoid structures in anticancer immunity. Nat. Rev. Cancer 24, 629–646 (2024).
Forero, A. et al. Expression of the MHC class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol. Res. 4, 390–399 (2016).
Zhang, Q. & Wu, S. Tertiary lymphoid structures are critical for cancer prognosis and therapeutic response. Front. Immunol. 13, 1063711 (2022).
Cox, D. R. & Snell, E. J. Analysis of Binary Data, 2nd edn (Chapman and Hall/CRC, 1970).
Lin, J.-R. et al. High-plex immunofluorescence imaging and traditional histology of the same tissue section for discovering image-based biomarkers. Nat. Cancer 4, 1036–1052 (2023).
Rivest, F. et al. Fully automated sequential immunofluorescence (seqIF) for hyperplex spatial proteomics. Sci. Rep. 13, 16994 (2023).
McCaffrey, E. F. et al. The immunometabolic topography of tuberculosis granulomas governs cellular organization and bacterial control. Preprint at bioRxiv https://doi.org/10.1101/2025.02.18.638923 (2025).
Piyadasa, H. et al. Multi-omic landscape of human gliomas from diagnosis to treatment and recurrence. Cancer Cell 44, 112–128 (2026).
Shan, S. et al. Transparent single-cell set classification with kernel mean embeddings. In Proc. 13th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (eds Bubak, M. et al.) (ACM, 2022).
Palla, G. et al. Squidpy: a scalable framework for spatial omics analysis. Nat. Methods 19, 171–178 (2022).
Fowlkes, E. B. & Mallows, C. L. A method for comparing two hierarchical clusterings. J. Am. Stat. Assoc. 78, 553–569 (1983).
Ranek, J. S. & Angelo, M. QUICHE simulation design. protocols.io https://doi.org/10.17504/protocols.io.6qpvrwjk2lmk/v1 (2025).
Liu, C. C. et al. Reproducible, high-dimensional imaging in archival human tissue by multiplexed ion beam imaging by time-of-flight (MIBI-TOF). Lab. Invest. 102, 762–770 (2022).
Bosse, M., Camacho, C., Bendall, S. & Angelo, M. MIBI and IHC solutions. protocols.io https://doi.org/10.17504/protocols.io.bhmej43e (2021).
Bosse, M., Bendall, S. & Angelo, M. IHC staining. protocols.io https://doi.org/10.17504/protocols.io.bf6ajrae (2021).
Bosse, M., Bendall, S. & Angelo, M. MIBI staining. protocols.io https://doi.org/10.17504/protocols.io.dm6gprk2dvzp/v5 (2022).
Camacho, C., Bosse, M., Bendall, S. & Angelo, M. Antibody lyophilization. protocols.io https://doi.org/10.17504/protocols.io.bhmgj43w (2021).
van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015).
Monti, S. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 52, 91–118 (2003).
Schiemann, R., Gherardini, P. F., Kageyama, R., Travers, M. & Kitch, L. Mantis viewer. Zenodo https://doi.org/10.5281/zenodo.4009579 (2020).
Wang, X. Q. et al. Data associated with ‘Spatial predictors of immunotherapy response in triple negative breast cancer’. Zenodo https://doi.org/10.5281/zenodo.7990870 (2023).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995).
Heumos, L. et al. Pertpy: an end-to-end framework for perturbation analysis. Nat. Methods https://doi.org/10.1038/s41592-025-02909-7 (2025).
Frangi, A. F., Niessen, W. J., Vincken, K. L. & Viergever, M. A. Multiscale vessel enhancement filtering. In Proc. Medical Image Computing and Computer-Assisted Intervention (eds Wells, W. M. et al.) (Springer, 1998).
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991).
Cox, D. R. Regression models and life-tables. J. R. Stat. Soc. 34, 187–202 (1972).
Kaplan, E. L. & Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457 (1958).
Ranek, J. S. et al. QUICHE reveals structural definitions of anti-tumor responses in triple negative breast cancer. BioStudies https://doi.org/10.6019/S-BIAD1507 (2024).
Ranek, J. S. et al. Quiche reveals structural definitions of anti-tumor responses in triple-negative breast cancer. Zenodo https://doi.org/10.5281/zenodo.14290163 (2024).
