Abstract
Understanding dissolved organic matter (DOM) transformation is crucial for comprehending soil biogeochemical cycling. However, the extent that soil microbes mediate DOM transformation at the molecular level, and whether this is regulated by fertilization remain largely unknown. Here we investigated soil DOM transformations under long-term fertilization using Fourier-transform ion cyclotron resonance mass spectrometry, high-throughput sequencing, and machine learning. Fertilization greatly promoted transformation potential of DOM molecules. Organic fertilization increased the mean transformation number of DOM molecules by 260% compared to no-fertilization, while chemical fertilization increased it by 193%. Machine learning indicated that intrinsic DOM molecular characteristics could predict transformation potential, especially for medium- or low-transformation-potential molecules. However, high-transformation-potential DOM molecules were more influenced by soil microorganisms. Our study provides a parameter to characterize potential transformation capacity of DOM molecules, the effects of different fertilization treatments on this potential, and highlights microbial contributions to molecular transformation processes, identifying the key microbial groups.

Similar content being viewed by others
Introduction
Paddy soils (0–100 cm) contain 18 Pg carbon worldwide, which is 10% higher than the average for soils generally. They account for 1.2% of the global carbon stock and 14.2% of the carbon stock in agricultural land1. The carbon cycle in rice paddies is not only affects soil fertility and crop productivity, but also an essential part of the global carbon cycle, which is important for understanding and regulating global climate change. Rice paddies contribute to greenhouse gas emissions, particularly methane (CH₄), because the anaerobic conditions created by flooded fields promote the activity of methanogenic microorganisms2. This increased microbial activity facilitates the decomposition of organic matter, leading to enhanced carbon transformation processes that result in higher methane emissions3. Dissolved organic matter (DOM) is an essential part of the soil carbon pool and plays an important role in soil biogeochemical processes, plant productivity, and human health4. The transformation of DOM molecules in rice paddies not only enhances soil quality by improving nutrient availability and microbial activity but also plays a crucial role in regulating greenhouse gas emissions, such as carbon dioxide and methane. This process directly influences ecosystem functions, including carbon sequestration and plant growth, thereby linking specific agricultural practices, such as fertilization and water management, to broader climate dynamics5. Previous studies on fertilization have generally demonstrated that DOM is influenced by fertilization6. The responses of DOM concentrations to fertilization vary across different locations globally, indicating that complex mechanisms may be at play7,8. Numerous studies have highlighted changes in DOM concentration following fertilization, with a secondary focus on shifts in its bioavailability of dissolved organic carbon9. However, because DOM molecules in soil ecosystems are highly dynamic and diverse, it remains highly challenging to comprehensively understand DOM transformations in soils.
Transformation potential refers to the ability of DOM molecules to undergo biochemical changes, which influences their persistence, degradation, and contribution to soil organic matter cycling10. Unlike the bioavailability of dissolved organic carbon, which specifically addresses the fraction of DOM that is readily bioavailable to microorganisms11, transformation potential encompasses broader biochemical reactions that include both labile and recalcitrant compounds10. It is assumed that when external conditions, such as nutrient levels or environmental factors, are altered, DOM molecules will undergo transformations12. Additionally, it is assumed that the ability of DOM transformation is closely linked to external drivers, meaning that changes in factors like fertilization or climate can influence molecular transformations13. Transformation potential is typically measured by analyzing the changes in molecular composition, often using advanced techniques like mass spectrometry14. However, there is a lack of consensus regarding the accurate characterization of the potential transformation capabilities of soil DOM molecules.
Previously, biochemical reaction categories have been mainly used as the smallest research unit to investigate the transformation potential of DOM molecules. However, characterizing DOM transformation potential based on biochemical reaction classification is not sufficiently precise. For instance, lignin is typically viewed as recalcitrant in degradation processes15. However, lignin-like compounds can interconvert16, and recent findings indicate that environmental factors influence lignin degradation, challenging the notion of it being absolutely recalcitrant17. Thus, relying on biochemical classifications can lead to imprecise conclusions about the transformation potential of different molecules within the same category. In previous research, the classification of the transformation potential of DOM molecules was based on their presence/absence under different states18 or based on molecular weight to investigate the DOM transformation after dividing molecules into different categories19. Nevertheless, it should be noted that the aforementioned categorizations may lack generalizability, which poses challenges in their application across diverse environmental contexts. Recently, Yu et al.20 introduced a novel approach known as reactomics, which enables the assessment of untargeted mass spectrometry (MS) profiles at the reaction level. The principle of reactomics is to infer the bioreactions between paired DOM molecules based on paired mass distance (PMD) information. Using this analysis, a previous study demonstrated that the microbe-mediated DOM transformations mainly include the oxidoreduction of CH–NH2 groups, transferring nitrogenous groups, and formation of the C–N bond in the wastewater treatment process21. Consequently, we assume that PMD-based reactomics may be a powerful tool for deciphering soil DOM transformation.
The interactions between DOM and microorganisms have consistently garnered attention22 since DOM in natural ecosystems is the main nutrient and energy source for microbes. As such, DOM is greatly consumed or transformed by microbes. However, it remains largely unknown how microorganisms mediate the transformation of DOM, since both the microbial community and DOM transformation are highly complex21, and the DOM transformation is difficult to track. Although some recent studies have attempted to disentangle the microbial-mediated DOM transformation using ecological models, e.g., network analysis22, they only provided some overall DOM–microbe interactions23, and the more refined relationships between microorganisms and DOM transformations were not revealed. For example, it remains uncertain whether microorganisms influence the potential transformation of specific molecules or if certain molecular reactions are directly mediated by given microorganisms.
