Abstract
The biogenic methane contributes obviously to global warming, and direct interspecies electron transfer (DIET) driven CO2 reductive methanogenesis is one of the important methanogenic pathways. Here we report DIET-driven methylotrophic methanogenesis in paddy soil and the prevalent Methanomassiliicoccus. M. luminyensis CZDD1 exhibited a 1.9-fold higher methanol-derived methanogenic rate when cocultured with Clostridium malenominatum than its H2-dependent monoculture. Coculturing with Geobacter metallireducens, a known extracellular electron producer, CZDD1 showed the same efficient methanol-derived methane production, thus identified DIET-based methylotrophic methanogenesis. Chronoamperometry detected efficient methane production by M. luminyensis CZDD1 and two paddy soils from methanol and dimethylarsenate in accompany with current consumption. Differential transcriptomics predicted a membrane-bound Fpo-like complex of CZDD1 for uptake of extracellular electrons. Co-occurrence of Methanomassiliicoccaceae with Geobacteraceae and Clostridiaceae was found in five Chinese paddy soils, and Methanomassiliicoccus is ubiquitously distributed among various anoxic environments. Therefore, DIET-driven methylotrophic methanogenesis can be an important mechanism in methane emission.
Similar content being viewed by others
Introduction
Flooded paddy soil contributes ~5–19% of the global methane mission1, as the anoxic soils harbor anaerobic methanogenic archaea and anaerobic bacteria, which convert the soil organic carbons to methane. It generally believes that aceticlastic and hydrogenotrophic methanogenesis are the primary pathways for methane emission in rice fields2,3, and methanogens affiliating with Methanosarcinaceae, Methanobacteriaceae, and Methanocellaceae (RC-I group) could implement the methanogenic activities4. While a recent metatranscriptomic analysis indicated active methylotrophic methanogenesis in a straw amended Philippine paddy soil5. This finding suggests that methylotrophic methanogenic pathway could also play important role in methane emission from paddy soils.
Recently, we discovered an alternative methylotrophic methanogenic pathway: methane production from demethylation of dimethylarsenate (DMAs) in the Chinese paddy soils containing higher contents of DMAs6. DMAs is an arsenic species causing rice straighthead disease and large yield losses7. Subsequently, we found that Methanomasillicoccus luminyensis CZDD1, a H2-dependent methylotrophic methanogen isolated from a DMAs-contaminated paddy soil, efficiently demethylated DMAs(V) and produced methane from methanol when cocultured with the co-isolated Clostridium malenominatum CZB58. Whereas M. luminyensis CZDD1 exhibits markedly lower efficiency of H2-dependent methylotrophic methanogenesis, like M. luminyensis B10 isolated from the human intestine, and is incapable of demethylating DMAs(V) on its own. This implies that other reductants than H2 could facilitate an efficient methylotrophic methanogenesis by M. luminyensis9,10.
Recently, direct interspecies electron transfer (DIET) has been reported to facilitate CO2 reductive methanogenesis between the EET bacterium Geobacter metallireducens and methanogens Methanosarcina barkeri as well as Methanosarcina acetivorans, those use the outer-surface multiheme c-type cytochrome for uptake of extracellular electrons11,12. While, DIET between G. metallireducens and the cytochrome C deficient Methanobacterium has also been reported13. Furthermore, DIET even activated the “dormant” CO2 reductive methanogenic pathway encoded by Methanotrix (Methanosaeta) harundinacea, an obligately aceticlastic methanogen incapable of CO2 reductive methanogenesis14. In addition of the Geobacter species, Clostridium spp. and Listeria monocytogenes are also demonstrated the extracellular electron transfer activity15,16,17,18. This indicates that DIET and EET can be prevalent in anaerobic ecosystems. However, aside from DIET-driven CO2 reductive methanogenesis, whether DIET and EET are involved in methylotrophic methanogenesis remains unknown.
This study aimed to explore the involvement of DIET and EET in methylotrophic methanogenesis using Methanomasillicoccus CZDD1 isolated from paddy soil as an object. Our findings revealed that strain CZDD1 exhibited much higher methanol-derived methanogenic rates when cocultured with the co-isolated Clostridium malenominatum CZB5 or with Geobacter metallireducens than its H2-dependent monoculture. Moreover, by using the cathodic electrons, Methanomasillicoccu CZDD1 not only efficiently produced methane from methanol, but also demethylated DMAs(V) itself, thus determined the paddy soil prevalent Methanomasillicoccus implemented EET- and DIET-based methylotrophic methanogenesis. Based on differential transcriptomic analysis, EET-based methanogenic pathway, and energy metabolism mode were explored. Additionally, Methanomassiliicoccaceae co-occurred with Geobacteraceae and Clostridiaceae in the investigated paddy soils; and the soil electrochemical cultures showed faster methane production. Consequently, this research provides compelling evidence for EET/DIET-mediated methylotrophic methanogenesis as a potentially widespread process in paddy soil environments.
Results
C. malenominatum CZB5 enables efficient H2-independent methylotrophic methanogenesis by M. luminyensis CZDD1
We first verified the co-isolated C. malenominatum CZB5 promotion on methylotrophic methanogenesis by M. luminyensis CZDD1. Similar to M. luminyensis B10, M. luminyensis CZDD1 grows and obtains energy exclusively via H2-dependent methylotrophic methanogenesis. Upon inoculating 10% of M. luminyensis CZDD1 culture (about 0.1 mg total cell protein) into a fresh medium containing methanol and H2, methane production started after 7-days with a production rate of 23.5 μmol·d−1 (Fig. 1A). However, when cocultured M. luminyensis CZDD1 with C. malenominatum CZB5 by inoculating the same amounts of them, methane production started after 1 day inoculation and achieved 1.9-fold higher methanogenic rate (43.75 μmol·d−1) without adding H2 (Fig. 1A). Indeed, H2 was produced from the C. malenominatum CZB5 monoculture and in the earlier period of the co-culture, but it disappeared as CH4 was produced in the coculture (Fig. 1B). The calculations revealed that the coculture produced more than 300 μmol CH4 but only consumed about 18 μmol H2, a far lower ratio than 1:1 stoichiometry of H2 to CH4 expected in H2-dependent methylotrophic methanogenesis. Therefore, this suggests other reductants were provided from CZB5 to M. luminyensis CZDD1. To verify that H2 is not involved in methanogenesis, a gas mixture of 10% carbon monoxide (CO) and 90% N2 at 0.2 MPa was used to inhibit NiFe hydrogenases19. This abolished the methane production in the M. luminyensis CZDD1 monoculture, but only reduced methanogenic rate to 30.05 μmol·day−1 of the coculture (Fig. 1A), while it was still higher than that of the CZDD1 monoculture. CO obviously reduced H2 production by C. malenominatum CZB5 monoculture (Fig. 1B) yet not affect its growth (Supplementary Fig. 1).