Acknowledgements
We would like to thank all patients. We thank E. McCaffrey, S. Park and K. Abel for their thoughtful discussions related to this work. This work was supported by National Institutes of Health grants 5U54CA20997105 (M.A.), 5DP5OD01982205 (M.A.), 1R01CA24063801A1 (M.A.), 5R01AG06827902 (M.A.), 5UH3CA24663303 (M.A.), 5R01CA22952904 (M.A.), 1U24CA22430901 (M.A.), 5R01AG05791504 (M.A.) and 5R01AG05628705 (M.A.), the Department of Defense grant W81XWH2110143 (M.A.), the Wellcome Trust (M.A.) and other funding from the Bill and Melinda Gates Foundation (M.A.), Cancer Research Institute (M.A.), Parker Center for Cancer Immunotherapy (M.A.) and Breast Cancer Research Foundation (M.A.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
J.S.R. and M.A. conceptualized this study. S.M. and M.Q.-F. provided samples and clinical data for the Spain TNBC cohort. R.B.W. provided samples and clinical data for the Stanford cohort. N.F.G., C.C.F. and M.G. generated the MIBI-TOF data. J.S.R. performed the data analysis and wrote the paper. C.S. developed the fiber segmentation algorithm. A.K. performed Rosetta image compensation. M.A. and S.C.B. supervised the project. All authors read and provided feedback on the paper.
Corresponding author
Ethics declarations
Competing interests
M.A. and S.C.B are named inventors on patent US20150287578A1, which covers the mass spectrometry approach used by MIBI to detect elemental reporters in tissue using secondary-ion mass spectrometry. M.A. and S.C.B. are board members of and shareholders in IonPath, which develops and manufactures the commercial MIBI platform. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Qualitative comparisons of spatial clustering methods on unstructured spatial topologies.
We benchmarked spatial methods on their ability to recover condition-specific cellular niches when varying the underlying spatial topology, niche size, or sample prevalence. (a-h) Performance of different spatial clustering approaches on detecting condition-specific differences in cellular organization on simulated data with unstructured spatial topologies. Visualizations show clustering results and differential enrichment for condition 1 where cell types A, C, and E were differentially-enriched and condition 2 where cell types B and D were differentially-enriched. Here, low niche size (1.6% sample), low prevalence (40% patient samples), high niche size (11.1% sample), and high prevalence indicates (100% patient samples).
Extended Data Fig. 2 Qualitative comparisons of spatial clustering methods on structured spatial topologies.
We benchmarked spatial methods on their ability to recover condition-specific cellular niches when varying the underlying spatial topology, niche size, or sample prevalence. (a-h) Performance of different spatial clustering approaches on detecting condition-specific differences in cellular organization on simulated data with structured spatial topologies. Visualizations show clustering results and differential enrichment for condition 1 where immune cells were differentially-enriched in the cancer core and condition 2 where immune cells were differentially-enriched in the cancer border. Here, low niche size (25 immune cells within 500 pixel radius), low prevalence (40% patient samples), high niche size (100 immune cells within 500 pixel radius), and high prevalence indicates (100% patient samples).
Extended Data Fig. 3 Performance of spatial clustering methods across different parameters.
(a-b) Heatmap shows the performance of spatial clustering methods, K-Means++ and CellCharter, on detecting differentially-enriched cellular niches on (a) unstructured spatial topologies and (b) structured spatial topologies across different clustering resolution parameters. (c-e) Multi-patient spatial proteomic data were simulated with structured spatial topologies (that is, niche size of 50 immune cells within 500 pixel radius and niche prevalence of 100% samples). QUICHE achieves similar (c) niche recall and (d) niche purity scores across a range of hyperparameters, including the number of nearest neighbors in spatial niche detection, the radius size in spatial niche detection, the number of niches sampled from each patient sample, and the number of k-nearest neighbors in niche similarity graph construction. (e) Runtime performance of QUICHE as you vary (left) the number of niches selected from each patient sample in a cohort with 100 samples or (right) the cohort size, where each patient sample has 1000 niches. The error bands show the mean +/- standard deviation over n=5 random trials. Source data is provided in the Source Data file.
