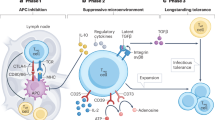
Abstract
Recent studies using single-cell RNA sequencing technology have uncovered several subpopulations of CD4+ T cells that accumulate with aging. These age-associated T cells are emerging as relevant players in the onset of inflammaging and tissue senescence. Here, based on information provided by single-cell RNA sequencing data, we present a flow cytometry panel that allows the identification of age-associated T cell subsets in systematic larger analysis in mice. We use this panel to evaluate at the single-cell level mitochondrial and senescence marks in the different age-associated CD4+ T cell subpopulations. Our analysis identifies a subpopulation of regulatory T (Treg) cells that is characterized by the extracellular expression of the co-inhibitory molecule killer cell lectin-like receptor subfamily G member 1 (KLRG1) and accumulates with aging in humans and mice. KLRG1-expressing Treg cells display senescence features such as mitochondrial alterations, increased expression of cell-cycle regulators and genomic DNA damage. Functionally, KLRG1+ Treg cells show a reduced suppressive activity in vivo accompanied by a pro-inflammatory phenotype.
Similar content being viewed by others
Main
With age, the immune system loses the ability to respond to infections, cancer or vaccination. Instead, it engages in autoimmune and pro-inflammatory responses, which favor tissue damage and the acquisition of a low-grade systemic chronic inflammation, known as inflammaging, increasing the risk of many age-related diseases1. Thus, the decline of immune function during aging represents a major clinical challenge in many disease conditions including autoimmune diseases, infectious diseases, cancer and neurodegenerative and cardiovascular disorders2,3,4. The use of single-cell RNA sequencing (scRNA-seq) has allowed a deeper characterization of the different subsets of CD4+ T cells that accumulate during aging. CD4+ age-associated T cells (TAAs) include effector/memory T cells; a population of regulatory T (Treg) cells expressing activation genes (denoted as activated Treg cells or aTreg cells); cells with an exhaustion signature (denoted as exhausted); and cells overexpressing genes associated with cytotoxicity that have previously been described in the context of viral infections and cancer as CD4+ cytotoxic T cells (denoted as cytotoxic)5.
Treg cells, a subtype of CD4+ T cells characterized by the expression of the forkhead box transcription factor (FOXP3), are required to maintain immune homeostasis and avoid excessive tissue damage preventing inflammatory and autoimmune diseases6. Understanding the molecular mechanisms behind age-associated changes in the Treg compartment is critical to understand the deterioration of the immune system during aging and the consequences for immunosenescence and inflammaging. Treg cells in aged mice are augmented in lymphoid organs (for example, spleen and lymph nodes)7,8,9 and other nonlymphoid tissues such as the visceral fat and lungs10, whereas they are reduced in muscle11. Treg cells also undergo molecular changes with aging, becoming more dependent on IL-15 signaling owing to a lower expression of the IL-2 receptor CD25, while exhibiting enhanced levels of CD122 (IL-2/IL-15 receptor chain β)12,13. Moreover, they acquire a memory-like profile during aging, characterized by the downregulation of CD62L and the upregulation of CD44 and CD69 (refs. 14,15). These aged effector–memory Treg cells display increased expression of canonical pro-inflammatory transcription factors such as T-BET and RORγT, and pro-inflammatory cytokines such as IFN-γ or IL-17A16. Treg cells with pro-inflammatory phenotypes have been denoted as fragile Treg cells in the context of cancer, although its function in antitumoral responses is still debated17,18.
Killer cell lectin-like receptor subfamily G, member 1 (KLRG1) is a co-inhibitory receptor expressed by natural killer (NK) cells and antigen-experienced T cells. The expression of KLRG1 increases dramatically with age in human blood samples19. In mice, KLRG1 has been used to identify memory precursor cells from effector T cells. In acute viral infection models, KLRG1 distinguishes short-lived effector CD8+ T cells (KLRG1hi) and memory precursor effector CD8+ T cells (KLRG1lo)20,21. Importantly, KLRG1 identifies senescent CD8+ T cells in humans and mice, as KLRG1+CD8+ T cells exhibit reduced cytokine production and senescence characteristics such as a reduced proliferative capacity19,22,23,24. Regarding CD4+ T cells, KLRG1 also identifies terminally differentiated cells25. Specifically in Treg cells, KLRG1 is considered a marker of effector suppressive Treg cells that display increased levels of activation markers (CD69, CD44, CD103 and CD39) and enhanced production of IL-10 (ref. 26).
Mitochondria are the key controller of cellular metabolism, but also play a crucial role as signaling hubs for inflammation and cell death. Mitochondrial function declines with age in different cell types and tissues27, including T cells28,29, affecting the mitochondrial ATP generation as well as reactive oxygen species (ROS) production and cellular signaling30. In addition, mitochondrial dysfunction is sufficient to drive cellular senescence in fibroblasts31,32. In contrast to conventional T cells, which engage glycolysis during immune response, Treg cells preferentially use mitochondrial respiration as source of ATP33 and the genetic induction of Treg-specific mitochondrial dysfunction leads to premature death by uncontrolled inflammation34,35. Here, by using multiparametic spectral flow cytometry, we investigate the specific subpopulations of T cells that accumulate mitochondrial alterations during aging. Among them, we identify a population of Treg cells characterized by the expression of KLRG1 (kTreg cells) and FOXP3 that displays senescence features such as nuclear DNA damage. Although the suppressive capacity of kTreg cells is maintained in vitro, it is compromised in vivo. kTreg cells produce pro-inflammatory cytokines and, importantly, are also increased among human peripheral blood mononuclear cells (PBMCs) with age.
Results
T cells with mitochondrial alterations accumulate with aging
To investigate age-associated mitochondrial dysfunction in T cells, we examined mitochondrial mass and mitochondrial membrane potential (MMP) by flow cytometry in circulating CD4+ T cells from C57BL/6 mice at different ages ranging from 2 to 21 months old by combining MitoTracker Green (MtG) and MitoTracker DeepRed (MtDR) staining. As a control, we used oligomycin that, by inhibiting the proton ATPase, increased the MMP and therefore the MtDR signal (Extended Data Fig. 1a). Both the MtG and MtDR signals declined during aging in circulating CD4+ T cells (Extended Data Fig. 1b,c), suggesting that the proportion of T cells with mitochondrial alterations increases in aged mice (Fig. 1a,b). We obtained similar results using two additional mitochondrial probes: MitoTracker Red CM-X-ROS (Fig. 1c,d) and tetramethylrhodamine methyl ester (TMRM) (Extended Data Fig. 1d). Interestingly, MtGloMtDRlo cells, which accumulate with aging, showed a reduced MtDR/MtG ratio, suggesting mitochondrial defects (Extended Data Fig. 1e). To confirm that MtGloMtDRlo cells have altered mitochondria, we sorted MtGhiMtDRhi and MtGloMtDRlo CD4+ T cells from aged mice and performed electron microscopy (Fig. 1e,f). MtGloMtDRlo cells showed an increased percentage of mitochondria with altered morphology36 identified as round, small and with unstructured or even lost mitochondria cristae (Fig. 1f,g).
a,b, Representative flow cytometry plots showing the simultaneous analysis of MtG and MtDR (a) and the percentage of cells with MtGhiMtDRhi, MtGhiMtDRlo and MtGloMtDRlo (b) in the circulating CD4+ T cells from 2-month-old (n = 5), 9-month-old (n = 5), 13-month-old (n = 5), 17-month-old (n = 5) and 21-month-old (n = 6) mice. c,d, Representative flow cytometry plots (c) and quantifications (d) of the simultaneous analysis of MtG and MitoTracker Red CMXROS in splenic CD4+ T cells from young (2-month-old) and aged (21-month-old) mice. e, Representative flow cytometry plots showing the sorting strategy of MtGhiMtDRhi and MtGloMtDRlo CD4+ T cells from old mice. f,g, Representative images of electron microscopy (f) and quantifications (g) of healthy and morphologically altered mitochondria in MtGhiMtDRhi (green) and MtGloMtDRlo (red) splenic CD4+ T cells from old mice (n = 90 and 300 cells from 2 different mice). h,i, Representative flow cytometry plots (h) and quantifications (i) of MtGhiMtDRhi and MtGloMtDRlo cells in circulating naive (CD62LhiCD44lo, blue) and effector (CD62LloCD44hi, purple) CD4+ T cells from 2-, 9-, 13-, 17- and 21-month-old mice (n = 6 mice per group). Each dot represents an individual mouse. Data are presented as mean values ± s.e.m. Statistical analysis was performed using one-way ANOVA with with post hoc Tukey’s correction (b), two-tailed unpaired Student’s t-test (d and i: MtDRhiMtGhi 2 months, 9 months, 17 months, 21 months; MtDRloMtGlo 2 months, 9 months, 21 months), two-tailed Welch’s t-test (i: MtDRhiMtGhi 13 months; MtDRloMtGlo 13 months, 17 months) or Mann–Whitney U test (g). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Because the T cell subsets dramatically change with age and the most notable changes are the loss of naive T cell subsets and the increase in the effector memory pool37, we assessed mitochondrial probes in naïve (CD62LhiCD44lo) and effector–memory (CD62LloCD44hi) CD4+ T cells in mice at different ages. We observed that, even in old mice, naive T cells were MtGhiMtDRhi, while effector–memory cells were characterized by MtGloMtDRlo, even in younger mice (Fig. 1h,i).