Pre-reduced modified MM medium was used in all experiments, and all the cultures were initiated with the same inoculation sizes and incubated at 37 °C. A, B M. luminyensis CZDD1 (M.l), C. malenominatum CZB5 (C.m) monocultures, and coculture of the two were all cultured in 375 μmol methanol. The M. luminyensis monoculture was purged with 0.1 MPa H2, and the C. malenominatum monoculture and the co-culture were purged with 0.1 MPa N2. 0.2 MPa of 10% CO and 90% N2 was added to cultures to inhibit [NiFe] hydrogenase activity. Methane (A) and H2 accumulations (B) were monitored. C Aggregations in the 6th subcultures of the coculture. D Using Cy3 (red)- and Cy5 (green)-labeled 16S rRNAs to specifically probe M. luminyensis and C. malenominatum, respectively, FISH was performed on the coculture aggregates (left). Bright field micrograph of the same aggregates (right). Bars indicated 25 μm. E, F Cocultures were initiated with 10% each of M. luminyensis (M.l) and G. metallireducens (G.m, E), or its pilA deletion mutant (ΔpilA, F) into 200 μmol methanol and 100 μmol ethanol. Methane in headspace, and methanol and ethanol in liquid culture were monitored. Data are averages and standard deviations of three replicates.
To identify the potential reductants other than H2 from C. malenominatum CZB5, its spent culture and the cell lysate were added to the M. luminyensis CZDD1 monoculture. However, none of the amendments enhanced the H2-dependent methylotrophic methanogenesis (Supplementary Fig. 2). Notably, visible cell aggregates with diameters ranging 100–200 µm were observed after 6 generations of the coculture, but not in the monoculture (Fig. 1C), suggesting cell-to-cell contact between the two microorganisms. Using Cy3- and Cy5-labeled M. luminyensis- and C. malenominatum-specific 16S rRNA fragments as the respective probes, fluorescence in situ hybridization (FISH) determined roughly similar cell numbers of the two types of cells in the aggregates (Fig. 1D). These experimental results suggested that DIET between C. malenominatum and M. luminyensis may facilitate methylotrophic methanogenesis.
DIET between M. luminyensis CZDD1 and Geobacter metallireducens promotes methylotrophic methanogenesis
To test DIET involvement in H2-independent methylotrophic methanogenesis by M.luminyensis CZDD1, Geobacter metallireducens, a bacterium with defined EET activity was used as an indicator. G. metallireducens is not able to oxidize ethanol to acetate, protons, and electron on its own (formula 1), while M. luminyensis cannot convert ethanol nor methanol to methane without a reductant. When M. luminyensis accepts the extracellular electrons from G. metallireducens, the coculture would reduce methanol to methane using ethanol as the electron donor (formula 2). A coculture was constructed by 10% inoculation of each strain, and after 12 days incubation it consumed 100 μmol ethanol and 200 μmol methanol and produced 160 μmol methane but not hydrogen (Fig. 1E), thus, about 80% of electrons from ethanol oxidation were channeled to methane production. However, no methane was detected in the co-culture of M. luminyensis CZDD1 with the deletion mutant of the G. metallireducens pilA (Fig. 1F), the gene encoding e-pili for transferring extracellular electrons. This demonstrates that M. luminyensis CZDD1 is capable of DIET-based methylotrophic methanogenesis. The coculture of G. metallireducens also exhibited much higher methanogenic rate and initiated methane production 1 day after inoculation (Fig. 1E) comparing with the H2-dependent monoculture of M. luminyensis CZDD1. This indicates that DIET is preferred to H2 as the electron donor in H2-dependent methylotrophic methanogenesis.
C. malenominatum CZB5 delivers extracellular electrons and M. luminyensis CZDD1 implements EET-based methylotrophic methanogenesis
To determine the EET activity of C. malenominatum CZB5, gradient contents of ferric citrate were amended to its monoculture. Ferrous iron production was measured after 1 h incubation, and 13.3 µM·min−1 Fe (III) reduction rate was detected on 7.5 mM ferric citrate (Fig. 2A), approximately half of Fe (III) reduction rate (30 µM·min-1) of Listeria monocytogenes16. Furthermore, C. malenominatum CZB5 was grown in an electrochemical reactor, and current was produced at a density of ~20 µA·cm−2 (Fig. 2B), similarly to that produced by Listeria monocytogenes. But no current was measured in the reactor without inoculation of the bacterium. These experiments confirm the extracellular electron transfer activity of C. malenominatum CZB5.
A C. malenominatum was 10% inoculated in pre-reduced MM medum containing the indicated concentrations of ferric citrate and incubated at 37 °C, and ferrous ion accumulation was determiend. B C. malenominatum (C.m) was 10% inoculated in the MM medium without sulfide and resazurin inside a cathode H-cell reactor with a potential of +0.4 V versus Ag/AgCl. An electrochemical culture without inoculation (-) served as abiotic control. The electrochemical cultures were mixed by a magnetic stir bar, and current production was monitored during incubation at 37 °C. Experiments were performed on three batches of culture, and one representative dataset is shown. C M. luminyensis was 10% (0.1 mg total cell protein) inoculated in a H-cell containing pre-reduced MM medium with 400 μmol methanol, and purged with 100% N2 gas. An uninoculated H-cell served as an abiotic control. The cathode potential was set at −0.4 V in the first 8-day incubation at 37 °C and then reduced to −0.6 V and −0.8 V as indicated. Productions of CH4 and H2 were monitored. D Current consumption and methane production were measured in the same electrochemical reactor as in (C) at −0.4 V potential of the cathode. A final concentration of 10 mM 2-Bromoethanesulfonate (BES) was added after 48 h as indicated by arrow. Two independent experiments were performed, and one representative is shown here.