Agricultural practices play a pivotal role in shaping the dynamics of carbon sources and sinks in the environmen24. As the largest anthropogenic wetlands on earth25, paddy fields serve as both carbon sources and sinks. The transformation and dynamics of DOM in these systems influence carbon sequestration and emissions, thereby impacting climate change. Specifically, the biochemical processes that govern DOM production, degradation, and turnover in paddy fields can either enhance or mitigate greenhouse gas emissions, highlighting their importance in the context of global climate change26. However, there is limited knowledge regarding the transformation potential of DOM molecules in paddy soils under different fertilization treatments and the mediating role of microorganisms. Using high-resolution MS, we obtained molecular formulas for DOM compounds in paddy soils under long-term different fertilization in this study. Based on PMD analysis of molecular formulas detected by Fourier-transform ion cyclotron resonance mass spectrometry (FT-ICR-MS), we determined the maximum transformation number (MTN) of DOM molecules and introduced the concept of molecular transformation potential (MTRP), which focuses on tracking changes in specific reactants and products to show the transformation potential of DOM under different conditions. Leveraging machine learning, we also studied the impact of molecular characteristics and microbes on MTRP. The aims of this study were to (i) identify a parameter that can serve as an indicator of the potential transformation capacity of DOM molecules, (ii) investigate the changes in the transformation potential of DOM molecules under different fertilization treatments, and elucidate the intrinsic factors driving these variations in paddy soils, and (iii) understand how microorganisms contribute to the role in enhancing MTRP.
Results and discussion
The physicochemicals of soil under different fertilization
The initial physicochemical properties of the untreated soil sample were as follows: pH 5.11, total nitrogen (TN) 0.47 g kg−1, total phosphorus (TP) 0.5 g kg−1, total potassium (TK) 16.9 g kg−1, available nitrogen (AN) 0.0063 g kg−1, available phosphorus (AP) 0.0033 g kg−1, available potassium (AK) 0.07 g kg−1, free iron oxide (Fe₂O₃) 20.31 g kg−1, free aluminum oxide (Al₂O₃) 4.60 g kg−1, and organic matter (OM) 8.94 g kg−1. After long-term exposure to different fertilization treatments, there were changes in the soil's physical and chemical properties. Compared to the control (no fertilization), under chemical fertilization treatments, the OM, TN, and AN content significantly increased, while AK, Fe₂O₃, and Al₂O₃ contents significantly decreased. Under organic fertilization treatments, the TP, TK, and AP contents significantly increased, while the Al₂O₃ content significantly decreased (Supplementary Fig. 1).
The molecular composition of DOM under different fertilization
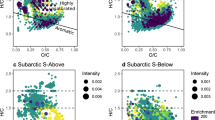
The FT–ICR–MS was employed to determine the molecular composition of DOM. There were 1503–6129 molecules detected under different fertilization treatments (Supplementary Table 1). A total of 7682 distinct molecules across the analyzed samples were also revealed. A van Krevelen diagram was used to visualize the compound distribution (Supplementary Fig. 2(a)–(c)), which distinctly divided soil DOM biochemical categories into seven regions: carbohydrate, condensed aromatic, lignin, lipid, protein/amino sugar, tannin, and other compounds. Lignin was the most abundant molecule type in the DOM originating from the long-term fertilization paddy soils, accounting for 59.4–60.6% of all assigned molecules (Supplementary Fig. 2(d)). The highest abundance of lignin was under no fertilization. Tannin and protein/amino sugar contents increased while lipid content decreased under fertilization treatments, and the highest abundances of both tannin and protein/amino sugar were under chemical fertilization, and the lowest abundance of lipids was under organic fertilization (Supplementary Fig. 2(d)). The principal coordinate analysis (PCoA) showed significant variation in DOM composition under different fertilization treatments (p < 0.001) (Supplementary Fig. 2(e)). The possible reasons follow: first, organic fertilization introduces allochthonous DOM, leading to an increase in tannins content and a decrease in lipid content within the DOM pool27, which is similar to our results. Second, the application of organic fertilizer has been shown to enhance microbial diversity and composition28, and the increase in microbial diversity is believed to contribute to the accumulation of recalcitrant compounds such as protein/amino sugar29. Moreover, DOM transformation is partly influenced by physical and chemical conditions30, which have different effects on the components of DOM31.
PMD-based reactomics of DOM under different fertilization
Based on reactomics analysis, we identified reactant–product pairs with specific PMD values20,21. We identified 131 PMDs as widely present among DOM molecules and quantified their relative intensities (Fig. 1). Low-molecular-weight PMDs were prevalent, indicating that DOM transformation in the soil was more likely to involve the addition or reduction of low-molecular-weight functional groups. Previous studies indicated that smaller, plant-derived molecules are more likely to undergo transformation into larger, more complex compounds during DOM processing, particularly through the addition or removal of low-molecular-weight functional groups14,21, consistent with our study. Compared to organic fertilization, both the CK and chemical fertilization treatments exhibited lower overall reaction levels. For each specific DOM molecule, we quantified its total PMD pair count (i.e., MTN) (Supplementary Data 1). To avoid bias caused by DOM molecule numbers, we calculated the MTN of different fertilization treatments after random sampling (high similarity between random samples, p = 0.98). We found the highest MTN values of DOM molecules under organic fertilization (mean = 54), followed by chemical fertilization (mean = 44) and then no fertilization (mean = 15). This suggested possibly enhanced DOM transformation under organic fertilization. Usually, organic fertilization results in a higher proportion of labile fractions32, which may contribute to the higher DOM molecular transformation. Previous studies also demonstrated that organic fertilization enhances soil organic matter activity by increasing mineral-associated organic matter-C turnover and reducing its mean residence time33, further supporting our findings.
Top 30 of the 131 PMDs that are widely present among DOM molecules (a) and their relative intensities in different fertilization treatments (b). CK no fertilization added and complete removal of crop straw, NK urea and potassium chloride, NP urea and calcium-magnesium phosphate, NPK urea, calcium-magnesium phosphate, and potassium chloride, NPKgm NPK chemical fertilization plus green manure, NPKpm NPK chemical fertilization plus pig manure, NPKst NPK chemical fertilization plus rice straw. Each bubble represents the occurrence frequency of a specific PMD under a given treatment, with bubble size corresponding to the frequency (value ranging from 1000 to 4000).