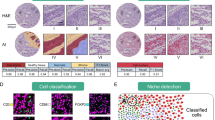
Extended Data Fig. 4 Preprocessing and quality control for the Spain TNBC cohort.
(a-e) Antibody panel validation. Single-plex images of profiled proteins in (a) control lymph node sample (b) control spleen sample and (c-e) three representative triple-negative breast cancer samples from the Spain TNBC cohort. (f) Image compensation with Rosetta. Representative images of several channels before and after background subtraction with the Rosetta algorithm. (g-j) Normalization and batch effect correction assessment. (g) Representative images show the unnormalized intensities from the start of acquisition (top left) to the end of acquisition (bottom right). (h) Representative images show the normalized intensities from the start of acquisition (top left) to the end of acquisition (bottom right) using median pulse height normalization. For compensation and normalization, representative of n=314 patient samples. (i) Heatmap shows the average change in normalized expression of protein markers across control tissues on different tissue microarrays. (j) Heatmap shows the average change in normalized expression across spatial locations of each tissue microarray. (k-m) Cell segmentation validation with Mesmer. (k) Representative images show the membrane markers (CD14, CD38, CD45, CK17, ECAD) used for cell segmentation, in addition to the nuclear markers (H3K9ac, H3K27me3). (l) Representative images of three tumors annotated by nuclear and membrane markers (top) and predicted cell segmentation masks (bottom). (m) Histogram shows the number of cells in each tumor image in the Spain TNBC cohort. (n) Morphological confirmation of cell phenotype maps. Hematoxylin and eosin (H&E) slides were digitized, annotated by a dedicated breast cancer pathologist, and demarcated with areas of interest. Each identified region was punched with a 1.5 mm core and placed onto a tissue microarray. Three illustrative examples of the H&E image of the entire core, a cropped inset of the portion of the core, and the corresponding cell phenotype map generated from the selected region. Source data is provided in the Source Data file.
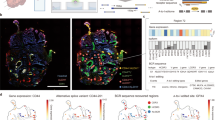
Extended Data Fig. 5 Stanford and NeoTRIP cohort overview.
(a-g) Stanford cohort. (a) Pie chart shows the number of patients that did (pink) or did not relapse (blue). (b) Histogram shows the number of cells in each tumor image. (c) Heatmap shows the average standardized expression of twenty cell phenotypes clustered according to protein expression of phenotypic markers. Barplots to the right show the total number of cells within each cell phenotype. (d) Barplots show the frequency of tumor (top), structural (middle) and immune lineages (bottom) across all primary tumor samples in this cohort. Samples were hierarchically clustered according all frequency subsets. Relapse status is denoted by color in the top row. (e) Violin plots show the top 15 differentially-enriched niche neighborhoods in patients who relapsed (red) or did not relapse (blue). Barplots to the right show the proportion of patients with a niche neighborhood in the respective patient groups. (f) Niche network for non-relapsing patients, where nodes represent cell types within recurrence-free niches and edge weights correspond to the number of unique patients with the corresponding interaction. Node size is proportional to connectedness, as measured by eigenvector centrality. (g) Niche network for relapsing patients, where nodes represent cell types within recurrence-associated niches and edge weights correspond to the number of unique patients with the corresponding interaction. Node size is proportional to connectedness, as measured by eigenvector centrality. (h-n) NeoTRIP cohort analysis, where pCR is complete pathologic response and RD is residual disease. Source data is provided in the Source Data file.
Extended Data Fig. 6 Differential cell type abundance and stepwise immune infiltration analysis across TNBC cohorts.