Next, the early stationary culture of M. luminyensis CZDD1 was inoculated into the methanol containing culture inside an electrochemical cell with −0.4 V of the cathode versus Ag/AgCl, a condition under which abiotic H2 would not be generated. Methane production was detected at Day 1 incubation and achieved a similar production rate (29.7 µmol·d−1) as CO-amended coculture. Further lowering the cathode potential to -0.8 V increased the methanogenic rate to 40.2 μmol·d−1 (Fig. 2C). Abiotic H2 was detected at the cathode potential of −0.8 V but not at −0.4 V (Fig. 2C). This result suggested that EET enables methylotrophic methanogenesis by M. luminyensis CZDD1 without H2 as an intermediate. To verify that M. luminyensis CZDD1 used the cathodic electrons for methanogenesis, methane production coupled current consumption was assayed. At a cathode potential of −0.4 V, M. luminyensis CZDD1 produced 14.2 μmol CH4 after 48 h and consumed about 69.9 μA·cm−2 current. The addition of bromoethane sulfonate (BES), a specific inhibitor of methyl-CoM reductase, simultaneously inhibited methane production and current consumption (Fig. 2D). This suggests that M. luminyensis CZDD1 can use the cathodic electrons to reduce methanol for methane production, i.e., performing an electromethanogenesis. Similar but lower activity was detected with human intestine M. luminyensis B10 (Supplementary Fig. 3), indicating that electromethanogenesis is a shared characteristic of the M. luminyensis strains.
EET enables methanogenesis from DMAs(V) by M. luminyensis CZDD1
Next, we explored whether EET could facilitate CH4 production from DMAs(V) by M. luminyensis CZDD1 alone. Low concentrations of DMAs(V) were used to preclude accumulation of the intermediate DMAs(III), a high toxic compound20. M. luminyensis CZDD1 was incubated with 1 μM or 2 μM 13CH3-DMAs(V) mixed with 15 mM methanol in electrochemical chambers at −0.4 V of the cathode potential as above. After a few days of incubation, 13CH4 was enriched, in contrast, no 13CO2 was enriched but maintained at the natural abundance (Fig. 3A, B). While no 13CH4 was detected in the H2 culture of M. luminyensis CZDD1 containing 1 μM DMAs(V) and 15 mM methanol even though up to 40 μM methane were determined (Fig. 3C). Therefore, M. luminyensis CZDD1 was capable utilizing cathodic electrons instead of H2 to produce CH4 from DMAs(V). Notably, a higher proportion of 13CH4 (average 200‰ vs. 60‰) was detected from the culture containing 1 μM 13C-DMAs(V) than that of 2 μM 13C-DMAs(V) (Fig. 3A). This difference could be attributed to the inhibition of DMAs (III) from DMAs (V) reduction on M. luminyensis.
M. luminyensis CZDD1 was cultured in 400 μmol methanol and 20 μM DMAs(V), and cultivated in the H-cell reactor at 37 °C. The electrochemical cultures were set at -0.4 V of the cathode potential and amended with 1 μM and 2 μM of 13C-DMAs(V), respectively. The H2-dependent culture was amended with 1 μM 13C-DMAs(V) and purged with 0.1 MPa H2 but without power supplied. 13CH4 (A) and 13CO2 (B) in the headspace of the cultures were determined using GC–IRMS, and methane (C) was determined by gas chromatography. Duplicated experiments were performed, and the means and standard deviations are shown.
Differential expressed M. luminyensis CZDD1 genes in the DIET
To explore the genes that may involve in DIET-methylotrophic methanogenesis by M. luminyensis CZDD1, differential transcriptomes between its hydrogen-depended monoculture and the coculture with C. malenominatum CZB5 were performed in their exponential cells, at day 2 of the coculture and day 14 of the monoculture, respectively. It found that among the 2636 transcribed genes, 253 were upregulated for more than 2-fold (P < 0.05) in the coculture, including 17 genes involved in methane metabolism; whereas mcrABDG that encodes methyl-CoM reductase complex, the methanogenic characteristic enzyme, remained unchanged. Noticeably, comparing with the H2-culture, expression of the genes encoding the protein complexes potentially involved in the electron flow inside M. luminyensis CZDD1 were all elevated in the coculture with C. malenominatum CZB5 (Fig. 4A), those include fpoABCDHIJKLMN encoding a Fpo-like complex, and hdrABC/mvhAGD encoding cytoplasmic heterodisulfide reductase-hydrogenase complex, and hdrD encoding a subunit of membrane-bound heterodisulfide reductase HdrD. This suggested an increased electron flux in the M. luminyensis CZDD1 cells when cocultured with C. malenominatum CZB5. In addition, genes encoding proteins containing Fe-S clusters, the brf encoding bacterioferritin, and the cell division protein FtsZ were also markedly upregulated in coculture (Fig. 4A and Supplementary dataset S1). However, echABCDE encoding the membrane-bound energy-converting hydrogenase were slightly reduced in coculture. In contrast to M. luminyensis B10, in which transcription of the echABCDEF was not detected21, higher transcription of echABCDE (RS01460-RS01475 and RS13925, Supplementary dataset S1) was determined in the H2-dependent culture of CZDD1. Consistent with the transcriptomics, quantitative RT-PCR determined a 100-fold decreased transcription of the M. luminyensis echC and echF, and about 3- and 4-fold higher transcription of hdrD and fpoC in the coculture than in the H2-dependent monoculture. (Fig. 4B), verifying the transcriptomic data and determined that Fpo-like complex could be the components for uptake of the extracellular electrons.
Heat plot representation of the differential expression (Log2) in the mid-exponential methylotrophic cultures of M. luminyensis grown respectively under 0.1 MPa H2 at day 14 (M.l + H2), and in coculture with C. malenominatum at day 2 (C.m + M.l) (A). Blue and red represent the minima and maxima fold, respectively. Quantitative RT-PCR of some differentially transcribed genes involved in methanogenesis in the M. luminyensis monoculture (red bar), coculture (blue bar) and electrochemical culture (orange bar) (B). Three replicate experiments were performed, and the averages and standard deviations are shown in (B). Statistics are estimated by T-test, and ***, ** and ns indicate very significant (P < 0.01), significant (P < 0.05) and not significant, respectively.