KLRG1 identifies age-associated regulatory T cells
To assess whether age-associated mitochondrial dysfunction equally affects all TAA subsets, we developed a multiparametric staining panel to identify the different age-associated CD4+ T cell subsets. Our panel included surface markers based on gene expression of surface proteins identified by scRNA-seq5,38, complemented with known markers for immunosenescence, such as KLRG1 (ref. 19). In previous scRNA-seq experiments, splenic T cells from young mice were mainly classified as naive, naive_ISG15 and rTreg cells, whereas T cells from old mice were characterized by the accumulation of TAAs such as aTreg, effector memory T (TEM) cells, exhausted and cytotoxic T cells5. Unbiased clusterization of spectral flow cytometry analysis of splenic CD4+ T cells from young (2-month-old) and old (21-month-old) mice resulted in seven subpopulations of T cells (Fig. 2a,b). Naive T cells were characterized by the expression of the adhesion molecule CD62L and the absence of the activation marker CD44. Three subsets of Treg cells were identified by the expression of CD25 and discriminated by different expression levels of CD62L, CD44 and KLRG1. The other CD4+ T cell clusters presented marks of differentiation (CD62Llo and CD44hi) and were identified as cytotoxic based on the expression of NK-like markers, such as NKG2A and NKG2D, or as exhausted, based on the expression of classical exhaustion markers such as PD-1 and TIM3 (Fig. 2c,d). Young mice mostly displayed naive and rTreg cells, while aTreg, kTreg, TEM, cytotoxic and exhausted cells were increased in spleen from old mice (Fig. 2e,f). Importantly, this panel discriminated six out of the seven subpopulations previously defined by scRNA-seq analyses; however, because the panel is based on surface markers, we could not identify the ISG15_naive T cell subpopulation (Extended Data Fig. 2a,b). Our analysis identified a TAA subset characterized by the expression of KLRG1 (Fig. 2b and Extended Data Fig. 2a,b). In fact, circulating KLRG1+ CD4+ T cells progressively accumulated with aging (Extended Data Fig. 2c). Notably, these cells co-expressed KLRG1 and CD25, suggesting that KLRG1+ CD4+ T cells are probably a subpopulation of Treg cells (Fig. 2c,d). To confirm whether these CD25+ KLRG1+ cells were Treg cells, we combined the panel with the intracellular marker FOXP3 and we found that most of KLRG1+ cells expressed CD25 and FOXP3 (83–87%) (denoted as kTreg cells), supporting their Treg identity (Fig. 2g). The percentages of splenic kTreg cells assessed by spectral flow cytometry gradually increased with age within both the Treg compartment and CD4+ T cells (Extended Data Fig. 2d,e).
a, UMAP representation of splenic CD4+ T cells from young (left) and old mice (right) analyzed by spectral flow cytometry. b, UMAP with Cluster-X overlay showing the distribution of the different clusters of CD4+ T cells identified by spectral flow cytometry (n = 4 mice per group). c, UMAP representation of the expression levels of representative markers used to identify TAAs by flow cytometry. d, The distribution of the gMFI of representative markers used to identify TAAs in CD4+ T cell subpopulations: naive (blue), rTreg (yellow), aTreg (orange), kTreg (red), TEM (light green), cytotoxic (brown) and exhausted (dark green) (n = 4 mice per group). e,f, Representative pie charts (e) and box plots (f) comparing the percentage of cells belonging to each T cell subset in young (2-month-old, n = 4) and old (22-month-old, n = 5) mice. g, The gating strategy to identify FOXP3+ cells within the kTreg cells in young (2-month-old) and old (22-month-old) mice (n = 3 per group). gMFI, geometric mean fluorescence intensity. Each dot represents an individual mouse. Box-and-whisker plots show the median, the maximum, the minimum and the 25th and 75th percentiles. Statistical analysis was performed using two-tailed unpaired Student’s t-test (f: naive) or two-tailed unpaired Welch’s t-test (f: rTreg, aTreg, kTreg, TEM, exhausted and cytotoxic). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
In addition, we reanalyzed existing and publicly available scRNA-seq data from young and old mice to separate the novel kTreg cluster5. By increasing the depth of the clusterization, we obtained an additional cluster, which accumulated during aging, within the aTreg subset (Extended Data Fig. 3a,b). Interestingly, the most determinant genes of this cluster were reminiscent of kTreg cells (Supplementary Table 1). These cells showed increased gene expression of Klrg1 and altered expression of genes related to Treg function compared with aTreg cells, such as Icos and Maf (Extended Data Fig. 3c). We also found that kTreg cells expressed higher levels of inflammatory genes such as those belonging to the s100a family (including S100a4, S100a6, S100a10, S100a11 and S100a13) and inflammation-related proteins such as galectins (Lgals1 and Lgals9), IL-18 receptor (Il18r) and IFN-γ receptor (Ifngr1) (Extended Data Fig. 3d).
Then, we investigated the tissue distribution of kTreg cells. To map their preferential location in the different tissues, we used the multiparametric flow cytometry panel to analyze T cells from different tissues including spleen, bone marrow, liver, colonic lamina propria, white adipose tissue and Peyer’s patches from young and old mice. kTreg cells were found in all these tissues (Fig. 3a), and interestingly, the highest percentage of kTreg cells was observed in the colonic lamina propria (Fig. 3b,c). Importantly, the absolute numbers and the percentage of kTreg cells were increased in different tissues during aging (Fig. 3d) and their percentage was increased in spleen, bone marrow, liver and Peyer’s patches but not in lamina propria and white adipose tissue (Fig. 3e). Altogether, we have developed a multiparametric flow cytometry method that allows the identification of TAA subsets in both lymphoid and nonlymphoid organs. Using this strategy, we have identified a subset of Treg cells characterized by the expression of KLRG1 that accumulates with aging.
a, UMAP with Cluster-X overlay showing the distribution of the different clusters of CD4+ T cells from spleen, bone marrow, liver and colonic lamina propria of young mice analyzed by spectral flow cytometry. b, UMAP with Cluster-X overlay showing the distribution of the different clusters of CD4+ T cells from young mice in different tissues (n = 5). c, Representative bar plots showing the percentage of cells belonging to each T cell subset in spleen, bone marrow, liver and colonic lamina propria from young mice (n = 5). d,e, Quantifications of absolute numbers of kTreg cells (d) and the percentage of kTreg cells among Treg cells (e) in different tissues in young (2-month-old, n = 5) and old (22-month-old, n = 4) mice. WAT, white adipose tissue. Box-and-whisker plots show the median, the maximum, the minimum and the 25th and 75th percentiles. Statistical analysis was performed using two-tailed unpaired Student’s t-test (d: spleen and WAT; e: bone marrow, liver, lamina propria and WAT) or two-tailed Welch’s t-test (d: bone marrow, lamina propria and Peyer’s patches; e: spleen and Peyer’s patches]. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
KLRG1+ Treg cells show mitochondrial alteration and senescence features
To evaluate if any of the CD4+ T cell subsets are more susceptible to the age-associated mitochondrial dysfunction, we combined our spectral flow cytometry panel together with the assessment of mitochondrial probes in young and old mice. Old CD4+ T cells showed an increased proportion of MtGloMtDRlo cells in all subsets compared with young CD4+ T cells (Fig. 4a,b). Strikingly, even within the same mouse, there is a remarkable difference in the percentage of MtGloMtDRlo in the distinct T cell subsets. While the naive cluster mainly exhibits low frequencies of MtGloMtDRlo cells, TEM cells displayed the major accumulation of MtGloMtDRlo cells in both young and old mice. Among Treg cells, kTreg cells showed an increased percentage of MtGloMtDRlo cells compared with other Treg subsets in both young and old mice (Fig. 4a,c), suggesting that kTreg cells are terminally differentiated Treg cells with mitochondrial alterations.
a, Representative flow cytometry plots showing the simultaneous analysis of MtG and MtDR in the different subsets of CD4+ T cells from 2- and 21-month-old mice: naive (blue), rTreg cells (yellow), aTreg cells (orange), kTreg cells (red), TEM (light green), cytotoxic (brown) and exhausted (dark green) (n = 6 mice per group). b, A comparison of the percentage of MtGloMtDRlo cells in each cluster of CD4+ T cells in young (2-month-old) and old (21-month-old) mice (n = 6 mice per group). c, A statistical comparison of the percentage of MtGloMtDRlo cells in the different clusters in young or old mice (n = 6 mice per group). Each dot represents an individual mouse. Data are presented as mean values ± s.e.m. Box-and-whisker plots show the median, the maximum, the minimum and the 25th and 75th percentiles. Statistical analysis was performed using two-tailed unpaired Student’s t-test (b: naive, rTreg, aTreg and TEM), Mann–Whitney U test (b: kTreg and exhausted) or Friedman test with post hoc Dunn’s correction (c). n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
To better characterize the different Treg subpopulations, we compared the transcriptomic fingerprint of rTreg, aTreg and kTreg cells isolated from young (2-month-old) and old (26-month-old) mice by bulk RNA-seq. This transcriptomic dataset allowed us to improve the resolution of the kTreg cell expression profile. The clusterization grouped the samples by their identity rather than the age of the mice (Fig. 5a,b). We then analyzed the differentially expressed genes between the two age-associated Treg subsets, aTreg and kTreg. We found Klrg1, Il1rl1 and Cd200r1 as the most significantly increased genes in kTreg cells. Other genes upregulated in kTreg cells included Foxp3, Gata3, Ahr, NK-related genes (Klra8), granzymes (Gzmb, Gzma and Gzmc) and chemokine receptors (Ccr3 and Ccr10) (Fig. 5c). The pathway analysis showed that kTreg cells exhibit increased expression of cellular senescence genes and P53 target genes, decreased expression of genes related to mitochondrial function and increased expression of inflammatory genes (Fig. 5d).
a–d, RNA sequencing of rTreg, aTreg and kTreg cells from young and old mice: PCA (a) and clusterization (b); volcano plot showing the DEGs between aTreg cells and kTreg cells (c); heat map of selected DEGs illustrating genes characteristic of identity, cellular senescence, P53 signaling, mitochondrial function and inflammation (d) (n = 4 per group). e,f, Quantification of the expression of P16 (e) and P21 (f) by flow cytometry in splenic CD4+ T cells from young (2 months old, n = 4) and old (21 months old, n = 3) mice measured as gMFI. g,h, Representative histograms and quantifications of the expression of P16 (g) and P21 (h) by flow cytometry in the different subsets of splenic Treg cells from young (n = 4) and old (n = 3) mice measured as gMFI: rTreg cells (yellow), aTreg cells (orange) and kTreg cells (red). i, Quantification of γH2AX gMFI in CD4+ T cells from young and old mice (n = 5 per group). j, Representative histogram and quantification of γH2AX by flow cytometry in the different subsets of Treg cells from young and old mice measured as gMFI (n = 5 per group). gMFI values are relative to the gMFI in naive CD4+ T cells. DEGs, differentially expressed genes; gMFI, geometric mean fluorescence intensity. Each dot represents an individual mouse. Data are presented as mean values ± s.e.m. Statistical analysis between young and old mice was performed using two-tailed unpaired Student’s t-test (e–h: aTreg; i and j) or two-tailed Welch’s t-test (h: rTreg and kTreg). Statistical analysis between subsets within the same animal was performed using RM one-way ANOVA with with post hoc Tukey’s correction (g, h and j). Asterisks refers to statistic comparisons between rTreg, aTreg and kTreg in the same age group; the number signs refer to statistic comparisons between young and old mice. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Given that a mitochondrial dysfunction is a trigger of cellular senescence31, and that the transcriptional fingerprint of kTreg cells showed increased expression of senescence-related genes, we interrogated whether kTreg cells display other features of cellular senescence beyond mitochondrial alterations. We analyzed the expression of the cell cycle regulators P16 and P21, which are increased in CD4+ T cells during aging (Fig. 5e,f). We compared the three different subsets of Treg cells and found that kTreg cells displayed the highest expression of P16 and P21 (Fig. 5g,h). Moreover, as an additional marker of cellular senescence, we analyzed the genomic DNA damage measured as the phosphorylation of the histone H2AX in the serine 139 (γH2AX), a marker of DNA double strand breaks that is increased in CD4+ T cells from old mice (Fig. 5i). Interestingly, kTreg cells also displayed increased DNA damage compared with aTreg cells (Fig. 5j).