DOM molecule transformation under different fertilization
We constructed multiple PMD networks to reveal the core DOM molecules involved in the transformations. The high proportion (accounting for 57.6%) of lignin molecules in the network indicated their predominant roles in soil DOM transformation (Fig. 2). For the same biochemical category, fertilization treatments changed the proportion of molecules contained in the network. Zou et al.34 also demonstrated that the application of fertilization can regulate changes in the molecular composition of organic matter, consistent with our findings. For example, the fertilization treatment altered the proportion of different types of molecules participating in transformations. The fertilization treatment increased the proportion of molecules of carbohydrate (CK, 1.5%; fertilization, 1.6–1.8%), lipids (CK, 2.1%; fertilization, 2.6–3.4%), and tannins (CK, 5.9%; fertilization, 7.9–8.7%), while reducing those of lignin (CK, 57.6%; fertilization, 57.2–56.5%) and protein/amino sugar (CK, 5.6%; fertilization, 4.5–4.7%) (Fig. 2). This increase in AP and TP due to fertilization enhanced the microbial breakdown of carbohydrates and tannins, leading to higher proportions of these molecules under fertilized conditions, aligning with the expected impact of phosphorus availability on the degradation rates of specific organic molecules35. This explains why the proportions of carbohydrates and tannins involved in transformations increased under fertilization treatments. Secondly, under the no-fertilization treatment, the Al₂O₃ content increased compared to fertilization. The Al₂O₃ can interact with lipids, tannins, and hydroxyl groups in carbohydrates, reducing their bioavailability and making them more resistant to transformation36,37. This explains why the proportions of lipids, tannins, and carbohydrates involved in transformations decreased under no-fertilization treatment, as higher Al₂O₃ levels made these molecules less bioavailable. Additionally, the increase in nitrogen content due to fertilization likely accelerates carbohydrate metabolism38. This further confirms the increase in the proportion of carbohydrates involved in transformations under fertilization treatments. Furthermore, the fertilization treatments increased total MTN (Fig. 3) in specific chemical categories based on the PMD network. Although the proportion of lignin molecules was lower under the fertilization treatments, this is likely because lignin, a major component of plant cell walls, is difficult to degrade and relies on specialized microbial communities and enzymes39. The increased organic matter content under fertilization supports a more diverse microbial community, which facilitates lignin degradation and enhances its overall transformation40. We selected the top 1% of molecules with higher MTN in each treatment as key molecules, which were primarily composed of CHO and CHON (Supplementary Fig. 3). The transformation reactions of molecules were improved under fertilization treatments (Supplementary Data 2). Application of organic (mean MTN 89) and chemical (mean MTN 101) fertilization resulted in higher connectivity in the molecular network compared to the control (mean MTN 44) (Supplementary Fig. 3).
a CK plots with no fertilization and complete removal of crop straw, b NP plots with urea and calcium–magnesium phosphate, c NK plots with urea and potassium chloride, d NPK plots with urea, calcium–magnesium phosphate, and potassium chloride, e NPKst plots with NPK chemical fertilization plus rice straw; f NPKpm plots with NPK chemical fertilization plus pig manure, and g NPKgm, plots with NPK chemical fertilization plus green manure.
CK no fertilization added and complete removal of crop straw, NK urea and potassium chloride, NP urea and calcium-magnesium phosphate, NPK urea, calcium-magnesium phosphate, and potassium chloride, NPKgm NPK chemical fertilization plus green manure, NPKpm NPK chemical fertilization plus pig manure, NPKst NPK chemical fertilization plus rice straw.
Impact of molecular characteristics on molecule transformation
We employed machine learning to predict MTRP classes based on molecular characteristics. The k-means algorithm classified the molecules into high (MTN > 70), medium (MTN > 39 and MTN ≤ 70), and low classes (MTN ≤ 39). The average MTRP belongs to the low class under CK treatment, middle class under chemical treatment, and high class under organic treatment, partly indicating the difference in MTRP under different fertilization treatments. The machine learning model achieved an accuracy of 85.47% on the test set, indicating a correlation between molecular characteristics and MTRP classes. We also found that the DOM molecules with low transformation potential consistently demonstrated the highest precision and recall by machine learning (precision: 91.7% in low transformation potential, 85.7% in medium transformation potential, and 82.3% in high transformation potential; recall: 89.1% in low transformation potential, 85.7% in medium transformation potential, and 80.3% in high transformation potential) (Fig. 4), suggesting that the molecular characteristics had higher efficiency in predicting the transformation potential of low-transformation-potential molecules. On the contrary, the transformation potential of high-transformation-potential molecules cannot be accurately predicted by molecular characteristics.
a NP plots with urea and calcium–magnesium phosphate, b NK plots with urea and potassium chloride, c NPK plots with urea, calcium-magnesium phosphate, and potassium chloride, d NPKst plots with NPK chemical fertilization plus rice straw, e NPKpm plots with NPK chemical fertilization plus pig manure, f NPKgm, plots with NPK chemical fertilization plus green manure. Each row of the confusion matrix represents the actual class, while each column represents the predicted class. “True” means the true transformation potential class of the molecule; “Predict” means the predicted transformation potential class of the molecule; and 1–3 represent the molecules with high, medium, and low transformation potential, respectively. The percentages in the blue squares represent the percentage of prediction correct under each classification, and the percentages in the brick red squares represent the percentage of prediction errors under each classification.
We calculated feature importance and found that molecular characteristics such as O/C and H/C ratios were closely related to MTRP (Supplementary Fig. 4(a)). To avoid the potential bias of a single algorithm, we employed multiple algorithms, which consistently supported the same conclusion (Supplementary Fig. 4(a)). Furthermore, the findings regarding the effect of intrinsic molecule characters on MTRP under different treatments were similar (Supplementary Fig. 4(b)). Therefore, we constructed a distribution plot of MTRP based on Gibbs free energy and O/C and H/C ratios (Supplementary Fig. 4(c)). The plot clearly demonstrated distinct ranges of MTRP classifications, suggesting that to some extent, the distribution of MTRP classifications is determined by Gibbs, O/C, and H/C ratios. Moreover, we observed a significant difference (p < 0.001) in the aromaticity index (AI) values of DOM molecules with different transformation potential (Supplementary Fig. 5). Similar to previous research, molecules with a high transformation potential had lower AI values than those with low transformation potential41 and, to some extent, validates the accuracy of the transformation potential classification derived from our clustering analysis.