Methanomassiliicoccaceae, Geobacteraceae, and Clostridiaceae cooccur in five paddy soils
Next, we surveyed the distributions of methanogens and bacteria in five paddy soils via 16S rRNA amplicon sequencing and found that the methanogen community primarily comprised of the families of Methanobacteraceae (37.9–67.2%), Methanosarcinaceae (4.7–24.6%) and Methanomassiliicoccaceae (4.3–21.9%) (Fig. 5A). The bacteria community consisted mainly of Peptostreptococcaceae (11.2–52.9%), Clostridiaceae (2.5–16.7%), Caloramatoraceae (2.3–8.6%) and Hungateiclostridiaceae (1.3–2.9%). The electroactive Geobacteraceae was also present from 0.2% to 6.3% (Fig. 5B). To find the potential electroactive bacteria that interplay with Methanomassiliicoccaceae, we analyzed the network among methanogens and bacteria inhabiting in the five paddy soils. Our analysis revealed a high co-occurrence of Clostridiaceae and Geobacteraceae with Methanomassiliicoccaceae, but not with Methanobacteraceae (Fig. 5C). The positive relationships of Methanomassiliicoccaceae with Clostridiaceae, and Geobacteraceae were further confirmed using Spearman analysis method (P < 0.01) (Fig. 5D). This suggests that Methanomassiliicoccaceae may implement methylotrophic methanogenesis in collaboration with Clostridiaceae or Geobacteraceae, presumably via DIET in paddy soils.
Relative abundances of Methanomassiliicoccaceae among methanogen community (A), and of bacteria (B) in the paddy soils of FY, QY, TC, SHY, CZ were estimated based on the specific 16S rRNA gene homologous. Six replicates of each paddy soil were used for 16S rRNA gene sequencing and analysis. C A co-occurrence network of the methanogen and bacteria families among five paddy soils (n = 30) was analyzed using WGCNA package. D The correlations between Methanomassiliicoccaceae and Clostridiaceae (light magenta), and Geobacteraceae (dark magenta) among the five paddy soils were analyzed using Spearman analysis method.
DIET or EET facilitates efficient methylotrophic methanogenesis from DMAs and methanol in paddy soils
Given that biochar enhanced DIET-based CH4 production from ethanol and acetate in paddy soils22, biochar was amended into the methanol containing soil enrichments. After two transfers of 0.5% (W/V) the average maximum methanogenic rate of 9.3 μmol·d−1 was assayed, 2–7-fold higher than that of the three soil enrichments without biochar (T-test, P < 0.01) (Supplementary Fig. 4A).
DIET facilitated CH4 production from DMAs was then determined in paddy soils of CZ, TC, and FY. The three soil samples were cultured in methanol amended with 13C-DMAs(V) and 30% carbon monoxide (CO) to inhibit Ni-Fe hydrogenase. Accumulation of hydrogen was monitored during the 17-day incubation at 30 °C, and CO amendment almost completely suppressed hydrogen production of paddy soils (Supplementary Fig. 4B). very high fractionations of 13CH4 with isotopic compositions ranging from 250‰ to 2000‰ were detected in all three soil slurries in the first week of incubation; however, 13CO2 fractionations were very low (Fig. 6A–C). This suggested that the methyl group of DMAs(V) could be mainly reduced to CH4 by extracellular electrons, namely DIET facilitated methane production from DMAs in the paddy soils. Noticeably, the ratios of 13CH4 appeared going down along incubation in two paddy soils, this could be due to 13CH4 dilution by increased total CH4 from methanol.
One gram of each paddy soils of CZ (A), TC (B), and FY (C) was inoculated into 5 mL methanol medium amened with 20 μM DMAs and 2 μM 13C-DMAs, and gas mixture including 30% CO to inhibit hydrogenase. 13CH4 (red bar) and 13CO2 (blue bar) in the headspace were determined using GC–IRMS. Each 9 g paddy soils sampled from CZ and FY were added to cathodic chambers of H-cell containing 90 mL modified MM medium with about 1800 μmol methanol. The cathodic potential was set at −0.4 V. Methane content (D) in the headspace of cathodic chambers with (closed symbols) or without (open symbols) power supply was monitored during 12 day-incubation at 30 °C. Current consumptions (E) of the cathodic chamber with power supply as A were monitored during 12 days incubation, and a chamber without inoculation of paddy soils was included as an abiotic control. Duplicate experiments were performed, and the means and standard deviations are shown. Relative abundances of methanogens of cathodic enrichments with (+) or without (−) power supply were determined via archaea specific 16S rRNA ampilcon sequeccing (F).
EET facilitated methylotrophic methanogenesis in paddy soil was investigated by culturing the CZ and FY soil in cathode chambers containing about 1800 μmol methanol and under −0.4 V of the cathode potential. It found that upon power supply, methane was produced 3 days earlier in CZ and FY soils (Fig. 6D); meanwhile, current consumption of about 200 μA·cm−2 was detected in the two soils incubated in cathodic chambers (Fig. 6E). Furthermore, Methanomassiliicoccaceae in CZ and in FY soils was also enriched by culturing in the cathodic chamber (Fig. 6F). This further determined that DIET- and EET-based methylotrophic methanogenesis occurs in paddy soils.
Discussion
This work reported DIET-driven methylotrophic methanogenesis in the paddy soil prevalent H2-dependent methylotrophic methanogen Methanomassiliicoccus luminyensis CZDD1. This methanogen utilizes extracellular electrons from co-existing electroactive bacteria like Geobacter and Clostridium to reduce methanol or DMAs for efficient methane production. Co-occurrence between Methanomassiliicoccaceae, and Geobacteriaceae and Clostridiaceae was found in the five tested paddy soils. Experiments of CO inhibited H2 metabolism and the cathodic cultures exhibited rapid methane production in paddy soils all determined the prevalence of DIET-driven methylotrophic methanogenesis, which can be an overlooked methane source.
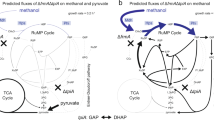
Methanogens affiliating with Methanomassiliicoccales conserve energy exclusively from reductive methylotrophic methanogenesis9. They lack cytochromes and active energy-converting hydrogenases but possess a membrane-bound Fpo-like complex. It predicted that the Fpo-like complex in Methanomasiliicoccus oxidizes reduced ferredoxin and directly interacts with membrane-associated heterodisulfide reductase subunit D (HdrD), and thus constitutes an energy-converting ferredoxin:heterodisulfide oxidoreductase complex to generate proton motive force21. Based on the physiological and transcriptomic data in this study, we proposed a putative model as depicted in Fig. 7. M. luminyensis could utilize extracellular electrons most likely taken up via archaella or the Fpo-like complex, and electrons inside cell would then be transferred to membrane-associated HdrD to reduce the heterodisulfide CoM-CoB. Subsequently, the reduced CoM-SH will serve as an acceptor for methyl groups during methane production.