(a-c) Differential cell type analysis across recurrence groups in the Spain TNBC cohort. (a) Boxplots show the frequency of cell phenotypes across patients that did (orange, n=109 patients) or did not relapse (green, n=205 patients). A two-sided Wilcoxon rank sum test was used to test for differences between patient groups. Multiple hypothesis testing correction was performed using Benjamini Hochberg. (b) Differential cell type abundance testing was performed using Milo. Boxplots show the log2 fold change in the abundance of cell type neighborhoods across recurrence groups. (c) A logistic regression classifier was trained on the proportion of cell types to predict relapse status using 5-fold cross validation. Boxplots show the average area under the receiver operator curve (AUC) values across 10 random trials. Permuted label performance is shown in gray. All boxplots show median +/- IQR range; whiskers show 1.5*IQR. (d-f) Differential cell type analysis across recurrence groups (no recurrence n=97patients, recurrence n=45 patients) in the Stanford TNBC cohort. (g-i). Differential cell type analysis across pathological complete response (pCR) groups (pCR n=53 patients, residual disease (RD) n=66 patients) in the NeoTRIP TNBC cohort. (j-m) Stepwise immune infiltration across TNBC cohorts. Heatmaps show the presence (yellow) or absence (blue) of immune cells across patients in the (j) Spain, (k) Stanford, and (l) NeoTRIP TNBC cohorts. (m) Heatmap shows immune infiltration with different thresholds for positivity (x-axis) across patients in the Spain (top), Stanford (middle) and NeoTRIP (bottom) TNBC cohorts. For each cohort, a two-sided Chi-square test was used to test for differences in the infiltration of cell type pairs (Mono Mac - CD8T, CD8T - CD4T, CD4T - B, B - NK). * indicates p-value < 0.05. Source data is provided in the Source Data file.
Extended Data Fig. 7 Tumor border and ECM analysis.
(a) Barplots show the average frequency of cell phenotypes within tumor regions across samples (stroma n=548, cancer border n=539). (b) Heatmap shows the change in cell type expression within tumor border-associated niche neighborhoods as compared to all cells within the tumor border. (c) Heatmap shows the change in cell type expression within stroma-associated niche neighborhoods as compared to all cells within the stroma. (d) Barplots show the average frequency of cell phenotypes within ECM alignment regions across samples (aligned n=548 samples, unaligned n=548 samples) (c-d) Tumor-immune border analysis. (e) Heatmap shows the change in cell type expression in cellular niche neighborhoods within aligned extracellular matrix (ECM) regions as compared to the entire cohort. (f) Heatmap shows the change in cell type expression in cellular niches within unaligned ECM regions as compared to the entire cohort. (g) Boxplots show normalized morphological measurements of cancer cells within aligned (n=215 patients) or unaligned (n=73 patients) ECM regions (median +/- interquartile range; whiskers indicate minima and maxima after removing outliers using Tukey’s fences). A two-sided Wilcoxon rank sum test was used to compare differences between groups. (h) Histogram shows the distribution of the frequency of cancer cell neighbors around each EMT-like cancer cell. (i) Boxplots show the normalized morphological measurements of cancer cells with either 60% (top panel, aligned n=191-205 patients, unaligned n=65-69 patients), 80% (middle panel, aligned n=189-200 patients, unaligned n=66-72 patients), or 95% (bottom panel, aligned n=166-183 patients, unaligned n=66-72 patients) of cancer neighbors (median +/- interquartile range; whiskers indicate minima and maxima after removing outliers using Tukey’s fences). A two-sided Wilcoxon rank sum test was used to compare differences between groups. (j) Dotplots shows the average normalized expression of associated proteins of cancer cells with either 60% (top panel), 80% (middle panel), or 95% (bottom panel) cancer neighbors within aligned or unaligned ECM regions. (k) Scatter plots show the standardized expression of Vimentin within cancer cells or fibroblast subsets across aligned or unaligned ECM regions. Dotted lines highlight the median expression across all cells of that cell type within the cohort. r indicates Pearson correlation. Source data is provided in the Source Data file.
Extended Data Fig. 8 Spain TNBC cohort analysis.