Relationships between microbial composition and DOM transformation potential
Given that the molecular characteristics had a relatively lower efficiency in predicting the transformation of high-transformation-potential molecules, we hypothesize that the high-transformation-potential molecules may be more affected by other factors such as soil microorganisms42,43. Therefore, differences in microbial communities under different fertilization treatments were also observed in our study (Supplementary Figs. 6 and 7). The redundancy analysis (RDA) indicated that soil AP, TP, and AK significantly influenced community composition of soil microorganisms, leading to shifts in microbial community structure under different fertilization treatments (Supplementary Fig. 8). On this basis, we conducted a Mantel test and Procrustes analysis to investigate the potential links between DOM molecules with different transformation potentials and soil microbial community composition (Fig. 5a–c, Supplementary Fig. 9). The high-transformation-potential, rather than medium- or low-transformation-potential, DOM molecules showed stronger associations with microorganisms (low-transformation-potential: r = 0.049, p = 0.078; medium-transformation-potential: r = 0.187, p = 0.028; and high-transformation-potential: r = 0.513, p = 0.006). Furthermore, the associations between microbial community and high-transformation-potential molecules were much stronger under organic fertilized soils than inorganic fertilized soils (r = 0.632, p = 0.004 and r = 0.395, p = 0.005, respectively). These findings clearly suggest that the high-transformation-potential molecules were more influenced by soil microorganisms, especially under organic fertilization treatments. Previous studies indicated that microbial diversity plays a crucial role in shaping the chemical diversity of soil DOM16,44. Our results showed that the stronger DOM molecule reactivity, the more it was affected by soil microbial communities, consistent with a previous finding of a much greater influence of microorganisms on reactive than on recalcitrant components27. This may be attributed to less accessibility of recalcitrant components for microbial utilization45.
Mantel test-based correlations between microbes and DOM molecules under (a) no-fertilization, b chemical fertilization, and c organic fertilization treatment. Low, Mid, and High mean high-transformation-potential DOM molecules classified by machine learning. Color gradient denotes Spearman’s correlation coefficients, edge color denotes the p-value, and edge width corresponds to Mantel’s r statistic for the corresponding distance correlations.
Microbe-mediated reactions involved in DOM transformation
Based on the 131 PMDs identified above (Fig. 1), we were able to infer the molecular transformation processes, which led to the identification of the enzymatic reactions involved in DOM transformation. Then we compared the enzymatic reactions with the KEGG database, which served as a bridge to identify the microbial taxa and enzymes involved in mediating the DOM transformation processes (Supplementary Data 3). In Fig. 6a, we illustrate that a larger proportion of operational taxonomic units (OTUs), a proxy for microbial species in biodiversity studies, enriched under organic fertilization treatment contributed functional genes necessary for DOM transformation. Among the 243 differential OTUs, 187 (76.95%) were enriched under the organic fertilization treatment and contributed to the functional genes required for DOM transformation. Similarly, among the 102 differential OTUs enriched under the chemical fertilization treatment, 62 OTUs (60.78%) contributed to the functional genes required for DOM transformation. In contrast, among the 356 differential OTUs under the CK treatment, only 105 OTUs (29.49%) contributed to the functional genes required for DOM transformation (Fig. 6a). These findings suggest an increased proportion of microorganisms that contributed to DOM transformations in fertilized soils.
a Venn diagram of OTUs enriched under different fertilization treatments and OTUs providing functional genes required for transformation. b Correlations between specific microbial taxa and enzymes involved in DOM transformation. c Microorganisms enriched under no fertilization (G1), chemical fertilization (G2), and organic fertilization (G3). Group A, OTUs that provide the genes needed for transformation; Group B, OTUs enriched under no-fertilization treatment; Group C, OTUs enriched under chemical fertilization treatment; and Group D, OTUs enriched under organic fertilization treatment. The numbers inside the colored squares of c represent the number of specific microbes at different levels of classification.
Figure 6b highlights the correlations between specific microbial taxa and enzymes involved in DOM transformation, while Fig. 6c shows the microbial groups enriched under different fertilization treatments. Under organic fertilization treatment, there was an increase in key microbial groups (Desulfobacterota and Gemmatimonadota) positively correlated with the metabolic pathways of molecules with high transformation potential. For example, caffeic acid O-methyltransferase catalyzes the methylation of oxygen atoms on the hydroxyl groups of phenylpropanoids, playing a critical role in the composition of monolignols, and is a key enzyme in the biosynthesis of lignin and lignans46. In contrast, no fertilization treatment increased the key microbial group (Chloroflexi), which was negatively correlated with the metabolic pathways of molecules with high transformation potential. For instance, lavonoid 3′-monooxygenase is mainly involved in reduction reactions, which is closely related to carbon degradation (Fig. 6b, c). Our study emphasized the importance of microbial roles in altering DOM MTRP. Thus, fertilization might be involved in molecular transformation through changes in certain microbes and regulating key functional genes17,47,48.
Conclusion
We conducted high-resolution MS measurements, combined with machine learning and data mining based on PMD reactomics, to augment our understanding of the transformation potential of DOM and its variation under different fertilization treatments mediated by microorganisms. Our study provides a critical parameter that can serve as an indicator of the transformation potential of DOM molecules, offering a practical tool for evaluating and managing paddy soil ecosystems, particularly in relation to carbon cycling and transformation processes under different fertilization treatments. The findings contribute to improving conceptual frameworks related to soil organic matter transformation and microbial interactions. Specifically, our results suggest that fertilization practices, especially organic fertilization, can promote the transformations of DOM molecules and that the intrinsic molecular characteristics such as O/C and H/C ratios can effectively be used to predict DOM transformation potential. The high-transformation-potential DOM molecules were more influenced by soil microorganisms such as Desulfobacterota and Gemmatimonadota. Despite some uncertainties, the PMD-based reactomics method can be applied to MS data, and it has been proven effective in water systems. Further research is needed to validate its applicability in diverse natural environments, with the aim of expanding its use to other ecological contexts in the future. This understanding will ultimately contribute to a more comprehensive grasp of the global carbon cycle.