Extracellular electrons from C. malenominatum or cathode are accepted by the M. luminyensis Pili and transferred to, or directed accepted by the membrane bound Fpo-like complex. Electrons are then transferred to membrane associated HdrD to reduce heterodisulfide CoM-S-S-CoB for methane production from methanol (MeOH) or trimethylamine (TMA). Protein icons shadowed in purple, blue and gray represent the encoding genes to be up- and down-regulated, and no change when M. luminyensis uses extracellular electron versus H2. MT methylotransferase, MCR methyl-CoM reductase.
Although hydrogen involvement in methylotrophic methanogenesis could not be excluded in the coculture and the cathode reactor due to the current inability to delete the hydrogenase genes in M. luminyensis CZDD1, the following experimental data verify DIET but not H2 as reductant used in methanogenesis, (i) CO inhibition of NiFe- hydrogenases did not abolish methane production in the coculture and paddy soil enrichments; (ii) BES inhibition of methanogenesis also reduced current consumption in the electrochemical culture of CZDD1; and (iii) electronic chamber incubated paddy soils exhibited rapid methane production than without power supply. Thus, we propose that reduction of methyl compounds by extracellular electron could represent the predominant pathway for methylotrophic methanogenesis in paddy soils and by the prevalent Methanomassiliicoccus. This also conforms to the speculation of Lovley and Holmes23, which indicates that the free H2 may not be the main electron vehicle during interspecies electron transfer. Comparing with other methanogens, M. luminyensis B10 has an extremely low H2 threshold24. Considering that the Methanomassiliicoccus members are ubiquitous in natural environments and animal intestinal tracts25,26,27,28,29, they could play a important role in methane emission via electromethanogenesis, particularly in the areas including rice paddy field having low levels of H2.
DIET can even activate the “dormant” methanogenic pathways or facilitate reductive chemical reactions. The genus Methanothrix comprises methanogens that exclusively perform aceticlastic methanogenesis despite carrying the complete gene suite for CO2 reductive methanogenesis but without hydrogenase. Rotaru et al. found that the electroactive bacterium Geobacter metallireducens enabled CO2 reductive methanogenesis by Methanotrix harundinacea14. In this study, we also found that the electrochemical culture, but not the H2-culture, of M. luminyensis CZDD1 could produce methane from DMAs (Fig. 3).
In conclusion, this study reports for the DIET-based methylotrophic methanogenesis in paddy soils and by the prevalent Methanomassiliicoccus luminyensis CZDD1, a strain isolated from a DMAs contaminated paddy soil. Strain CZDD1 preferentially uses extracellular electrons for methylotrophic methanogenesis, in particular from DMAs, a toxic chemical to rice. This study not only reveals a new electromethanogenic pathway, but also found that DIET/EET can activate the inactive methanogenic pathways harbored in methanogens. Based on the ubiquitous distribution of Methanomassiliicoccales and methylotrophic methanogenic pathway in natural environments, electromethanogenesis including DIET-driven methylotrophic methanogenesis could be an important methane source especially from paddy field.
Materials and methods
Strains and cultivation conditions
Both M. luminyensis CZDD1 and C. malenominatum CZB5 were isolated from paddy soils8. Monocultures of M. luminyensis CZDD1 and C. malenominatum CZB5, as well as coculture of M. luminyensis CZDD1 and C. malenominatum CZB5 were routinely cultivated in modified MM medium. Pre-reduced and modified MM medium10, which in 1L includes 0.5 g KH2PO4, 0.4 g MgSO4 ∙ 7H2O, 5 g NaCl, 1 g NH4Cl, 0.05 g CaCl2 2H2O,1.6 g sodium acetate, 1 g yeast extract, 1 g tryptone and 4 g NaHCO3. Geobacter metallireducens ATCC53774 and its pili-deficient mutant ΔpilA were routinely cultured in a medium containing 20 mM ferric citrate and 20 mM ethanol11.
To culture C. malenominatum CZB5, nitrogen gas (N2) was introduced into the headspace at a pressure of 0.1 MPa. In contrast, for the cultivation of M. luminyensis CZDD1, a combination of 15 mM methanol and 0.1 MPa hydrogen gas (H2) was utilized in the headspace. The co-culture of strains CZDD1 and CZB5 was conducted in modified MM medium supplemented with 15 mM methanol and maintained under a nitrogen atmosphere at 0.1 MPa. Cocultures of M. luminyensis either with G. metallireducens wild type or ΔpilA strain were initiated with by 10% inoculation (0.1 mg total cell protein) of each strain into the same modified MM medium containing 20 mM ethanol and 40 mM methanol and under 0.1 MPa N2 phase. To assess the inhibitory effect of carbon monoxide on methane production, a gas mixture consisting of 10% CO and 90% N2 was introduced into the co-culture. While a gas mixture of 10% CO and 90% H2 was introduced into monoculture at a pressure of 0.2 MPa. Monocultures pulsed with 100% H2 and coculture with 100% N2 without any CO pulsed served as control. All cultures were incubated at a temperature of 30 °C.
Total cell protein determination
The M. luminyensis CZDD1 cells intended for inoculation were initially quantified based on the total cell protein. Cell pellets were resuspended with 1 mL PBS buffer and subsequently grinded in liquid nitrogen. After liquid nitrogen disruption, cells were centrifugated at 5000 × g at 4 °C for 15 min. The supernatant was collected to determine total cell protein using the BCA Protein Assay Kit (Thermo Fisher Scientific, Germany).
Fluorescent in situ hybridization
The mid-exponential phase of the co-culture of M. luminyensis CZDD1 and C. malenominatum CZB5 was harvested by centrifugation at 3500 × g for 10 min. The resulting cell pellets were rinsed with PBS, fixed in 4% paraformaldehyde for 12 h at 4 °C, and subsequently dehydrated with a series of gradient ethanol concentrations ranging from 50 to 100%. Fluorescent in situ hybridization (FISH) was carried out in a 20 mM Tris buffer (pH 7.4) supplemented with 900 mM NaCl, 0.01% SDS (w/v), and 40% formamide (v/v) at 46 °C for 2 h. The probes utilized were 5′-[CY3] GCTGCCTCCCGTAGGAGT-3′, which specifically target the 16S rRNA of C. malenominatum and 5′-[CY5] GTGCTCCCCCGCCAATTCCT-3′ designed to target the 16S rRNA of M. luminyensis. Fluorescent cells were observed under LSM 800 confocal microscope (Leica Microsystems, Buffalo Grove, IL, USA).