KLRG1+ Treg cells express ST2 and are induced by IL-33 in vivo
In light of the increased expression of Gata3 in kTreg cells (Fig. 5c), and to further characterize the kTreg subset, we compared the expression of different transcription factors related to T cell function by flow cytometry. This analysis revealed that kTreg cells expressed higher levels of FOXP3 compared with the other Treg subsets (Fig. 6a and Extended Data Fig. 4), whereas the levels of RORγT and T-BET were similar to aTreg cells. Importantly, kTreg cells expressed higher levels of GATA3 than aTreg cells in both young and old mice (Fig. 6a and Extended Data Fig. 4), supporting the RNA-seq results. TOX and MAF expression was higher in kTreg cells compared with rTreg and aTreg cells, but this difference was not observed in old mice (Fig. 6a and Extended Data Fig. 4).
a, Representative histograms and quantifications of the expression of different transcription factors by flow cytometry in splenic rTreg (yellow), aTreg (orange) and kTreg (red) cells from old (21-month-old) mice measured as gMFI (n = 6 mice per group). gMFI values are relative to the MFI in rTreg cells (n = 6 per group). b, Representative histograms and quantifications of ST2 expression by flow cytometry in the different clusters of splenic CD4+ T cells from young (2-month-old) and old (21-month-old) mice: naive (blue), rTreg (yellow), aTreg (orange), kTreg (red), TEM (light green), cytotoxic (brown) and exhausted (dark green) cells measured as gMFI (n = 6 mice per group). c–g, In vivo differentiation of kTreg cells: young control mice (2 months old) were treated with IL-2 + IL-33 or NaCl for 6 days and analyzed on day 7 after injection (n = 3 mice per group) (c); representative flow cytometry plot and quantification of the proportion of splenic Treg cells in young mice treated with NaCl or IL-2 and IL-33 (d); representative flow cytometry plot and quantification of the proportion of KLRG1+ Treg cells in young mice treated with NaCl or IL-2 and IL-33 (e); quantification of the absolute numbers of KLRG1+ Treg cells in young mice treated or not treated with IL-2 and IL-33 (n = 3 mice per group) (f); quantification of the percentage of each Treg cluster in young mice treated with NaCl or IL-2 and IL-33 (n = 3 mice per group) (g). gMFI, geometric mean fluorescence intensity. Each dot represents an individual mouse. Data are presented as mean values ± s.e.m. Box-and-whisker plots show the median, the maximum, the minimum and the 25th and 75th percentiles. Statistical analysis was performed using Friedman test with post hoc Dunn’s correction (a and b: old), RM one-way ANOVA with post hoc Tukey’s correction (b: young), two-tailed unpaired Student’s t-test (d, e and g) or two-tailed Welch’s t-test (f). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
GATA3 is essential for the regulatory function of Treg cells39,40. Given that GATA3-expressing Treg cells are characterized by the expression of the IL-33 receptor ST2 (refs. 41,42), we analyzed the expression of ST2 in the different CD4+ T cell clusters. We observed that kTreg cells expressed higher levels of ST2 than the rest of T cell subsets in both young and old mice (Fig. 6b). Based on that, we tried to induce the differentiation of kTreg cells in vivo by administration of IL-33 in young mice. Before IL-33 administration, we injected IL-2, which induces the expansion of Treg cells due to their expression of the IL-2 receptor CD25 (ref. 43) (Fig. 6c). Upon this treatment, we found an expansion of Treg cells in the injected mice (Fig. 6c) and an increase in the percentage and absolute number of kTreg cells (Fig. 6e,f). Interestingly, this treatment seems to induce the differentiation of aTreg cells into kTreg cells (Fig. 6g), suggesting that kTreg cells rely on ST2–IL-33 signaling for differentiation.
KLRG1+ Treg cells exhibit a pro-inflammatory phenotype
To investigate the function of kTreg cells, we analyzed the suppressive function of these cells both in vitro and in vivo. First, we isolated Treg cells from old mice, sorted them on the basis of KLRG1 expression and co-cultured them with naive T cells loaded with CellTrace Violet at a 1:2 ratio (1 Treg:2 naive). Both KLRG1+ and KLGR1− Treg cells showed a similar suppressive capacity in vitro (Extended Data Fig. 5a,b). Next, we performed an in vivo suppression assay by injecting naive CD45.1 CD4+ T cells alone or in combination with CD45.2 KLRG1− or KLRG1+ Treg cells into T cell-deficient mice (CD3ε−/− CD45.2) (Fig. 7a). While KLRG1− Treg cells were able to restrain the weight loss induced by naive T cells, mice injected with naive T cells together with KLRG1+ Treg cells lost weight to a similar extent as mice exclusively injected with naive T cells (Fig. 7b). Four months after the adoptive transfer, CD45.1 cells were more abundant in mice injected with KLRG1+ Treg cells (Fig. 7c). In addition, we found that transferred CD45.1 CD4+ T cells were more activated when co-transferred with KLRG1+ Treg cells, as shown by their increased expression of CCL5 and PD-1, than when co-transferred with KLRG1− Treg cells (Fig. 7d). Importantly, we observed an increased prevalence of fibrotic colon and fecal blood in mice injected with naive T cells alone or together with KLRG1+ Treg cells than in mice injected with naive T cells together with KLRG1− Treg cells (Fig. 7e,f). Altogether, these data suggest that the function and/or viability of KLRG1+ Treg cells in vivo is compromised.
a–f, In vivo assessment of the suppressive activity of KLRG1+ Treg cells. CD3ε−/− T cell-deficient mice were exclusively injected with naive CD45.1 CD4+ T cells (n = 4) or in combination with either CD45.2 KLRG1− Treg cells (n = 5) or CD45.2 KLRG1+ Treg cells (n = 5) isolated from young mice treated with IL-2 + IL-33. Noninjected CD3ε−/− (n = 3) are represented for the sake of comparison. The injected mice were monitored for 4 months and analyzed. a, Schematic diagram depicting the in vivo suppression assay. b, The body weight of CD3ε−/− mice injected with naive CD4+ T cells alone or in combination with KLRG1− or KLRG1+ Treg cells. The weight of each animal was normalized to its own weight before the inoculation. c, The percentage of transferred CD45.1 naive (striped color) or CD45.2 Treg (flat color) cells analyzed by flow cytometry in CD3ε−/− mice injected with naive and/or Treg cells. d, Quantification of the expression of CCL5 and PD-1 by flow cytometry in the injected CD45.1 CD4+ T cells measured as gMFI. e,f, The percentage of mice with fibrotic colon (e) or fecal blood (f) in mice injected with naive and/or Treg cells. g, Representative histograms and quantifications of the expression of different cytokines by flow cytometry in splenic rTreg (yellow), aTreg (orange) and kTreg (red) cells from old (21-month-old) mice measured as gMFI. gMFI values are relative to the MFI in rTreg cells (n = 6). h,i, Analysis of the secretion of different cytokines by the Treg subsets using multiplex. Treg subsets were sorted from young mice injected with IL-2 + IL-33 and incubated during 24 h in 10% FBS complete RPMI medium before the assay (n = 3). Heat map (h) and statistical comparisons of the secretion of different cytokines by rTreg, aTreg and kTreg cells. Heat map values are relative to the maximum of each cytokine. Quantification (i) of SASP-related cytokines secreted by rTreg, aTreg and kTreg cells. gMFI, geometric mean fluorescence intensity. Each dot represents an individual mouse. Data are presented as mean values ± s.e.m. Statistical analysis was performed using one-way ANOVA with post hoc Tukey’s correction (c: CD45.1; d and e), Krukal–Wallis test with post hoc Dunn’s correction (c: CD45.2), Friedman test with post hoc Dunn’s correction (g–i). n.s., not significant; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
To further explore the activity of kTreg cells, we assessed their capacity to produce cytokines by ex vivo intracellular flow cytometry. We found that kTreg cells from old mice expressed higher amounts of the anti-inflammatory cytokine IL-10 than the rest of the Treg subsets (Fig. 7g). We found no differences in the production of IL-17 or IFN-γ by kTreg cells compared with aTreg cells, but kTreg cells produced more TNF (Fig. 7g). Similar results were observed when comparing Treg subsets from young mice (Extended Data Fig. 5c). Furthermore, we measured the cytokine secretion of the different Treg subsets. We induced kTreg expansion in vivo by IL-33 administration and sorted rTreg, aTreg and kTreg cells. After 24 h in vitro, we assessed the cytokine release by multiplex analysis of the culture media and we observed that, in agreement with our flow cytometry data, kTreg cells secreted higher amounts of IL-10 together with pro-inflammatory cytokines than the other Treg subsets (Fig. 7h). Notably, kTreg cells produced higher levels of senescence-associated secretory phenotype (SASP)-related factors including IL-1β, IL-6, IL-13, IFN-γ, GM-CSF, CXCL2, CXCL1, CCL1 and CCL3 than rTreg or aTreg cells (Fig. 7i). Noteworthy, upon activation, aTreg cells secreted similar or even higher levels of most cytokines than kTreg cells. However, kTreg cells maintained increased production of IL-10, IL-6, GM-CSF and CXCL2 (Extended Data Fig. 5d,e). Finally, to investigate the function of kTreg cells in vivo, we adoptively transferred KLRG1− and KLRG1+ Treg cells into CD3ε−/− mice (Extended Data Fig. 5f). In agreement with the increased production of pro-inflammatory cytokines, we found that mice transferred with KLRG1+ Treg cells showed splenomegaly together with increased percentages of circulating neutrophils and basophils compared with noninjected or mice injected with KLRG1− Treg cells (Extended Data Fig. 5g,h).
KLRG1+ Treg cells increase in human blood samples during aging
We wondered whether the kTreg population is also increased in humans during aging. We used flow cytometry to analyze the presence of kTreg cells in PBMCs from a human cohort of 144 healthy volunteers separated into two groups: young (ranging from 18 to 25 years old) and senior (over 55 years old) (see Extended Data Fig. 6 for the gating strategy). We found an increased frequency of kTregs in the senior population (14.56% of Treg cells) compared with young individuals (6.35% of Treg cells) (Fig. 8).
a, Representative flow cytometry plots of the gating of KLRG1+ Treg cells in PBMCs samples from young and senior volunteers. b–d, Quantifications of the percentage of KLRG1+ Treg cells among Treg cells (b), among CD4+ cells (c) and among total lymphocytes (d) in young (18–25 years old, n = 42) and senior (≥55 years old, n = 75) individuals by flow cytometry. Each dot represents an individual. Data are presented as mean values ± s.e.m. Statistical analysis was performed using two-tailed unpaired Welch’s t-test (b) or Mann–Whitney U test (c and d). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Altogether, by using deep flow cytometry analysis, we have identified a subset of Treg cells characterized by the expression of KLRG1 and ST2. Besides typical Treg cytokines such as IL-10, kTreg cells produce other SASP-related cytokines and chemokines such as IL-1β, IL-6, IL-13, IFN-γ, GM-CSF, CXCL2, CXCL1, CCL1 and CCL3. kTreg cells also harbor senescence features such as mitochondrial decline, expression of cell cycle inhibitors and DNA damage. These kTreg cells with senescence features are induced in response to IL-33 and accumulate with aging in different tissues, but they are especially abundant in the gut in mice. Importantly, circulating kTreg cells are increased in humans during aging.