Materials and methods
Experimental site and soil sampling
The soil samples were collected from a long-term paddy field experiment site in Yingtan City, Jiangxi Province, China (28°15′30″N, 116°55′30″E). This long-term test plot was established in 1990 and was reclaimed from local red soil wasteland, so the climatic conditions, soil matrices, and initial fertility were identical, and the main factor contributing to the differences in soil properties was the 31 years of long-term differential fertilization. The site is characterized by a subtropical monsoon climate, with an average annual temperature of 17.6 °C and an average annual precipitation of 1795 mm. All soils, classified as typical Ultisols (USDA Soil Taxonomy), were derived from Quaternary red clay. Soil samples were collected in November 2021 from the 0 to 20 cm depth range, with three replicates for each of the seven different fertilization treatments: (1) CK, plots with no fertilization added and complete removal of crop straw; (2) NP, plots with 115 kg ha−1 N and 68 kg ha−1 P2O5 per season; (3) NK, plots with 115 kg ha−1 urea and 41 kg ha−1 K2O per season; (4) NPK, plots with 115 kg ha−1 N, 68 kg ha−1 P2O5, and 41 kg ha−1 K2O per season; (5) NPKst, plots with NPK chemical fertilization plus 2500 kg ha−1 rice straw per season; (6) NPKpm, plots with NPK chemical fertilization plus 2500 kg ha−1 pig manure per season; and (7) NPKgm, plots with NPK chemical fertilization plus 2500 kg ha−1 green manure (Astragalus sinicus L.) per season. The C contents in rice straw, pig manure, and green manure were 387, 267, and 441 g kg−1 respectively. Accordingly, the N contents were 9.88, 21.22, and 28.22 g kg−1 respectively. These treatments were designed to represent shifts in fertilization practices over different historical stages in China, with a focus on several organic fertilization regimes that are representative in southern regions. These seven treatments can be grouped into three categories: no fertilization (1), chemical fertilization (2–4), and organic fertilization (5–7). After soil sampling, all samples were stored at 4 °C for subsequent processing and analysis.
Soil chemical property measuring
Soil pH was determined using a pH meter in a 1:2.5 of soil:water suspension. The total nitrogen (TN) content in soil was determined using the Kjeldahl method. The available nitrogen (AN) was determined using the alkali hydrolysis and micro diffusion method. Total phosphorus (TP) and available phosphorus (AP) were determined by the vanadium-molybdate photometric method, and total potassium (TK) and available potassium (AK) were determined by inductively coupled plasma-atomic emission spectrometry. Soil organic matter (OM) was determined using the potassium dichromate volumetric method. Free iron oxide (Fe₂O₃) and free aluminum oxide (Al₂O₃) in the soil were determined using atomic absorption spectroscopy.
Fourier-transform ion cyclotron resonance MS (FT-ICR-MS) analysis
The DOM was extracted from soil samples (6 g) using a soil-to-water ratio of 1:5 and ultrapure water. Ultrapure water used for all experiments and solutions had a resistivity of 18.2 MΩ cm at 25 °C and a total organic carbon content lower than 5 ppb. The mixture was subjected to shaking for 12 h at room temperature on a horizontal shaker. Subsequently, the solutions were centrifuged at 1200×g for 10 min and filtered through a 0.45 μm membrane filter. For clean-up, HPLC methanol (10 mL) and acidified ultrapure water (10 mL, pH 2) were passed through PPL cartridges (Agilent Technologies, Santa Clara, CA, USA). The DOM solution was then loaded onto the PPL cartridges by gravity flow. Following that, DOM was collected from the cartridges using 10 mL of methanol (HPLC grade; Merck, Darmstadt, Germany). The collected DOM eluates were stored at −20 °C in darkness prior to electrospray ionization FT–ICR–MS analysis.
A deuterated octadecanoic acid compound was added to the samples as an internal standard, with a dosage of 15 μL (5 × 10−7 mol L−1) per milliliter of the sample. The FT–ICR MS instrument (Bruker, Billerica, MA, USA) utilized a 9.4 T actively shielded superconducting magnet in negative-ion mode. Each sample was injected into the ESI source at a flow rate of 180 μL h−1 using a syringe pump. The polarization voltage was set at 4.0 kV, while the capillary column introduction and outlet voltage were 4.5 kV and 320 V, respectively. Ions were accumulated in the hexapole for 0.001 s before being transferred to the ICR cell. The mass-to-charge ratio (m/z) range analyzed was 150–800 Da. A time-domain signal acquisition with a 4 M word size was selected. The signal-to-noise ratio and dynamic range were enhanced through the accumulation of 128 domain FT–ICR transients.
The data obtained were processed using the Data Analysis software (Bruker Daltonics version 4.2). The raw spectra were converted into a list of mass-to-charge ratio (m/z) values using the FTMS peak picker algorithm, with a signal-to-noise (S/N) threshold of 6 and an absolute intensity threshold of 100. To minimize cumulative errors, all peaks from the entire dataset were aligned with each other, ensuring the elimination of potential mass shifts. The molecular formulas of the identified mass peaks were determined using custom software designed for this purpose49. The assigned molecular formulas were classified into distinct categories based on the ratios of oxygen to carbon (O/C) and hydrogen to carbon (H/C) as follows: lipids for H/C = 1.5–2.0, O/C = 0–0.3; aliphatic/proteins for H/C = 1.5–2.2, O/C = 0.3–0.67; lignin/CRAM-like for H/C = 0.7–1.5, O/C = 0.1–0.67; carbohydrates for H/C = 1.5–2.4, O/C = 0.67–1.2; unsaturated hydrocarbons for H/C = 0.7–1.5, O/C = 0–0.1; aromatic structures for H/C = 0.2–0.7, O/C = 0–0.67; and tannin for H/C = 0.6–1.5, O/C = 0.67–1.016.