Ferrozine assay of ferric iron reductive activity of C. malenominatum
Ferric citrate was supplemented into the C. malenominatum CZB5 culture at final concentrations of 2.5 mM, 5 mM, and 7.5 mM during the late-log phase and incubated at 37 °C. A solution of 0.1% phenanthroline was added to samples from the culture, and absorbance was measured spectrophotometrically at a wavelength of 562 nm15. A standard curve for ferrous iron concentration was generated using FeCl2. All experiments were conducted in triplicate.
Characterization of the bio-electrochemical cultures
Chronoamperometry was carried out using a Bio-logic science instrument potentiostat CHI 1000C (CH Instrument, TX). Electrochemical experiments were set up in the H cell reactor, in which 150 mL-anode and cathode chambers were separated by a Nafion 117 proton-exchange membrane (Dupont, USA), and equipped with an Ag/AgCl reference electrode (CH Instruments), a Pt wire counter electrode (Alfa Aesar), and a 6.35-mm-thick graphite felt working electrode with a 16-mm radius (Alfa Aesar), in addition of two sampling ports. For onset of the electrochemical experiment, H cell reactor was first autoclaved and then flushed with 100% N2 gas for no less than 15 min to achieve an oxygen-free headspace. Next, the anode and cathode compartments were filled with 80 mL sterilized and pre-reduced modified MM medium omitting sulfide, and resazurin inside an anaerobic box (Thermo Fisher Scientific, Germany). The H-cell reactors were continuously purged with N2 gas to maintain anaerobic condition. The mid-exponential culture of C. malenominatum CZB5 was 10% inoculated into the MM medium within the chamber equipped with working electrode with an applied cathode potential set at +0.4 V versus Ag/AgCl. The mid-exponential culture of M. luminyensis CZDD1 was 10% inoculated in MM medium containing 15 mM methanol in the chamber under the cathode potential sequentially set at −0.4 V, −0.6 V and −0.8 V versus Ag/AgCl. The electrochemical culture was mixed with a magnetic bar during incubation at 37 °C. H cell reactors were connected to potential station, and currents are reported as a function of the geometric surface area of the electrode.
13C-labeled DMAs(V) was synthesized according to Chen et al.6. To determine 13C-DMAs(V) derived methanogenesis by M. luminyensis CZDD1, it was cultured in 15 mM methanol and 20 μM DMAs(V) and cultivated in a cathodic chamber of H-cell reactor. The electrochemical culture was supplied with a potential of −0.4 V versus Ag/AgCl and supplemented with 1 μM and 2 μM of 13C-DMAs(V), respectively. In the H2-dependent culture, 1 μM 13C-DMAs (V) and 0.1 MPa H2 was added but without power supply. The currents of the H-cell reactors are reported as a function of the geometric surface area of the electrode.
Measurements of methane, hydrogen, and 13C/12C ratios of methane and carbon Dioxide
Methane was measured using GC-14B gas chromatograph (Shimadzu, Japan) equipped with flame ionization detector and C18 column as described previously30. The temperature parameters were used as follows: column at 50 °C, injector at 80 °C, and detector at 130 °C. Hydrogen was measured using GC-4100 (GWAI) equipped with a TCD detector and column temperature parameters were used as follows: column at 80 °C, injector at 150 °C, and detector at 90 °C. The 13C/12C isotope ratios of methane and carbon dioxide in headspace were determined using GC–IRMS (Isotope Ratio Mass Spectroscopy, Thermo Fisher Scientific, Germany) according to Ai et al.31.
RNA library preparation and Illumina Hiseq sequencing
The mid-exponential cells were harvested from M. luminyensis CZDD1 H2-dependent culture and its co-culture with C. malenominatum CZB5 and the electrochemical culture with cathode potential set at -0.8 V. Total RNA was extracted using the RNAiso Plus kit (Takara, Dalian, China) according to the manufacturer’s instructions. RNA quality was determined using 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). High-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 23S:16S ≥ 1.0, >10 μg) is used to construct sequencing library.
Total RNA of 5 μg was used to construct strand-specific RNA-sequencing libraries using the TruSeq RNA sample preparation Kit from Illumina (San Diego, CA). Briefly, rRNA was removed by the RiboZero rRNA removal kit (Epicenter), and then RNA was fragmented using fragmentation buffer. cDNA synthesis, end repair, A-base addition, and ligation of the Illumina-indexed adapters were performed according to Illumina’s protocol. Libraries were then size selected for cDNA target fragments of 200–300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, paired-end libraries were sequenced on Illumina Nova Seq 6000 (150bp*2, Shanghai BIOZERON Co., Ltd).
Reads quality control and mapping
The raw paired end reads were trimmed and quality controlled by Trimmomatic with parameters (SLIDINGWINDOW:4:15MINLEN:75) (version0.36 http://www.Usadellab.org/cms/uploads/supplementary/Trimmomatic). The clean reads were aligned to the reference genome with orientation mode using Rockhopper http://cs.wellesley.edu/~btjaden/Rockhopper/) software. As input, Rockhopper takes RNA sequencing reads generated by high-throughput sequencing technology. This software was used to calculate gene expression levels with default parameters.
Differential expression analysis and function enrichment
To identify DEGs (differential expression genes) between two samples, transcript abundances of genes were calculated based on Fragments Per Kilobase of read per Million mapped reads (RPKM). EdgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html) was used for differential expression analysis. DEGs between two samples were selected based on the following criteria: the logarithmic fold change was greater than 2, and the false discovery rate was less than 0.05. To understand the functions of the differential expressed genes, GO functional enrichment and KEGG pathway analysis were carried out by Goatools (https://github.com/tanghaibao/Goatools) and Kobas (http://kobas.cbi.pku.edu.cn/kobas3) respectively. DEGs were significantly enriched in GO terms and metabolic pathways when their Bonferroni-corrected P value was less than 0.05.