(a) Heatmap shows the average standardized functional expression of twenty cell phenotypes across all patients that did not relapse. (b) Heatmap shows the average standardized functional expression of twenty cell phenotypes across all patients that relapsed. (c-e) Prevalence and representative accuracy of QUICHE niche neighborhoods in the Spain TNBC cohort. (c) Scatterplot shows the average abundance of recurrence-associated QUICHE niche neighborhoods vs. patient prevalence. Gray inset shows the QUICHE niche neighborhoods analyzed in panels b-c. (d) Scatterplot shows the representative area under the receiver operator curve (AUC) scores of QUICHE niche neighborhoods as you iteratively remove the most prevalent niches. (e) Scatterplot shows the representative AUC scores of QUICHE niche neighborhoods stratified by patient prevalence (niche neighborhoods in < 10% patients, within 10-20%, and within 20-35% patient samples). (f) Four representative patient tumor images, where cells are annotated according to cell type (top) or tumor regions (bottom). (g) Heatmap shows the correlation between the abundance of quiche niche neighborhoods and TLS status across patients that did (n=37 samples) or did not (n=259 samples) have tertiary lymphoid structures (TLS). P-values and correlations were computed using a two-sided point-biserial correlation test. Multiple hypothesis testing correction was performed using Benjamini Hochberg. * indicates FDR < 0.05. (h) Violin plots show the top differentially enriched QUICHE niche neighborhoods in patients who relapsed (red) or did not relapse (blue) in the Spain cohort. QUICHE generalized linear models included TLS status, T aggregate status, and recurrence status as covariates. Barplots to the right show the proportion of patients with a QUICHE niche neighborhood in the respective patient groups. (i) Boxplots show the distribution of collagen fiber metrics across patients that did (1, n=109 patients) or did not relapse (0, n=205 patients) (median +/- interquartile range; whiskers show 1.5*IQR). p-values were computed using a two-sided Wilcoxon rank sum test. Multiple hypothesis testing correction was performed using Benjamini Hochberg. (j) Heatmap shows the Spearman rank correlation between collagen fiber metrics and predicted recurrence-associated QUICHE niche neighborhoods. (k) Barplot shows the number of required channels for each differentially enriched QUICHE niche neighborhood. Source data is provided in the Source Data file.
Extended Data Fig. 9 Validation of outcome-associated QUICHE analysis on TNBC cohorts.
A Cox proportional hazards model was trained on either the abundance of (a) QUICHE niche neighborhoods, (b) cell types, or (c) K-Means++ spatial clusters in the Spain TNBC cohort to predict recurrence risk in the independent Stanford TNBC cohort. For each method, patients were stratified into two groups based on the median risk score. p-values were calculated using a two-sided Wilcoxon log-rank test to compare recurrence free survival differences between high- and low-risk groups. Left panels show the Kaplan-Meier curves comparing risk groups in the Stanford cohort for each method. Middle panels show the stability of the Log-rank test statistic using a bootstrap resampling approach, where the Stanford cohort was randomly resampled with replacement over 10,000 iterations. Two-sided p-values were calculated from bootstrapping. Right panels show robustness of the models using a permutation test, where relapse labels were randomly shuffled within the Stanford cohort without replacement over 10,000 iterations, while keeping the risk groups fixed. Two-sided p-values were calculated from a permutation test. (d) Scatter plots show the number of significant QUICHE niche neighborhoods as a function of the total number of cells in each patient sample for Spain (left), Stanford (middle), and NeoTRIP (right) TNBC cohorts. r indicates the Pearson correlation coefficient. (e) Boxplots show the average distance between cells within an annotated QUICHE niche neighborhood (intra distance) as compared the remaining annotated niche neighborhoods (inter distance) in the Spain (left, inter: n=25 niches, intra: n=25 niches), Stanford (middle, inter: n=60 niches, intra: n=60 niches), and NeoTRIP (right, inter: n=26 niches, intra: n=26 niches) TNBC cohorts (median +/- interquartile range; whiskers show 1.5*IQR). Intra-distances are lower than inter-distances validating pseudobulk niche labeling performance. (f) Boxplots plots show the log2 fold change in the abundance of niche neighborhoods across permuted outcome labels (left) and spatial FDR values (right) in the Spain (left), Stanford (middle), and NeoTRIP (right) TNBC cohorts (median +/- IQR; whiskers show 1.5*IQR). Source data is provided in the Source Data file.
Supplementary information
Supplementary Table 1
Supplementary Tables 1–5.
Source data
Source Data
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ranek, J.S., Greenwald, N.F., Goldston, M. et al. The automated computational workflow QUICHE reveals structural definitions of antitumor responses in triple-negative breast cancer. Nat Cancer (2026). https://doi.org/10.1038/s43018-026-01122-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43018-026-01122-5