Reactomics analysis and PMD network analysis
The PMD-based Reactomics was used to characterize the transformation process of DOM molecules. It was previously reported that reactant and product pairs in a reaction were formed by exchanging functional groups or atoms50. FT–ICR–MS can be used to analyze DOM molecules under different conditions to identify variations in molecular composition. Based on this, PMD analysis helps to identify specific molecular transformation events (such as oxidation, condensation, and hydrogenation), thereby revealing the molecular transformation pathways of DOM. For instance, a reaction generally occurs with the loss or addition of some typical groups such as CH2, O, or NH. The PMD, calculated by the mass difference between the reactant and product, can reflect such reaction rules. The aggregate of PMDs in one sample with complex DOM molecules may present the overall reaction profiles, called PMD-based Reactomics20. By performing PMD-based metabolomics analysis using the R package ‘pmd’ in R version 4.2.3, we constructed a peak list and applied the ‘GlobaStd’ function to identify independent peaks within the list. The ‘getrda’ function was used for targeted analysis of MS data. Subsequently, the ‘getstd’ function was employed to remove adducts, neutral losses, and common fragment ions50. By comparing against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database46, representative PMD values were identified. For example, we chose the pathways related to the carbon cycle, including carbon fixation (sdhD, ACO, and acsB) and carbon degradation (pulA, celC, and CBH2). Each reaction from these pathways was split into several reactant–product pairs. The PMD was calculated from the mass difference of each reactant–product pair. For each specific molecule, we quantified its total PMD pair count, termed the maximum transformation number (MTN). We posit that MTN reflects the molecule’s molecular transformation potential (MTRP). The ‘getreact’ function was then utilized to quantify the abundance of ion pairs in different samples20. In order to avoid the bias of MTN caused by the difference in total molecule numbers under different fertilization treatments, we calculated the MTN by randomly sampling the smallest 1503 molecules (equal to the numbers under CK treatment) with 1000 bootstraps under different treatments.
The PMD-based network was built by searching for the specific PMD values in DOM molecules in each sample. Each molecule in the PMD-based network had a connection count, representing potential transformation pairs.
Evaluation of transformation potential by machine learning
Cluster analysis based on machine learning was employed to evaluate the transformation potential of each molecule. First, the MTN and molecular characteristics were organized into a characteristic matrix, where each row represented a molecule, and each column represented a specific characteristic and MTN. The characteristic matrix was standardized using the Z-score normalization method to eliminate the dimensional differences among different characteristics and MTN. The MTN was weighted tenfold to characterize the transformation potential of DOM molecules. We employed the k-means algorithm to categorize MTRP into three classes (low, medium, and high), selecting three molecules at random as initial cluster centers through iterative optimization of the cluster center positions, either until convergence or after a predetermined number of iterations. The distance of each molecule to its cluster center (D) was calculated, taking into account the weighting factor of the number of transformations, and this was assigned to the nearest cluster center:
where \(D({x}_{i},{x}_{j})\) is the weighted Euclidean distance between molecules \({x}_{i}\) and \({x}_{j}\), n is the number of characteristics, \({w}_{k}\) is the weight factor of characteristic k, and \({x}_{{ik}}\) and \({x}_{{jk}}\) represent the values of samples \({x}_{i}\) and \({x}_{j}\) on characteristic k, respectively.
For each cluster, the weighted mean of all molecules within the cluster was calculated, and the weighted mean then assigned as the new cluster centroid (C). This process was iterated until the positions of the cluster centroids no longer changed or the maximum number of iterations was reached:
where \({C}_{k}\) is the centroid of the kth cluster, \({N}_{k}\) is the number of data points in the kth cluster, \({w}_{i}\) is the weight factor of data point \({x}_{i}\), and \({x}_{{ik}}\) is the value of data point \({x}_{i}\) on feature k.
According to the final cluster centroids, MTRPs of molecules with different MTN were clustered into low, medium, and high classes, and each molecule was assigned to its respective cluster based on the MTN under different fertilization treatments.
Evaluation of impact factors of transformation potential by machine learning
Machine learning can also be used to investigate the impact of intrinsic molecular characteristics on MTRP. Therefore, we utilized a supervised machine learning approach using the random forest (RF) algorithm with particle swarm optimization (PSO). Molecular characteristics were used as input characteristics, and MTN was used as the data labels to construct the datasets. In this study, seven datasets were constructed for three-class classification (Supplementary Table 1), and the datasets were randomly split into training and testing sets in an 8:2 ratio. We used 10-fold cross-validation in the low-level dataset to avoid the classifier training underfitting the low-level dataset. The PSO algorithm was employed to optimize the hyperparameters of the RF model, including n estimators, bootstrap, oob_score, max_depth, min_samples_leaf, and max_leaf_nodes. By computing the corresponding fitness values and continuously updating the velocities and positions of particles, the optimal fitness value was achieved, resulting in the determination of the best hyperparameters for the RF model. This approach enhanced the convergence speed and prediction performance of the RF model.
To gain a more detailed understanding of the impact of individual molecular characteristics on MTRP, we computed feature importance in the PSO-RF model. Calculation of feature importance in the PSO-RF model is typically based on two factors: the contribution of the characteristic to the node splits in the decision trees and the importance of the characteristic in the randomization process.
For each decision tree during the training process, a different training set was generated by sampling with a replacement from the original training set. At each node, a subset of characteristics was randomly selected from all the characteristics. The selected characteristics were then used to split the node in a way that maximized the purity of the target variable. The contribution of each characteristic to the splits was recorded as a weight or importance value. Subsequently, feature importance was aggregated. For each characteristic, all decision trees in the RF were traversed, and for each tree, the contribution of the characteristic at each node was accumulated. Finally, the contribution values were averaged across all decision trees to obtain the overall importance of the characteristic.
DNA extraction and sequencing analysis
Soil microbial genomic DNA was extracted from 0.5 g of fresh soil using a FastDNA™ SPIN Kit (MP Biomedicals, Santa Ana, CA, USA). The DNA sequencing was conducted on an Illumina NovaSeq platform51.