Quantitative reverse transcription PCR (RT-qPCR)
RNA extract used for sequencing above was treated with RNase-Free DNase (Takara) to remove contaminated DNA, and cDNA was synthesized using the M-MLV Reverse Transcriptase cDNA Synthesis Kit (Takara) following the manufacturer’s instructions. Using the primers listed in Supplementary Table S1. Quantified PCR was performed using SYBR Premix Ex Taq Kit (Takara Bio Inc., Japan), and carried out on ABI Prism 7000 sequence detection system (Applied Biosystems USA). Each qPCR mixture contained 12.5 μL SYBR qPCR mix (TOYOBO), 5 μL cDNA (250 ng), 100 nM of each primer, and double-distilled H2O to a final volume of 25 μL. PCR was initiated at 95 °C for denaturation for 30 s and 35 cycles as follows: denature at 95 °C for 10 s, annealing at the Tm listed in Supplementary Table S1 for 30 s, and elongation at 72 °C for 30 s. Fluorescence data were collected during elongation. To estimate mRNA copies of the tested genes, a standard curve of each gene was generated by quantitative PCR on 10- fold serially diluted plasmid DNA cloned with the gene. 16S rRNA gene was used as the biomass reference. Triplicate samples were determined, and each measurement was repeated at least three times.
Paddy soil sampling
Soils (0–20 cm) were collected from five paddy fields in China, two from As-contaminated sites of Qiyang (QY, 26°75′06″N, 111°84′72″E) and Chenzhou (CZ, 25°57′83″N,113°02′31″E) in Hunan province, and the other three were from Xinyang (XY, 32°11′32″N, 114°40′20″E) in Henan province, Tancheng (TC, 34°29′45″N, 118°20′16″E) in Shandong province and Shuyang (SHY, 34°10′36″N,118°51′55″E) in Jiangsu province, the sites with rice straighthead disease. No permission was required for collecting soils from five paddy fields in China. The soil samples were air-dried and ground to <0.149 mm for further analysis.
Paddy soil enrichment
Enrichment cultures were constructed by inoculating 0.5 g of paddy soils collected from CZ, TC, and FY into 12.5 mL anaerobic tube containing 5 mL pre-reduced medium with 20 mM methanol and supplied with or without 0.025 g (DW) activated charcoal (Sigma, C9157-500G) inside an anaerobic box (Thermo Fisher Scientific, Germany). For CO inhibition, enrichment cultures were supplied with additional 0.2 MPa gas mixture of 30% CO and 70% N2 in compared to a control pulsed with 100% N2 at 0.2 MPa. For DMAs enrichment, additional 20 μM DMAs and 2 μM 13C-DMAs were added. All enrichments were using 20 mM methanol as methanogenic substrate and incubated at 30 °C for 40 days.
For electrochemical enrichment, each 9 g paddy soils sampled from CZ and FY were added to cathodic chambers of H-cell containing 90 mL modified MM medium with about 1800 μmol methanol inside an anaerobic box (Thermo Fisher Scientific, Germany). The cathodic potential was set at −0.4 V versus Ag/AgCl. H-cell reactors were connected to potential station, and currents are reported as a function of the geometric surface area of the electrode. Methane content in the headspace of cathodic chambers with or without power supply was monitored during 12 day-incubation at 30 °C. Current consumptions of the cathodic chamber with power supply were monitored for 12 days incubation, and a chamber without inoculation of paddy soils was included as an abiotic control.
DNA extraction and 16S rRNA gene amplicon sequencing
DNA was extracted from ~0.5 g of paddy soil slurry samples or 1 mL enrichment using the MoBio Power soil DNA Isolation Kit (MoBio, Carlsbad, CA, United States) according to the manufacturer’s protocol. Purified DNA was quantified using NanoDropND-100 Spectrophotometer (Gene Company), and the qualities of extracted DNA were examined by 1% agarose gel electrophoresis.
The bacterial and archaeal community structures of in situ paddy soil or enrichment cultures were analyzed using high-throughput sequencing based on bacterial and archaeal 16S rRNA genes. The V4–V5 regions of bacterial and archaeal 16S rRNA were amplified using the primers Arch519F/915R and Bac515F/907R, respectively. Subsequently, the amplicons were purified, quantified, and sequenced on a Mieseq platform (Illumina; PE250 mode) by Shanghai Biozeron Biotech Co., LTD. Raw sequences underwent quality filtering and assembly before being processed using Quantitative Insights Into Microbial Ecology (QIIME, V1.9.0 http://qiime.org/scripts/assign_taxonomy.html). Effective sequences were then grouped into operational taxonomic units (OTUs) based on a similarity score of 97% using UPARSE (version 7.1 http://drive5.com/uparse/). Representative sequences were selected for each OTU, and their taxonomic classification was identified by the UCLUST algorithm (http://www.drive5.com/usearch/manual/uclust_algo.html) against SILVA (SSU138.1) 16S rRNA database with a confidence threshold of 80%.
Statistical analysis
Co-occurrence networks were constructed by using the WGCNA package based on the Spearman’s correlation methanogens and bacteria. Only OTUs with relative abundance within top 30% were used in the analyses. In networks, OTUs represent nodes, while correlations between OTUs are displayed as edges. The parameters describing network topological characteristics used in this study included node number (the number of OTUs), edge number (the number of connections among all nodes), betweenness (the number of times a node acts as a bridge along the shortest path between two other nodes). The correlation relationship between Methanomassiliicoccales and Clostridiaceae or and Geobacteraceae is visualized using R package of the ggpubr, STAT-Cor uses Spearman's coefficient of correlation calculation as measure of emphasizing the level of correlation32.
Data availability
The genomic sequences of and transcriptomic data of the C. malenominatum CZB5 and M. luminyensis CZDD1 have been deposited in NCBI SRA database under the accession numbers of PRJNA1032237 and PRJNA1028373, respectively. Supplementary Data 1 and Supplementary Data 2 have been uploaded in figshare with https://doi.org/10.6084/m9.figshare.28681487 and https://doi.org/10.6084/m9.figshare.28681454 respectively.
References
Bao, Q. et al. Effect of nitrogen fertilizer and/or rice straw amendment on methanogenic archaeal communities and methane production from a rice paddy soil. Appl. Microbiol. Biotechnol. 100, 5989–5998 (2016).
Conrad, R., Erkel, C. & Liesack, W. Rice Cluster I methanogens, an important group of Archaea producing greenhouse gas in soil. Curr. Opin. Biotechnol. 17, 262–267 (2006).
Ding, L. J. et al. Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol. Ecol. 95, fiz040 (2019).
Grosskopf, R., Janssen, P. H. & Liesack, W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 64, 960–969 (1998).
Li, X., Bei, Q., Rabiei Nematabad, M., Peng, J. & Liesack, W. Time-shifted expression of acetoclastic and methylotrophic methanogenesis by a single Methanosarcina genomospecies predominates the methanogen dynamics in Philippine rice field soil. Microbiome 12, 39 (2024).