Discussion
T cells play a critical role in recognizing and depleting damage or infected cells and sustaining tissue homeostasis preventing inflammation and tissue deterioration21. However, like every cell type in the body, T cells undergo changes as they age, leading to diminished function and compromising tissue homeostasis. Recently, scRNA-seq technology has enabled the decoding of the heterogeneity of age-associated CD4+ and CD8+ T cells that accumulate over time5,38. Using this information, we have developed a multiparametric panel of 20 antibodies, which allows routine discrimination of T cell heterogeneity during aging by spectral flow cytometry. With this strategy, we have mapped the different TAAs that accumulate during aging in different tissues and we have identified KLRG1-expressing Treg cells as a subset accumulating in blood, lymphoid and nonlymphoid tissues during natural aging and harboring aging features, including mitochondrial alterations.
Functional Treg cells are essential to restore tissue homeostasis and to prevent autoimmunity and inflammation. Although it is well established that Treg cells are augmented in several organs during aging in both humans and mice, whether their function is compromised with aging is still a matter of debate. Some authors showed that young and old Treg cells have equivalent suppressive capacity44,45, whereas others showed an age-related increase in Treg function linked to increased risk of malignancies and infections in older adults7,14. By contrast, there is also evidence of a deterioration of Treg function during aging. Old Treg cells are not able to suppress IL-17+ T cells upon chronic inflammation44, failing to control hypersensitivity46 and producing reduced amounts of IL-10 (ref. 47).
KLRG1 is a marker of highly differentiated T cells, and it has been considered as a marker of immunosenescence21. In addition, a subset of terminally differentiated Treg cells that express KLRG1 has been identified as short-lived Treg cells expressing higher levels of suppressive (that is, CTLA-4 and IL-10) but also inflammatory molecules26 located in nonlymphoid barriers, metabolic tissues or at sites of inflammation9,48. For example, KLRG1+ Treg cells contribute to insulin resistance49 and IFN-γ- and IL-10-secreting KLRG1+ Treg cells also infiltrate the central nervous system during experimental autoimmune encephalomyelitis50. Using our flow cytometry panel, we mapped the presence of kTreg cells in different tissues including spleen, bone marrow, liver, colonic lamina propria, white adipose tissue, Peyer’s patches and blood during aging. Our results demonstrate that kTreg cells increase during aging in most of the analyzed tissues and that the colonic lamina propria is a preferential location for kTreg cells, reaching 50% of the total Treg pool. Importantly, Treg cells are critical for gut homeostasis51,52, which is essential for healthy aging.
Our results also support that kTreg cells express the IL-33 receptor ST2 and that their differentiation depends on IL-33. IL-33 is an interleukin-1-like cytokine that plays a critical role in mucosal immunity, favoring Th2 differentiation53. IL-33 is increased in the adipose tissue with aging54, and it induces thymic involution and T cell dysfunction55. Further research is required to define whether IL-33 levels change in lymphoid tissues or in the circulation during aging or if the differentiation of kTreg cells depends on other factors.
By combining the multiparametric panel with mitochondrial probes, we assessed which of the different T cell subsets that accumulate during aging show mitochondrial dysfunction. We found that, whereas naive T cells are MtGhiMtDRhi, even in old mice, TAAs show increased percentage of cells with low MtG and MtDR stainings. Within Treg cells, we found that kTreg cells show higher percentage of MtGloMtDRlo, indicating that they harbor reduced mitochondrial mass and MMP. The MMP is an important indicator of mitochondrial integrity and function. During the mitochondrial respiration, electrons are ejected from the mitochondrial matrix to the intermembrane space to generate an electrochemical gradient. An optimal MMP is required for most of the mitochondrial functions including the generation of ATP, the uptake of ions, the synthesis of iron–sulfur clusters and others. When the MMP is not recovered, unpolarized mitochondria are degraded by mitophagy56. The simultaneous assessment of mitochondrial mass and MMP by flow cytometry have been extensively used to characterize the health status of mitochondria in different tissues, including T cells, under different conditions such as tumor environment57,58,59,60. Nevertheless, changes in the molecular composition of mitochondria, particularly in cardiolipins, lead to a reduction in the binding of MitoTracker probes in intraepithelial lymphocytes that, besides their low signal of MtG and MtDR, showed a similar mitochondria number on electron microscopy images61. Importantly, a reduction in the staining of these probes could also be explained by changes in the plasma membrane potential or by the expression of multidrug resistance transporters that expel the fluorescent probes62. To confirm that the changes that we observed in MtG and MDR stating are related to mitochondria, we sorted MtGhiMtDRhi and MtGloMtDRlo CD4+ T cells and assessed mitochondrial morphology by electron microscopy. Electron microscopy analysis shows that MtGloMtDRlo cells have mitochondrial changes including signs of cristae remodeling, supporting that MtGloMtDRlo T cells have mitochondrial alterations. However, additional research is needed to determine whether the MtGloMtDRlo T cell population also has structural or biochemical changes in the composition of mitochondria, in the plasma membrane potential and/or in the expression of multidrug transporters that could explain the dramatic reduction in MtG and MtDR staining. Importantly, beyond the key role of mitochondrial metabolism for the suppressive function of Treg cells35, a proper mitochondrial metabolism of Treg cells is essential for maintaining liver homeostasis during aging. Deleting Altre, a long noncoding RNA found in Treg cells and upregulated with aging, leads to mitochondrial dysfunction, causing liver inflammation, fibrosis and cancer in aged mice63. Therefore, the increase in kTreg cells with impaired mitochondria could be detrimental with aging. However, the function of kTreg cells during aging and age-associated diseases requires further exploration.
Our findings demonstrate that, besides mitochondrial alterations, kTreg cells harbor other signs of senescence. A recent study found that Treg cells undergo senescence faster than conventional T cells through a ROS-dependent mechanism64. In addition, both mitochondrial decline and cellular senescence are tightly interconnected31,32. An important number of senescent-associated changes are dependent on mitochondrial function, especially the pro-inflammatory phenotype. Therefore, the pro-inflammatory phenotype observed in kTreg cells could be a consequence of their mitochondrial dysfunction. The mitochondrial damage leads to the release of mitochondrial DNA (mtDNA) to the cytosol. mtDNA fragments trigger the activation of the cGAS–STING pathway, leading to the production of type I interferons, such as IFN-β, and pro-inflammatory cytokines, such as TNF or IL-6 (ref. 65), which are part of the SASP66. Cytosolic mtDNA, especially the oxidized mtDNA, also activates NLRP3 promoting the assembly of the inflammasome, a structure that leads to the activation of caspase I, therefore leading to the release of active IL-18 and IL-1β by the pyroptotic cell death67. As kTreg cells harbor mitochondrial damage, they could also experience a release of mtDNA that would trigger their pro-inflammatory phenotype. Moreover, the mitochondrial dysfunction also contributes to lysosomal stress favoring a pro-inflammatory phenotype in CD4+ T cells68.
Surprisingly, there is recent evidence supporting the idea that senescence could be a reversible state. Chemotherapy-induced senescent cells can be released from senescence and reenter into the cell cycle69. In addition, tissue senescence can be transitorily induced to promote tissue regeneration in Hydractinia symbiolongicarpus70. Regarding CD4+ T cells, a recent study demonstrated that the expression of the senescence marker KLRG1 is transient in a subset of resident Treg cells, despite their maintenance of an activated phenotype71. Because the specific ablation of mitochondria from senescent cells is sufficient to reverse many features of the senescent phenotype72, mitochondria are a promising target for interventions aimed at reducing the harmful effects of senescence in aging tissues. Interestingly, mitochondria dysfunction in exhausted CD8+ T cells can be reversed by scavenging ROS or boosting mitochondrial function with nicotinamide riboside57,73,74, suggesting that mitochondrial stress is reversible and that mitochondria could be key to restoring T cell function. Future research should clarify whether mitochondrial damage and other features of senescence are targetable in kTreg cells.
Sustaining a proper function of T cells during aging is essential for healthy aging. Unraveling the changes in different T cell compartments will help to better understand the underlying mechanisms of age-associated immunological decline and, thus, to develop therapeutic interventions that enhance immunity in older adults.
Methods
Animal procedures
C57BL/6J HccRsd mice were purchased from Envigo or generated at the Centro de Biología Molecular Severo Ochoa (Madrid, Spain) animal facilities. All mice required for this study were bred and aged in specific-pathogen-free conditions in the animal facility of Centro de Biología Molecular Severo Ochoa (Madrid, Spain). All mice were housed in ventilated cages within animal rooms maintained under a 12 h–12 h light-dark cycle. Animal rooms were temperature and humidity controlled. Standard diet and water were available ad libitum. Both male and female mice were used in this study.
All the procedures with animals were previously evaluated and approved (PROEX 287/16 and PROEX 52.1/23) by the ethics commitee on animal experimentation of the CBMSO, the authorized commitee of the Spanish National Research Council or the Universidad Autónoma de Madrid, and the regional government (Comunidad de Madrid). All mice were checked for any macroscopic abnormalities (according to the Jackson guide ‘AGED C57BL/6J MICE FOR RESEARCH STUDIES’). Mice were used at different ages: young (less than 4 months of age), adult (4–20 months of age) and old (over 20 months of age).
CD3ε−/− mice (JAX stock no. 004177) were kindly provided by Prof. B. Alarcón (CBM). CD45.1 (JAX stock no. 002014) mice were kindly provided by Dr. C. Cobaleda (CBM). Both strains were in C57BL/7 background.
IL-2 + IL-33 treatment
The IL-2 immune complex was prepared as previously described43. In brief, 1 µg of IL-2 (Peprotech, #212-12) was mixed with 5 µg of α-IL-2 (BioLegend, #503706) per mouse and incubated at room temperature (RT) for 30 min. Each mouse was intraperitoneally injected for three consecutive days. As a control, mice received only 200 μl of NaCl 0.9%. From the third day, IL-2 was combined with IL-33. 1 µg of IL-33 (BioLegend, #580506) was injected in 200 μl of NaCl per mouse for five consecutive days. As a control, mice received only 200 μl of NaCl.