The PCR amplification process involved specific primers targeting the 16S rRNA gene for bacterial communities. The PCR conditions included an initial denaturation step at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 45 s, with a final extension at 72 °C for 10 min.
Data preprocessing included quality filtering, trimming, and merging of sequencing reads. The filtered sequences were assigned taxonomy using a reference database. The resulting sequences were then analyzed for microbial diversity and community composition.
Data analysis
All statistical analyses were performed using R (v4.2.3). The Mantel test was conducted with the R package “phyloseq” to evaluate correlations between the compositions of microorganisms and DOM molecules52. Using the R package ‘vegan’, Redundancy Analysis (RDA) examined microbial-soil physicochemical properties relationships, while Procrustes analysis assessed microbial-DOM consistency. Principal coordinate analysis (PCoA), based on Bray–Curtis dissimilarity, was used to explore microbial community structures. Linear discriminant analysis effect size (LEfSe) was used to identify microbial features distinguishing different treatments, using the “microeco” R package. For microbial community gene abundance prediction, PICRUSt was used53, and a final list of latent function genes along with their abundances was generated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The Fourier transform ion cyclotron resonance mass spectrometry data, along with Supplementary Data 1, 2, and 3, have been deposited in the Dryad Digital Repository (DOI: 10.5061/dryad.pvmcvdnw4). The raw sequence data reported in this paper are available in the NCBI Sequence Read Archive repository under the BioProject ID of PRJNA1134104.
References
Liu, Y. L. et al. Rice paddy soils are a quantitatively important carbon store according to a global synthesis. Commun. Earth Environ. 2, 154 (2021).
Cai, Y. F., Zheng, Y., Bodelier, P. L. E., Conrad, R. & Jia, Z. J. Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat. Commun. 7, 11728 (2016).
Mo, D. M. et al. Warming increases CH4 emissions from rice paddies through shifts in methanogenic and methanotrophic communities. Soil Biol. Biochem. 200, 109639 (2025).
Khan, S., Chao, C., Waqas, M., Arp, H. P. H. & Zhu, Y. G. Sewage Sludge Biochar Influence upon Rice (Oryza sativa L) yield, metal bioaccumulation and greenhouse gas emissions from acidic paddy soil. Environ. Sci. Technol. 47, 8624–8632 (2013).
Li, S. D. et al. Spatiotemporal response of dissolved organic matter diversity to natural and anthropogenic forces along the whole mainstream of the Yangtze River (vol 234, 119812, 2023). Water Res. 235, 119812 (2023).
McDowell, W. H. et al. Effects of chronic nitrogen amendment on dissolved organic matter and inorganic nitrogen in soil solution. For. Ecol. Manag. 196, 29–41 (2004).
Jaffé, R. et al Spatial and temporal variations in DOM composition in ecosystems: The importance of long-term monitoring of optical properties. J. Geophys. Res. 113, G04032 (2008).
Magill, A. H. et al. Long-term nitrogen additions and nitrogen saturation in two temperate forests. Ecosystems 3, 238–253 (2000).
Liu, F. T. & Wang, D. Dissolved organic carbon concentration and biodegradability across the global rivers: A meta-analysis. Sci. Total Environ. 818, 151828 (2022).
Dalzell, B. J., Minor, E. C. & Mopper, K. M. Photodegradation of estuarine dissolved organic matter: a multi-method assessment of DOM transformation. Org. Geochem. 40, 243–257 (2009).
Marschner, B. & Kalbitz, K. Controls of bioavailability and biodegradability of dissolved organic matter in soils. Geoderma 113, 211–235 (2003).
Nebbioso, A. & Piccolo, A. Molecular characterization of dissolved organic matter (DOM): a critical review. Anal. Bioanal. Chem. 405, 109–124 (2013).
Cheng, Y. X. et al. Persistence of dissolved organic matter in sediments influenced by environmental factors: implication for nutrition and carbon cycle. J. Environ. Manag. 363, 121387 (2024).
Roth, V. N. et al. Persistence of dissolved organic matter explained by molecular changes during its passage through soil. Nat. Geosci. 12, 755 (2019).
Abdel-Hamid, A. M., Solbiati, J. O. & Cann, I. K. O. Insights into lignin degradation and its potential industrial applications. Adv. Appl. Microbiol. 82, 1–28 (2013).
Wu, M. et al. The chemodiversity of paddy soil dissolved organic matter is shaped and homogenized by bacterial communities that are orchestrated by geographic distance and fertilizations. Soil Biol. Biochem. 161, 108374 (2021).
Li, J., Yang, Y., Mo, F. & Liu, Y. Continuous manure application strengthens the associations between soil microbial function and crop production: Evidence from a 7-year multisite field experiment on the Guanzhong Plain. Agr. Ecosyst. Environ. 338, 108082 (2022).
Liu, J. B. et al. Comprehensive understanding of DOM reactivity in anaerobic fermentation of persulfate-pretreated sewage sludge via FT-ICR mass spectrometry and reactomics analysis. Water Res. 229, 119488 (2023).
Lin, Y. H. et al. Molecular probing of dissolved organic matter and its transformation in a woolen textile wastewater treatment station. J. Hazard. Mater. 457, 131807 (2023).
Yu, M. & Petrick, L. Untargeted high-resolution paired mass distance data mining for retrieving general chemical relationships. Commun. Chem. 3, 157 (2020).
Wang, L. Y. et al. Microbial roles in dissolved organic matter transformation in full-scale wastewater treatment processes revealed by reactomics and comparative genomics. Environ. Sci. Technol. 55, 11294–11307 (2021).
Hu, A. et al. Ecological networks of dissolved organic matter and microorganisms under global change. Nat. Commun. 13, 3600 (2022).
Li, H. Y. et al. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales (vol 6, 187, 2018). Microbiome 8, 169 (2020).
Wu, H. et al. The clean nitrogen removal process based on solid carbon sources: research progress and outlook. J. Clean. Prod. 383, 135508 (2023).
Kögel-Knabner, I. et al. Biogeochemistry of paddy soils. Geoderma 157, 1–14 (2010).