Chen, C. et al. Sulfate-reducing bacteria and methanogens are involved in arsenic methylation and demethylation in paddy soils. ISME J. 13, 2523–2535 (2019).
Limmer, M. A., Wise, P., Dykes, G. E. & Seyfferth, A. L. Silicon decreases dimethylarsinic acid concentration in rice grain and mitigates straighthead disorder. Environ. Sci. Technol. 52, 4809–4816 (2018).
Chen, C., Li, L., Wang, Y., Dong, X. & Zhao, F. J. Methylotrophic methanogens and bacteria synergistically demethylate dimethylarsenate in paddy soil and alleviate rice straighthead disease. ISME J. 17, 1851–1861 (2023).
Kroninger, L., Gottschling, J. & Deppenmeier, U. Growth characteristics of Methanomassiliicoccus luminyensis and expression of methyltransferase encoding genes. Archaea 2017, 2756573 (2017).
Dridi, B., Fardeau, M. L., Ollivier, B., Raoult, D. & Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 62, 1902–1907 (2012).
Rotaru, A. E. et al. Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl. Environ. Microbiol. 80, 4599–4605 (2014).
Holmes, D. E., Zhou, J., Ueki, T., Woodard, T. & Lovley, D. R. Mechanisms for electron uptake by Methanosarcina acetivorans during direct interspecies electron transfer. mBio 12, e0234421 (2021).
Zheng, S., Liu, F., Wang, B., Zhang, Y. & Lovley, D. R. Methanobacterium capable of direct interspecies electron transfer. Environ. Sci. Technol. 54, 15347–15354 (2020).
Rotaru, A. E. et al. A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energ. Environ. Sci. 7, 408–415 (2014).
Light, S. H. et al. A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144 (2018).
Light, S. H. et al. Extracellular electron transfer powers flavinylated extracellular reductases in Gram-positive bacteria. Proc. Natl. Acad. Sci. USA 116, 26892–26899 (2019).
Naradasu, D., Miran, W., Sakamoto, M. & Okamoto, A. Isolation and characterization of human gut bacteria capable of extracellular electron transport by electrochemical techniques. Front. Microbiol. 9, 3267 (2018).
Schwab, L., Rago, L., Koch, C. & Harnisch, F. Identification of Clostridium cochlearium as an electroactive microorganism from the mouse gut microbiome. Bioelectrochemistry 130, 107334 (2019).
Pandelia, M. E., Ogata, H., Currell, L. J., Flores, M. & Lubitz, W. Inhibition of the [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F by carbon monoxide: an FTIR and EPR spectroscopic study. Biochim. Biophys. Acta 1797, 304–313 (2010).
Chen, C. et al. Reduction of dimethylarsenate to highly toxic dimethylarsenite in paddy soil and rice plants. Environ. Sci. Technol. 57, 822–830 (2023).
Kroninger, L. et al. Energy conservation in the gut microbe Methanomassiliicoccus luminyensis is based on membrane-bound ferredoxin oxidation coupled to heterodisulfide reduction. FEBS J. 286, 3831–3843 (2019).
Liu, F. H. et al. Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci. 5, 8982–8989 (2012).
Lovley, D. R. & Holmes, D. E. Electromicrobiology: the ecophysiology of phylogenetically diverse electroactive microorganisms. Nat. Rev. Microbiol. 20, 5–19 (2022).
Feldewert, C., Lang, K. & Brune, A. The hydrogen threshold of obligately methyl-reducing methanogens. FEMS Microbiol. Lett. 367, fnaa137 (2020).
Narrowe, A. B. et al. Uncovering the diversity and activity of methylotrophic methanogens in freshwater wetland soils. mSystems 4, e00320–19 (2019).
Nobu, M. K., Narihiro, T., Kuroda, K., Mei, R. & Liu, W. T. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 10, 2478–2487 (2016).
Sollinger, A. & Urich, T. Methylotrophic methanogens everywhere—physiology and ecology of novel players in global methane cycling. Biochem. Soc. Trans. 47, 1895–1907 (2019).
Vanwonterghem, I. et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat. Microbiol. 1, 16170 (2016).
Borrel, G. et al. Phylogenomic data support a seventh order of methylotrophic methanogens and provide insights into the evolution of methanogenesis. Genome Biol. Evol. 5, 1769–1780 (2013).
Zhang, G., Jiang, N., Liu, X. & Dong, X. Methanogenesis from methanol at low temperatures by a novel psychrophilic methanogen, "Methanolobus psychrophilus" sp. nov., prevalent in Zoige wetland of the Tibetan plateau. Appl. Environ. Microbiol. 74, 6114–6120 (2008).
Ai, G., Zhu, J., Dong, X. & Sun, T. Simultaneous characterization of methane and carbon dioxide produced by cultured methanogens using gas chromatography/isotope ratio mass spectrometry and gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 27, 1935–1944 (2013).
Li, R., Wang, Y., Hu, H., Tan, Y. & Ma, Y. Metagenomic analysis reveals unexplored diversity of archaeal virome in the human gut. Nat. Commun. 13, 7978 (2022).
Acknowledgements
We thank Professor Yahai Lu for talking about electrochemical experiment and CO inhibition, and Mr Yuhong Zhong for providing strain G. metallireducens. This work was supported by National Natural Science Foundation of China (92251302, 91851211, 32070061, and 42377014), the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (2019QZKK0304) and Opening Fund of the State Key Laboratory of Environmental Geochemistry (SKLEG2024207).
Author information
Authors and Affiliations
Contributions
X.Z.D. conceptualized the projects, designed experiments, and acquired funding. L.Y.L. designed and performed the experiment. X.P.T. and Q.Z. performed electrochemical experiment, X.M.W. performed isotope measuring experiment, L.Q. performed cultivation experiment, C.C. synthesized 13C labeled DMAs, J.L., K.X., F.J.Z., and Y.F.W., all give suggestions for experiment and paper writing. L.Y.L. and X.Z.D. wrote the manuscript and all authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Earth and Environment thanks Sarah Zecchin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Somaparna Ghosh [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, L., Tian, X., Wang, X. et al. Extracellular electron transfer based methylotrophic methanogenesis in paddy soil and the prevalent Methanomassiliicoccus. Commun Earth Environ 6, 297 (2025). https://doi.org/10.1038/s43247-025-02276-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43247-025-02276-3