Adoptive transfer of Treg cells
For the study of the function of kTreg cells in vivo, KLRG1− and KLRG1+ Treg cells (CD4+CD25+) were isolated by fluorescence-activated cell sorting (FACS) from IL-33-treated mice and injected into CD3ε−/− young mice for three consecutive days. Approximately 8 × 105 total Treg cells were injected per mouse. Mice were weighed and monitored every 3 days for 4 months after the adoptive transfer and then were euthanized and the spleens were analyzed.
Suppression assays
For the in vivo suppression assays, KLRG1− and KLRG1+ Treg cells (CD4+CD25+) were isolated by FACS from IL-33-treated mice. Naive CD4+ T cells were purified by magnetic isolation (Stemcell Technologies, #19765) from young CD45.1 mice. A total of 6 × 105 naive CD4+ T cells alone or in combination with 1.5 × 105 KLRG1− or KLRG1+ Treg cells were intravenously injected into CD3ε−/− young mice. The animals were weighed and monitored every 3 days for 4 months after the adoptive transfer and then were euthanized and the colonic lamina propria was analyzed.
For the in vitro suppression assays, KLRG1− and KLRG1+ Treg cells (CD4+CD25+) were isolated by FACS from old mice. Naive CD4+ T cells were purified by magnetic isolation from young CD45.1 mice. CD45.1+ naive CD4+ T cells were labeled with 1 µM of CellTrace Violet (Invitrogen, C34571) in PBS for 30 min at 37 °C. A total of 2.5 × 104 naive Treg cells were incubated with 1.25 Treg cells for 3 days. Cells were stimulated with Mouse T-Activator Dynabeads (Gibco, 11452D) following the manufacturer’s instructions. The percentage of suppression was determined as follows: 100 − (% of proliferating cells with Treg cells)/(% of proliferating cells without Treg cells).
Human samples
Volunteer recruitment was performed through the GENYAL Clinical Trials Platform of IMDEA Alimentación (Madrid, Spain). This study was approved by the institutional Research Ethics Committee (IMDEA Food Foundation, IMD PI-052 and IMD PI-055) and performed in accordance with the principles of research involving human subjects stated in the Declaration of Helsinki (1964). All patients were clearly informed about the study methodology and provided written informed consent.
Peripheral blood samples were collected from a total of 117 volunteers belonging to two different population groups, whose inclusion criteria were as follows:
-
Young healthy population (n = 42): 14 male and 28 female healthy volunteers aged between 18 and 25 years.
-
Senior population (n = 75): 28 male and 47 female volunteers over 55 years of age.
Exclusion criteria were common for both study groups and included: decreased cognitive function, pregnancy or breastfeeding, severe chronic health conditions (for example, chronic kidney, liver and heart disease), immunodeficiencies and autoimmune diseases, and immunosuppressive or psychotropic pharmacological treatment. The results shown in the study apply to both sexes. The experimental groups have been defined by the age or the participants, and no sex- and gender-based analyses have been performed. Volunteers received financial compensation after the study.
Tissue processing for flow cytometry
Mice were euthanized with CO2 followed by perfusion with cold PBS. The indicated tissues were extracted and processed as specified.
Spleen and lymph nodes
Lymph nodes were collected from inguinal, mesenteric, cervical and axillar areas. Spleen and lymph nodes were mashed and filtered through a 70-μm cell strainer. The cell suspension was centrifuged at 300g for 5 min at 4 °C. Red blood cells were removed using 5 ml of erythrocyte lysis buffer (ammonium chloride 0.15 M, sodium bicarbonate 0.01 M and EDTA 0.0001 M) for 5 min. Cells were washed, centrifuged, resuspended and counted.
Blood
Blood was extracted from either the facial vein or the heart in living or euthanized mice, respectively. The cell suspension was centrifuged at 300g for 5 min at 4 °C. Red blood cells were removed resuspending the cells in 5 ml of erythrocyte lysis buffer for 5 min. Cells were washed, centrifuged, resuspended and stained.
Colonic lamina propria
T cells from colonic lamina propria were isolated as previously reported75. Colon samples between the cecum and rectum were obtained and cleaned from fat and feces. Tissues were cut longitudinally, washed with cold PBS and then cut transversally into 1-cm-long fragments, mixed in prewarmed 5 mM EDTA, 14 mH HEPES, 10% FBS PBS and incubated under shaking at 180 rpm for 30 min at 37 °C. After washing with PBS, tissue pieces were then minced and mixed in prewarmed 25 mM HEPES, 10% FBS RPMI supplemented with 300 U ml−1 collagenase type VIII (Sigma, C2139) under shaking at 180 rpm for 45 min at 37 °C. The digested tissue was filtered through a 70-μm cell strainer, washed with 5 mM EDTA, 14 mH HEPES, 10% FBS PBS and centrifuged at 650g for 5 min at RT. To further enrich in leukocytes, supernatants were centrifuged in a 40%/70% Percoll gradient (Sigma, GE17-0891-01) at 750g for 20 min at RT with minimum acceleration and without brake. Isolated cells were washed with PBS and resuspended in 2% FBS RPMI for counting.
White adipose tissue
Gonadal white adipose tissue was obtained from the mouse abdominal cavity and mixed in 2 mg ml−1 BSA, 2% FBS RPMI supplemented with 2 mg ml−1 collagenase type II (Sigma, C6885) under shaking at 180 rpm for 40 min at 37 °C. The digested tissue was vertically rested to separate fat from the aqueous phases, which were obtained using an 18 G syringe. Then, the cell suspensions were filtered through a 70-μm cell strainer and washed with 2% FBS RPMI. Finally, erythrocytes were removed by incubation with a lysis buffer for 5 min at 4 °C, washed with 1 mM EDTA PBS and finally resuspended in 1 ml of 1 mM EDTA 2% FBS PBS for counting.
Peyer’s patches
Peyer’s patches were collected from the intestine and mashed into a 70-μm cell strainer. The cell suspension was centrifuged at 400g for 5 min at 4 °C. Finally, the cell pellets were resuspended in 1 ml of 2% FBS RPMI for counting.
Liver
Liver was collected and cut into prewarmed 25 mM HEPES, 10% FBS RPMI supplemented with 0.4 mg ml−1 collagenase type VIII (Sigma, C2139) under shaking at 180 rpm for 45 min at 37 °C. Digested tissue was filtered through a 70-μm cell strainer and centrifuged at 350g for 5 min at 4 °C. Red blood cells were removed using 5 ml of erythrocyte lysis buffer for 5 min. To further enrich in leukocytes, supernatants were centrifuged in a 40%/70% Percoll gradient (Sigma, GE17-0891-01) at 1,250g for 30 min at RT with acceleration on 6 and without brake. Isolated cells were washed with PBS and resuspended in ml 2% FBS RPMI for counting.
Bone marrow
Femurs and tibias were collected and the cells from the bone marrow were obtained by centrifuging the bones at 6,000g for 1 min. Red blood cells were removed using 5 ml of erythrocyte lysis buffer for 5 min. Cells were washed and resuspended in 2% FBS PBS for counting.
Human samples
Human blood samples were collected by venipuncture in an overnight fasting state. Three milliliters of blood were collected in TransFix/EDTA vacuum blood collection tubes (Cytomark) and preserved until the day of staining and cell acquisition.
Flow cytometry
To differentiate between live and dead, the cells were first stained with the Zombie NIR Fixable Viability Kit (BioLegend, 423106, 1:3,000), the Zombie Yellow Fixable Viability Kit (BioLegend, 423104, 1:3,000) or the Ghost Dye Violet 540 (Tonbo Biosciences, 13-0879, 1:3,000) for 20 min at 4 °C. Then, the cells were washed with FACS staining buffer (PBS supplemented with 2% fetal bovine serum and 1 mM EDTA) and incubated with Fc receptor blocker purified rat anti-mouse anti-CD16/CD32 (BD Biosciences, 553142, 1:200) for 20 min at 4 °C. Cells were then incubated with primary antibodies for 20 min at 4 °C and were washed twice with FACS staining buffer. The following antibodies were diluted in Brilliant Stain Buffer (BD Biosciences, 566349) for surface antigen staining:
Antigen | Fluorochrome | Dilution | Clone | Supplier | Catalog |
|---|---|---|---|---|---|
CD38 | Pacific Blue | 1:500 | 90 | BioLegend | 102720 |
TIM3 | BV480 | 1:200 | 5D12/TIM-3 | BD Biosciences | 747618 |
CD244.2 (2B4) | BV510 | 1:200 | 2B4 | BD Biosciences | 740115 |
CD44 | BV570 | 1:200 | IM7 | BioLegend | 103037 |
CD69 | BV650 | 1:100 | H1.2F3 | BioLegend | 104541 |
CD62L | BV711 | 1:400 | MEL-14 | BioLegend | 104445 |
CD95 | BV750 | 1:100 | Jo2 | BD Biosciences | 747413 |
KLRG1 | BV785 | 1:200 | 2F1 | BioLegend | 138429 |
CD223 (LAG3) | BB515 | 1:200 | C9B7W | BD Biosciences | 566210 |
CD49d | PerCP-Cy5.5 | 1:100 | R1-2 | BioLegend | 103619 |
PD-1 | PerCP-eFluor710 | 1:200 | J43 | ThermoFisher | 46-9985-80 |
ST2 | PerCP-eFluor710 | 1:200 | RMST2-33 | ThermoFisher | 46-9333-82 |
NKG2A | PE | 1:200 | 16A11 | BioLegend | 142804 |
NKG2D | PE-Dazzle594 | 1:200 | CX5 | BioLegend | 130214 |
CD25 | PE Cy5 | 1:400 | PC61 | BioLegend | 102007 |
CD8 | PE-Fire700 | 1:1000 | 53-6.7 | BioLegend | 100792 |
CD28 | APC | 1:50 | E18 | BioLegend | 122016 |
CD153 | R718 | 1:200 | RM153 | BD Biosciences | 751871 |
CD27 | APC-Cy7 | 1:400 | LG.3A10 | BioLegend | 124226 |
CD4 | APC-Fire810 | 1:1000 | Gk1.5 | BioLegend | 100480 |
For intracellular staining, after staining for membrane markers, the cells were fixed and permeabilized using the FOXP3/Transcription Factor Staining Kit (eBioscience, 00-5523-00) for 20 min at RT and darkness. To assess cytokine production, cells were stimulated for 4 h with 50 ng ml−1 phorbol 12-myristate 13-acetate (PMA) (ThermoFisher, 356150010) and 1 μg ml−1 ionomycin (Tocris, 1704) in the presence of brefeldin A (eBiosciences, 00-4506-51). Cells were then stained the following intracellular antibodies:
Antibody | Fluorochrome | Dilution | Clone | Supplier | Catalog |
|---|---|---|---|---|---|
IL-10 | Pacific Blue | 1:100 | JES5-16E3 | BioLegend | 505020 |
IL-17 | PE | 1:100 | TC11-18H10.1 | BioLegend | 506904 |
TNF | APC | 1:100 | MP6-XT22 | BioLegend | 506308 |
IFN-γ | Spark NIR 685 | 1:100 | XMG1.2 | BioLegend | 505861 |
MAF | eFluor450 | 1:100 | sym0F1 | ThermoFisher | 48-9855-41 |
FOXP3 | FITC | 1:200 | FJK-165 | ThermoFisher | 11-5773-82 |
RORγt | PE | 1:100 | Q31-378 | BD Biosciences | 562607 |
T-BET | AlexaFluor594 | 1:100 | 4B10 | BioLegend | 644834 |
T-BET | APC | 1:100 | 4B10 | BioLegend | 17-5825-82 |
TOX | eFluor660 | 1:200 | TXRX10 | ThermoFisher | 50-6502-80 |
γH2AX | – | 1:300 | 20E3 | CellSignaling | 9718 |
P16 | – | 1:200 | Polyclonal | ThermoFisher | PA5-119712 |
P21 | Alexa Fluor 647 | 1:400 | Polyclonal | Abcam | ab237265 |
Donkey anti-rabbit | Alexa Fluor 647 | 1:500 | Polyclonal | ThermoFisher | A-31573 |
All flow cytometry experiments from mouse samples were performed with four-laser (violet, blue, yellow-green and red) or five-laser Aurora analyzers (Cytek Biosciences). Data were analyzed with the FlowJo v10.5.3 software (BD Biosciences). Gating strategies were set on the basis of fluorescence minus one controls, unstained samples and unstimulated samples (when needed). All the samples in the experiment excluded dead cells, clumps and debris.