Luo, Y. et al. Rice rhizodeposition promotes the build-up of organic carbon in soil via fungal necromass. Soil Biol. Biochem. 160, 108345 (2021).
Ling, L. et al. Biochar induces mineralization of soil recalcitrant components by activation of biochar responsive bacteria groups. Soil Biol. Biochem. 172, 108778 (2022).
Shu, X. Y. et al. Organic amendments enhance soil microbial diversity, microbial functionality and crop yields: A meta-analysis. Sci. Total Environ. 829, 154627 (2022).
Jaffé, R. et al. Dissolved Organic Matter in Headwater Streams: Compositional Variability across Climatic Regions of North America. Geochim Cosmochim. Ac 94, 95–108 (2012).
Kajan, K., Osterholz, H., Stegen, J., Udovic, M. G. & Orlic, S. Mechanisms shaping dissolved organic matter and microbial community in lake ecosystems. Water Res. 245, 120653 (2023).
Hou, J. Y. et al. Molecular and optical signatures of photochemical transformation of dissolved organic matter: Nonnegligible role of suspended particulate matter in urban river. Sci. Total Environ. 903, 166842 (2023).
Lange, M. et al. Plant diversity enhances production and downward transport of biodegradable dissolved organic matter. J. Ecol. 109, 1284–1297 (2021).
Mayer, M. et al. Dynamic stability of mineral-associated organic matter: enhanced stability and turnover through organic fertilization in a temperate agricultural topsoil. Soil Biol. Biochem. 184, 109095 (2023).
Zou, Z. C. et al. Decadal application of mineral fertilizers alters the molecular composition and origins of organic matter in particulate and mineral-associated fractions. Soil Biol. Biochem. 182, 109042 (2023).
Luo, R. Y. et al. Phosphorus addition decreases plant lignin but increases microbial necromass contribution to soil organic carbon in a subalpine forest. Glob. Change Biol. 28, 4194–4210 (2022).
Liu, S. et al. Dysfunction of the 4-coumarate:coenzyme A ligase 4CL4 impacts aluminum resistance and lignin accumulation in rice. Plant J. 104, 1233–1250 (2020).
Yang, Z. T., Ohno, T. & Singh, B. Effect of land use change on molecular composition and concentration of organic matter in an oxisol. Environ. Sci. Technol. 58, 10095–10107 (2024).
Wang, W. X. et al. Vertical stratification of dissolved organic matter linked to distinct microbial communities in subtropic estuarine sediments. Front. Microbiol 12, 697860 (2021).
Zhao, S. T. et al. Lignin bioconversion based on genome mining for ligninolytic genes in QL-Z3. Biotechnol. Biofuels Bioprod. 17, 25 (2024).
Jia, P. Y. et al. Combined use of biochar and microbial agent can promote lignocellulose degradation and humic acid formation during sewage sludge-reed straw composting. Bioresour. Technol. 370, 128525 (2023).
Zherebker, A., Rukhovich, G. D., Sarycheva, A., Lechtenfeld, O. J. & Nikolaev, E. N. Aromaticity index with improved estimation of carboxyl group contribution for biogeochemical studies. Environ. Sci. Technol. 56, 2729–2737 (2022).
Chen, W. X., Gao, Q. Q., Hu, H. Y., Shao, T. W. & Zhou, C. F. Microbial control of soil DOM transformation during the vegetation restoration in the Loess Plateau. Plant Soil 504, 385–404 (2024).
Niu, Y. L. et al. Microbial transformation mechanisms of particulate organic carbon to mineral-associated organic carbon at the chemical molecular level: highlighting the effects of ambient temperature and soil moisture. Soil Biol. Biochem. 195, 109454 (2024).
Chen, Q. et al. Correspondence between DOM molecules and microbial community in a subtropical coastal estuary on a spatiotemporal scale. Environ. Int. 154, 106558 (2021).
Spence, A. et al. The degradation characteristics of microbial biomass in soil. Geochim. Cosmochim. Acta 75, 2571–2581 (2011).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462 (2016).
Labouyrie, M. et al. Patterns in soil microbial diversity across Europe (vol 14, 3311, 2023). Nat. Commun. 14, 3311 (2023).
Liao, J. J., Dou, Y. X., Yang, X. & An, S. S. Soil microbial community and their functional genes during grassland restoration. J. Environ. Manag. 325, 116488 (2023).
Xia, M. & Liu, M. Fourier transform ion cyclotron resonance (FT ICR) mass spectrometry data [Dataset]. Dryad. https://doi.org/10.5061/dryad.pvmcvdnw4 (2025).
Yu, M., Olkowicz, M. & Pawliszyn, J. Structure/reaction directed analysis for LC-MS based untargeted analysis. Anal. Chim. Acta 1050, 16–24 (2019).
Lee, C. K., Barbier, B. A., Bottos, E. M., McDonald, I. R. & Cary, S. C. The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities. Isme J. 6, 1046–1057 (2012).
McMurdie, P. J. & Holmes, S. phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. Plos ONE 8, e61217 (2013).
Douglas, G. M. et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688 (2020).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (42177294, 42267046, and 42167049), the China Agriculture Research Systems of MOF and MARA (CARS-22), the Key Research and Development Program of Jiangxi Province (20212BBF63007). No specific permissions were required for the described field studies and sampling activities, as the long-term paddy field experiment is managed by the authors themselves. The field location is not privately owned or protected in any way, and the studies did not involve endangered or protected species.
Author information
Authors and Affiliations
Contributions
Mingming Xia conceived the initial idea, analyzed the data, plotted the figures, and wrote the initial manuscript; Pengfa Li and Ming Liu led the research and improved the manuscript; Jia Liu, Wenjing Qin, Qingjun Dai, Meng Wu, Zhongpei Li, and Daming Li discussed the results and reviewed the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth & Environment thanks Taqi Raza and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Somaparna Ghosh. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xia, M., Li, P., Liu, J. et al. Long-term fertilization promotes the microbial-mediated transformation of soil dissolved organic matter. Commun Earth Environ 6, 114 (2025). https://doi.org/10.1038/s43247-025-02032-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43247-025-02032-7