In human samples, each tube contained at least 2 × 106 human whole blood cells. Cells were labeled by incubation with appropriate fluorescence-conjugated antibodies for 15 min at RT in the dark. Cells were then lysed with 2 ml of FACS lysing solution (BD Biosciences) for 10 min and centrifuged at 500g for 5 min at RT. Then, the cells were washed with 5 ml of PBS. The following antibodies were used for surface antigen staining:
Antibody | Fluorochrome | Dilution | Clone | Supplier | Catalog |
|---|---|---|---|---|---|
CD8 | BUV496 | 1:100 | RPA-T8 | BD Biosciences | 612942 |
CD25 | BUV615 | 1:100 | 2A3 | BD Biosciences | 612996 |
CD19 | BUV661 | 1:100 | 1D3 | BD Biosciences | 612971 |
CD45 | BV510 | 1:100 | HI30 | BD Biosciences | 563204 |
CD4 | BV786 | 1:100 | SK3 | BD Biosciences | 664528 |
CD127 | PE | 1:100 | HIL-7R-M21 | BD Biosciences | 561028 |
CD3 | APC | 1:100 | HIT3a | BD Biosciences | 555342 |
KLRG1 | APC-Cy7 | 1:100 | 2F1/KLRG1 | BioLegend | 138426 |
Experiments with human samples were performed in a BD FACSymphony A5 SORP flow cytometer (BD Biosciences). To generate comparable results among patients and over time, the photomultiplier voltages were adjusted to unlabeled lysed whole blood cells to obtain optimal photomultiplier tube voltages for the resolution of dim cell populations. The target values resulting of the optimization were used for subsequent calibrations to maintain instrument standardization. When possible, at least 40,000 events of CD4 population were acquired to reach the maximum Treg events. Data were analyzed using the FlowJoTM v.10 software.
The gating strategy for identifying kTreg cells is illustrated in the Extended Data Fig. 6. In brief, first, doublets and debris were excluded in forward scatter (FSC) – side scatter (SSC) dot plots. Then, a region in CD45 and SSC parameters were used to discriminate leukocytes. Next, lymphocytes were gated in a SSC – CD4 dot plot. Lymphoid cells were further cleaned in a SSC – CD45 plot. After that, CD3+CD19− T cells were gated and, among them, CD4+ T lymphocytes were distinguished. The Treg cells were identified, within the CD4+ T cells, by gating the CD25hiCD27lo population. Then, the expression of KLRG1 was analyzed in Treg cells. To have a negative control to properly gate KLRG1+ cells, the gate was established in B cells.
Analysis of mitochondrial fitness
Analysis of mitochondrial mass and MMP was performed by flow cytometry in cells labeled for 30 min with 50 nM MitoTracker Green FM (Invitrogen, M7514), 200 nM MitoTracker DeepRed FM (Invitrogen, M22426), 40 nM Image-iT TMRM (Invitrogen, I34361) or 50 nM MitoTracker Red CMXRos (Invitrogen, M7512) in RPMI medium with no FBS in a 37 °C and 5% CO2 incubator before extracellular staining. After incubation, cells were washed with FACS staining buffer and incubated with antibodies for extracellular staining.
Dimensional reduction and clustering analysis of flow cytometry data
Dimensional reduction and clustering analysis of flow cytometry data was done using OMIQ (Dotmatics). First, nonlymphocyte cells, doublets and dead cells were excluded on the basis of viability staining and FSC and SSC parameters. Then, 15,000 CD4+ cells from each sample were subsampled for further analysis. For dimensional reduction, the Uniform Manifold Approximation and Projection (UMAP) algorithm was applied with the following parameters: neighbors = 15, minimum distance = 0.4, components = 2, learning rate = 1, epochs = 200. For unbiased clustering, the Cluster-X algorithm was applied with alpha = 0.001. Mean fluorescence intensity for each marker projected on the UMAP plots was used to infer the cluster’s identity, and similar clusters were combined.
RNA-seq
rTreg, aTreg and kTreg cells were isolated from young and old mice by FACS. RNA was extracted with the RNeasy Micro Kit (Qiagen, 74004) following manufacturer’s instructions. Sample quality was measured using the Qubit 3.0 fluorimeter for sample concentration and Agilent 5400 for fragment analysis. SMART-Seq V4 Ultra Low Input RNA kit for Sequencing 480 Rxns (Takarabio, 634893) was used for efficient cDNA synthesis and library preparation. After amplification, cDNA was purified using AMPure XP beads (Beckman Coulter, A63882) to remove any contaminants and ensure high-quality cDNA. The library was checked with Qubit and real-time PCR for quantification and bioanalyzer for size distribution detection. Quantified libraries were pooled and sequenced on Illumina Illumina Novaseq X plus using a PE150 (pair-end 150 base pair) strategy to produce 6G of data.Fastq files quality check was performed using FastQC v0.11.9. RNA-seq reads were mapped to the Mus musculus reference genome, GRCm39, using Hisat2 v2.2.1 software. Reads were then preprocessed with SAMtools v1.13 to transform Sequence Alignment/Map files into Binary Alignment/Map files and sorted. The number of reads covered by each gene is calculated by HTSeq-Count v1.99.2. Downstream data analysis was performed with R v4.4.1. Analysis of differentially expressed genes was performed using DESeq2 v1.44.0. Genes with P < 0.05 and |log2fold change| >log2(1.5) were determined to show statistically significant differences in group comparison. Over-representation analysis and gene set enrichment analysis were performed using clusterProfiler v4.12.0 package in GO, KEGG, WikiPathways, Reactome and the Hallmarks of the Molecular signatures database. Principal component analysis (PCA) plots and volcano plot were visualized using ggplot2 v3.5.1. Heat maps were visualized using heatmap v1.0.12.
Analysis of cytokine secretion by Treg subsets using Multiplex
rTreg, aTreg or kTreg cells were isolated from IL-33-treated young mice by FACS. A total of 5.5 × 105 cells were incubated in 200 µl of complete RPMI medium supplemented with 10% FBS in resting conditions or in presence of Mouse T-Activator Dynabeads for 24 h. The cells were centrifuged at 300g for 5 min, and the supernatant was used for cytokine detection with magnetic bead technology (Invitrogen, Cytokine & Chemokine 26-Plex Mouse ProcartaPlex Panel 1).
Electron microscopy
For electron microscopy, splenocytes from old mice were labeled for 30 min with 50 nM MitoTracker Green FM and 200 nM Mitotracker DeepRed FM before extracellular staining. Cells were then labeled with a fluorochrome-conjugated antibody against CD4, and MtGhiMtDRhi and MtGloMtDRlo alive CD4+ T cells were isolated by FACS. After sorting, cells were fixed with 4% paraformaldehyde, 2% glutaraldehyde diluted in 0.1 M phosphate buffer. Cells were postfixed with 1% osmiun tetroxide and 1% potassium ferrocyanide for 60 min. After washing, incubation with 0.15% tannic acid in buffer phosphate 0.1 M for 1 min was achieved. After washing, cells were counterstained with uranyl acetate 2% for 1 h. Then, cells were dehydrated with lowering concentrations of ethanol and embedded in resin EPON. The preparations were examined with a transmission electron microscope (JEM1400 Flash, Jeol), and images were acquired with a CMOS Oneview camera (Gatan).
Reanalysis of scRNA-seq data
scRNA-seq data were extracted from a previously published work5 and reanalyzed with the Seurat package v4.2.0 in R v4.1.3. Variable genes were identified with the FindVariablesFeatures function across the range of expression values and used to perform a PCA with the RunPCA function. Clustering was performed with the FindClusters function with the Leiden algorithm and the first 20 principal components. Cluster identification was done with the FindMarkers function in each subset with a minimum log fold change of 0.25 and a P value <10-3. The kTreg cluster was separated by using the FindSubCluster function in the aTreg cluster and then identified with the FindMarkers function.
Statistics and reproducibility analysis
Unless otherwise specified, n represents the number of individual biological replicates and is represented in graphs as one dot per sample. All the data are extracted from one representative experiment (n ≥ 3 per group) of a minimum of two separate experiments. Flow cytometry plots are representative of at least three replicates. No statistical method was used to predetermine sample size, but a minimum of three samples were used per experimental group and condition. Data collection and analysis were not performed blind to the age of the mice.
For statistical analysis, GraphPad Prism (version 9) was used. Data are expressed as mean ± s.e.m., unless otherwise indicated. Outliers were identified by the ROUT method (5%) and removed. Comparisons for two groups were calculated using unpaired two-tailed Student’s t-tests (for two groups meeting the normal distribution criteria) or Mann–Whitney U test (for two groups without normal distribution) according to the Shapiro–Wilk normality test. When comparing different populations within the same mouse, comparisons were calculated using two-tailed paired Student’s t-tests (for two groups meeting the normal distribution criteria) or Wilcoxon test (for two groups without normal distribution) according to the Shapiro–Wilk normality test. Comparisons for more than two groups were calculated using one-way analysis of variance (ANOVA) with Tukey’s correction for multiple comparisons (for three or more groups meeting the normal distribution criteria) or Kruskal–Wallis test with Dunn’s correction for multiple comparisons (for three or more groups without normal distribution) according to the Shapiro–Wilk normality test. When comparing at least three distinct populations within the same mouse, comparisons were calculated using repeated measures (RM) one-way ANOVA with Tukey’s correction for multiple comparisons (for samples meeting the normal distribution criteria) or Friedman test with Dunn’s correction for multiple comparisons (for at least three groups without normal distribution) according to the Shapiro–Wilk normality test.
The statistical significance is represented as follows: *P < 0.05, **0.05 < P < 0.01; ***0.01 < P < 0.001, ****P < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the Article and its Supplementary Information. The scRNA-seq dataset reanalyzed is publicly available (Single Cell Portal; accession number SCP490)5. The RNA-seq dataset generated during the current study is available in the Gene Expression Omnibus repository (GSE279926). The rest of the data were originally generated.
References
Franceschi, C. & Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A 69, S4–S9 (2014).
Ferrucci, L. & Fabbri, E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 15, 505–522 (2018).
Goronzy, J. J. & Weyand, C. M. Immune aging and autoimmunity. Cell. Mol. Life Sci. 69, 1615–1623 (2012).
Carrasco, E. et al. The role of T cells in age-related diseases. Nat. Rev. Immunol. 22, 97–111 (2022).
Elyahu, Y. et al. Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci. Adv. 5, eaaw8330 (2019).
Sakaguchi, S., Yamaguchi, T., Nomura, T. & Ono, M. Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008).
Sharma, S., Dominguez, A. L. & Lustgarten, J. High accumulation of T regulatory cells prevents the activation of immune responses in aged animals. J. Immunol. 177, 8348–8355 (2006).
Trujillo-Vargas, C. M. et al. Immune phenotype of the CD4+ T cells in the aged lymphoid organs and lacrimal glands. GeroScience 44, 2105–2128 (2022).
Soto-Heredero, G., Gómez de las Heras, M. M., Escrig-Larena, J. I. & Mittelbrunn, M. Extremely differentiated T cell subsets contribute to tissue deterioration during aging. Annu. Rev. Immunol. 41, 181–205 (2023).
Lorenzo, E. C. et al. Senescence-induced changes in CD4 T cell differentiation can be alleviated by treatment with senolytics. Aging Cell 21, e13525 (2022).
Kuswanto, W. et al. Poor repair of skeletal muscle in aging mice reflects a defect in local, interleukin-33-dependent accumulation of regulatory T cells. Immunity 44, 355–367 (2016).
Tsaknaridis, L. et al. Functional assay for human CD4+CD25+ Treg cells reveals an age-dependent loss of suppressive activity. J. Neurosci. Res. 74, 296–308 (2003).
Raynor, J. et al. IL-15 fosters age-driven regulatory T cell accrual in the face of declining IL-2 levels. Front. Immunol. 4, 161 (2013).
Lages, C. S. et al. Functional regulatory T cells accumulate in aged hosts and promote chronic infectious disease reactivation. J. Immunol. 181, 1835–1848 (2008).
Raynor, J. et al. IL-6 and ICOS antagonize Bim and promote regulatory T cell accrual with age. J. Immunol. 195, 944–952 (2015).
Morales-Nebreda, L. et al. Aging imparts cell-autonomous dysfunction to regulatory T cells during recovery from influenza pneumonia. JCI Insight 6, e141690 (2021).
Overacre-Delgoffe, A. E. et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell 169, 1130–1141 (2017).
Zagorulya, M. et al. Tissue-specific abundance of interferon-gamma drives regulatory T cells to restrain DC1-mediated priming of cytotoxic T cells against lung cancer. Immunity 56, 386–405 (2023).
Henson, S. M. et al. KLRG1 signaling induces defective Akt (ser473) phosphorylation and proliferative dysfunction of highly differentiated CD8+ T cells. Blood 113, 6619–6628 (2009).
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007).
Henson, S. M. & Akbar, A. N. KLRG1–more than a marker for T cell senescence. Age 31, 285–291 (2009).
Voehringer, D. et al. Viral infections induce abundant numbers of senescent CD8 T cells. J. Immunol. 167, 4838–4843 (2001).
Schwartzkopff, S. et al. Tumor-associated E-cadherin mutations affect binding to the killer cell lectin-like receptor G1 in humans. J. Immunol. 179, 1022–1029 (2007).
Li, L., Wan, S., Tao, K., Wang, G. & Zhao, E. KLRG1 restricts memory T cell antitumor immunity. Oncotarget 7, 61670–61678 (2016).
Ramello, M. C. et al. Polyfunctional KLRG-1+CD57+ senescent CD4+ T cells infiltrate tumors and are expanded in peripheral blood from breast cancer patients. Front. Immunol. 12, 713132 (2021).
Cheng, G. et al. IL-2 receptor signaling is essential for the development of Klrg1+ terminally differentiated T regulatory cells. J. Immunol. 189, 1780–1791 (2012).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Ron-Harel, N. et al. Defective respiration and one-carbon metabolism contribute to impaired naïve T cell activation in aged mice. Proc. Natl Acad. Sci. USA 115, 13347–13352 (2018).
Bektas, A. et al. Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging 11, 9234–9263 (2019).
Steinert, E. M., Vasan, K. & Chandel, N. S. Mitochondrial metabolism regulation of T cell-mediated immunity. Annu. Rev. Immunol. 39, 395–416 (2021).
Velarde, M. C., Flynn, J. M., Day, N. U., Melov, S. & Campisi, J. Mitochondrial oxidative stress caused by Sod2 deficiency promotes cellular senescence and aging phenotypes in the skin. Aging 4, 3–12 (2012).
Wiley, C. D. et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Chapman, N. M. et al. mTOR coordinates transcriptional programs and mitochondrial metabolism of activated Treg subsets to protect tissue homeostasis. Nat. Commun. 9, 2095 (2018).
Weinberg, S. E. et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499 (2019).
Venditti, P., Di Stefano, L. & Di Meo, S. Mitochondrial metabolism of reactive oxygen species. Mitochondrion 13, 71–82 (2013).
Mittelbrunn, M. & Kroemer, G. Hallmarks of T cell aging. Nat. Immunol. 22, 687–698 (2021).
Mogilenko, D. A. et al. Comprehensive profiling of an aging immune system reveals clonal GZMK+ CD8+ T cells as conserved hallmark of inflammaging. Immunity 54, 99–115 (2021).
Wang, Y., Su, M. A. & Wan, Y. Y. An essential role of the transcription factor GATA-3 for the function of regulatory T cells. Immunity 35, 337–348 (2011).
Wohlfert, E. A. et al. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121, 4503–4515 (2011).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Siede, J. et al. IL-33 receptor-expressing regulatory T cells are highly activated, Th2 biased and suppress CD4 T cell proliferation through IL-10 and TGFβ release. PLoS ONE 11, e0161507 (2016).
Boyman, O., Kovar, M., Rubinstein, M. P., Surh, C. D. & Sprent, J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311, 1924–1927 (2006).
Sun, L. et al. Aged regulatory T cells protect from autoimmune inflammation despite reduced STAT3 activation and decreased constraint of IL-17 producing T cells. Aging Cell 11, 509–519 (2012).
Garg, S. K. et al. Aging is associated with increased regulatory T-cell function. Aging Cell 13, 441–448 (2014).
Zhao, L. et al. Changes of CD4+CD25+Foxp3+ regulatory T cells in aged Balb/c mice. J. Leukoc. Biol. 81, 1386–1394 (2007).
Hwang, K.-A., Kim, H.-R. & Kang, I. Aging and human CD4+ regulatory T cells. Mech. Ageing Dev. 130, 509–517 (2009).
Delacher, M. et al. Precursors for nonlymphoid-tissue Treg cells reside in secondary lymphoid organs and are programmed by the transcription factor BATF. Immunity 52, 295–312 (2020).
Beppu, L. Y. et al. Tregs facilitate obesity and insulin resistance via a Blimp-1/IL-10 axis. JCI Insight 6, e140644 (2021).
Tauro, S., Nguyen, P., Li, B. & Geiger, T. L. Diversification and senescence of Foxp3+ regulatory T cells during experimental autoimmune encephalomyelitis. Eur. J. Immunol. 43, 1195–1207 (2013).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Cebula, A. et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 497, 258–262 (2013).
Kaur, H., Kaur, G. & Ali, S. A. IL-33’s role in the gut immune system: a comprehensive review of its crosstalk and regulation. Life Sci. 327, 121868 (2023).
Goldberg, E. L. et al. IL-33 causes thermogenic failure in aging by expanding dysfunctional adipose ILC2. Cell Metab. 33, 2277–2287 (2021).
Xu, L. et al. IL-33 induces thymic involution-associated naive T cell aging and impairs host control of severe infection. Nat. Commun. 13, 6881 (2022).
Monzel, A. S., Enríquez, J. A. & Picard, M. Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nat. Metab. 5, 546–562 (2023).
Yu, Y.-R. et al. Disturbed mitochondrial dynamics in CD8+ TILs reinforce T cell exhaustion. Nat. Immunol. 21, 1540–1551 (2020).
Monteiro, L., de, B., Davanzo, G. G., de Aguiar, C. F. & Moraes-Vieira, P. M. M. Using flow cytometry for mitochondrial assays. MethodsX 7, 100938 (2020).
Montes de Oca, M. et al. IL-27 signalling regulates glycolysis in Th1 cells to limit immunopathology during infection. PLoS Pathog. 16, e1008994 (2020).
Chen, P.-M. et al. CD38 reduces mitochondrial fitness and cytotoxic T cell response against viral infection in lupus patients by suppressing mitophagy. Sci. Adv. 8, eabo4271 (2022).
Konjar, Š. et al. Mitochondria maintain controlled activation state of epithelial-resident T lymphocytes. Sci. Immunol. 3, eaan2543 (2018).
Kowaltowski, A. J. & Abdulkader, F. How and when to measure mitochondrial inner membrane potentials. Biophys. J. 123, 4150–4157 (2024).
Ding, C. et al. A Treg-specific long noncoding RNA maintains immune-metabolic homeostasis in aging liver. Nat. Aging 3, 813–828 (2023).
Guo, Z. et al. DCAF1 regulates Treg senescence via the ROS axis during immunological aging. J. Clin. Invest. 130, 5893–5908 (2020).
Sun, Z. & Hornung, V. cGAS–STING signaling. Curr. Biol. 32, R730–R734 (2022).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Linder, A. & Hornung, V. Inflammasomes in T cells. J. Mol. Biol. 434, 167275 (2022).
Baixauli, F. et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 22, 485–498 (2015).
Milanovic, M. et al. Senescence-associated reprogramming promotes cancer stemness. Nature 553, 96–100 (2018).
Salinas-Saavedra, M. et al. Senescence-induced cellular reprogramming drives cnidarian whole-body regeneration. Cell Rep. 42, 112687 (2023).
Burton, O. T. et al. The tissue-resident regulatory T cell pool is shaped by transient multi-tissue migration and a conserved residency program. Immunity 57, 1586–1602 (2024).
Correia-Melo, C. et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 35, 724–742 (2016).
Vardhana, S. A. et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21, 1022–1033 (2020).
Scharping, N. E. et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215 (2021).
Valle-Noguera, A., Gómez-Sánchez, M. J., Girard-Madoux, M. J. H. & Cruz-Adalia, A. Optimized protocol for characterization of mouse gut innate lymphoid cells. Front. Immunol. 11, 563414 (2020).
Acknowledgements
Funded by the European Union. Views and opinions expressed are, however, those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them. This study was supported by European Research Council grant ERC-2021-CoG 101044248-LetTBe, and the Y2020/BIO-6350 NutriSION-CM synergy grant from Comunidad de Madrid, Ministerio de Ciencia e Innovacion, Spain (grants PID2022-141169OB-I00 and PID2022-138295OB-I00). GENYAL Clinical Trials Platform of IMDEA Alimentación led by A. Ramírez de Molina and R. Ramos Ruiz conducted the volunteer recruitment and human clinical trial. Flow cytometry of human samples was conducted by Flow Cytometry Unit, CNIC, being the cytometer part of the grant EQC2018-005009-P funded by MCIN/AEI/10.13039/501100011033 and by ‘ERDF A way of making Europe’. G.S.-H. is supported by a FPI-UAM grant (Universidad Autónoma de Madrid). E.G.-R. was funded by a Juan de la Cierva grant (IJC2018-036850-I; Universidad Autónoma de Madrid). M.G. and J.I.E.-L. are supported by FPU grants (FPU19/02576 and FPU20/04066, respectively), both from Ministerio de Ciencia, Innovación y Universidades (Spain). E.C. is supported by SI4/PJI/2024-00166 from Comunidad de Madrid and UAM (Spain). S.D.-P. was funded by a PIPF grant (PIPF-2022/SAL-GL-25208), from Comunidad de Madrid (Spain). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
G.S.-H., E.G.-R., E.C. and M.M. set up the research project and designed the experiments. G.S.-H., E.G.-R., E.C., J.I.E.-L., M.M.G., S.D.P., I.F.Q. and E.M.B. conducted most of the experiments and acquired the data. M.J.R. performed and analyzed the data in Fig. 1f,g. V.Z. provided technical and scientific support. C.M.F.-D. and M.B.A.-F. performed the experiments and analyzed the data in Fig. 8 under the supervision of A.R. A.F.A., D.A., S.J., A.d.S., F.S.-C. and C.T. performed bioinformatic analysis. G.S.-H. assembled the figures. G.S.-H. and M.M. wrote the manuscript. M.M. obtained financial support and provided supervision.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Jorg Goronzy and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 CD4+ T cells progressively lose mitochondrial mass and mitochondrial membrane potential during aging.
(a) Representative plots of mitochondrial mass and mitochondrial membrane potential analyzed by flow cytometry in CD4+ T cells treated or not treated with oligomycin. Cells were gated as MtGhiMtDRhi, MtGhiMtDRlo and MtGloMtDRlo. Mitochondrial mass analyzed as gMFI of MtG (b) and mitochondrial membrane potential analyzed as gMFI of MtDR (c) measured by flow cytometry in circulating CD4+ T cells from mouse from 2- (n = 5), 9- (n = 5), 13- (n = 5), 17- (n = 5) and 21- (n = 6) month-old mice. (d) Representative flow cytometry plots and quantifications of the simultaneous analysis of MtG and TMRM in splenic CD4+ T cells from young (2-month-old, n = 5) and old (21-month-old, n = 4) mice. (e) Quantification of the MtDR/MtG ratio in MtGhiMtDRhi and MtGloMtDRlo CD4+ T cells from mice at different ages (n = 6 mice per group). gMFI: Geometric Mean Fluorescence Intensity; MtG: Mitotracker Green; MtDR: Mitotracker DeepRed. Each dot represents an individual mouse. Data are presented as mean values ± SEM. Statistical analysis was performed using one-way ANOVA with post hoc Tukey’s correction (b), Kruskal-Wallis test with post hoc Dunn’s correction (c), two-tailed Student’s t test [(d): TMRMhiMtGhi; (e)] or two-tailed unpaired Welch’s t test [(d): TMRMloMtGlo] (b). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Extended Data Fig. 2 Identification of age-associated T cells by scRNAseq and by spectral flow cytometry.
(a) UMAP representation of CD4+ T cells from young (left) and old mice (right) analyzed by scRNAseq (top) or spectral flow cytometry (bottom). (b) UMAP with clustering overlay showing the distribution of the different clusters of CD4+ T cells identified by scRNAseq or by spectral flow cytometry: naive (blue), naïve_ISG15 (dark blue), rTregs (yellow), aTregs (orange), kTregs (red), TEM (light green), cytotoxic (brown) and exhausted (dark green) (n = 4 mice per group). (c) Percentage of circulating KLRG1+ cells in CD4+ T cells from 2- (n = 6), 9- (n = 6), 13- (n = 5), 17- (n = 6) and 21- (n = 6) month-old mice assessed by flow cytometry. (d-e) Percentage of splenic KLRG1+ cells in CD4+FOXP3+CD25+ T cells (d) or in CD4+ T cells (e) from 2- (n = 3), 9- (n = 4), 17- (n = 4) and 21- (n = 4) month-old mice. Each dot represents an individual mouse. Data are presented as mean values ± SEM. Statistical analysis was performed using one-way ANOVA with post hoc Tukey’s correction [(c), (d), (e)]. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Extended Data Fig. 3 Reanalysis of publicly available scRNAseq data identifies kTregs with a pro-inflammatory signature.
(a) UMAP representation and clustering of a new analysis of publicly available scRNAseq data from CD4+ T cells from young and old mice. Clusters are colored as naïve (blue), rTregs (yellow), aTregs (orange), kTregs (red), TEM (light green), cytotoxic (brown) and exhausted (dark green). (b) Quantification of the percentage of kTreg cells in young and old mice (n = 4 mice per group). (c) Comparative expression of genes related to Treg function in aTreg and kTreg (n = 4 mice per group). (d) Violin plots showing the expression of several pro-inflammatory markers in each cluster of CD4+ T cells (n = 4 mice per group). Each dot represents an individual mouse. Box-and-whisker plots show the median, the maximum, the minimum and 25th and 75th percentiles. Statistical analysis was performed using two-tailed unpaired Welch’s t test (b). Exact P values and additional statistical parameters can be found in the source data.
Extended Data Fig. 4 Analysis of the expression of different transcription factors in the Treg subsets.
Representative histograms and quantifications of the expression of different transcription factors by flow cytometry in splenic rTreg (yellow), aTreg (orange) and kTregs (red) from young (2-month-old) mice (n = 6 mice per group) measured as gMFI. gMFI values are relative to the MFI in rTregs (n = 6 per group). gMFI: Geometric Mean Fluorescence Intensity. Each dot represents an individual mouse. Data are presented as mean values ± SEM. Statistical analysis was performed using Friedman test with post hoc Dunn’s correction. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Extended Data Fig. 5 KLRG1+ Tregs show a pro-inflammatory phenotype and induce inflammation.
(a-b) In vitro assessment of the suppressive activity of KLRG1+ Tregs. KLRG1- or KLRG1+ Tregs were sorted from old (22-month-old) and co-cultured with CD45.1 naïve CD4+ T responder cells labelled with CellTrace Violet at 2:1 ratio (2 naïve:1 Treg) for 72 h in the presence of α-CD3/α-CD28 (n = 3 mice). (a) Representative histogram of the CellTrace Violet signal measured by flow cytometry in CD45.1 CD4+ T conventional responder cells after 72 h of culture. (b) Quantification of the capacity of KLRG1- and KLRG1+ Treg cells to suppress the proliferation of CD45.1 CD4+ responder T cells. The percentage of suppression was determined as: 100 − (% of proliferating cells with Tregs)/(% of proliferating cells without Tregs). (c) Representative histograms and quantifications of the expression of different cytokines by flow cytometry in splenic rTreg (yellow), aTreg (orange) and kTregs (red) from old (21-month-old) mice measured as gMFI. gMFI values are relative to the MFI in rTregs (n = 6 mice per group). (d-e) Analysis of the secretion of different cytokines by the Treg subsets in activating conditions using multiplex. Treg subsets were sorted from young mice injected with IL-2 + IL-33 and incubated during 24 h in the presence of α-CD3/α-CD28 before the assay (n = 3 mice). (d) Heatmap and statistical comparisons of the secretion of different cytokines by rTreg, aTreg and kTregs. Heatmap values are relative to the maximum of each cytokine. (e) Quantification of IL-10, IL-6, GM-CSF and CXCL2 secreted by activated rTreg, aTreg and kTregs. (f-h) In vivo assessment of the pro-inflammatory activity of KLRG1+ Tregs: CD3ε-/- T cell deficient mice were injected with splenic KLRG1- or KLRG1+ Tregs isolated from young mice injected with IL-2 + IL-33. The mice were analyzed four months after the adoptive transfer (n = 3 mice per group). (f) Schematic diagram depicting the adoptive transfer of kTregs. (g) Spleen weight from CD3ε-/- mice non-injected or injected with KLRG1- or KLRG1+ T cells. (h) Quantification of the percentage of circulating neutrophils or basophils in CD3ε-/- mice non-injected or injected with KLRG1- or KLRG1+ T cells. gMFI: Geometric Mean Fluorescence Intensity. Each dot represents an individual mouse. Data are presented as mean values ± SEM. Statistical analysis was performed using two-tailed unpaired Student’s t test (b), Friedman test with post hoc Dunn’s correction [(c), (e): IL-6], RM one-way ANOVA with post hoc Tukey’s correction [(d); (e): IL10, GM-CSF, CXCL2] or ANOVA with post hoc Tukey’s correction [(g), (h)]. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Exact P values and additional statistical parameters can be found in the source data.
Extended Data Fig. 6 Gating strategy to identify KLRG1+ Tregs in human PBMCs.
Gating strategy followed to identify KLRG1+ Tregs (CD45+CD3+CD4+CD8-CD25+IL7R-KLRG1+) in human PBMCs.
Supplementary information
Supplementary Table 1
Differentially expressed genes in the kTreg cluster.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Soto-Heredero, G., Gabandé-Rodríguez, E., Carrasco, E. et al. KLRG1 identifies regulatory T cells with mitochondrial alterations that accumulate with aging. Nat Aging 5, 799–815 (2025). https://doi.org/10.1038/s43587-025-00855-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00855-9
This article is cited by
-
Systemic inflammation as a central player in the initiation and development of Alzheimer’s disease
Immunity & Ageing (2025)
-
Immunosenescence and cancer: molecular hallmarks, tumor microenvironment remodeling, and age-specific immunotherapy challenges
Journal of Hematology & Oncology (2025)
-
Aging reshapes the adaptive immune system from healer to saboteur
Nature Aging (2025)