Abstract
Background
Genetic variants play a pivotal role in the initiation and progression of many diseases, including cancer. Detecting these variants is the first step in understanding their contribution to disease mechanisms. RNA sequencing (RNA-Seq) has become a crucial assay in cancer research, offering insights beyond those provided by DNA sequencing. This study introduces VarRNA, a novel method that utilizes RNA-Seq data to classify single nucleotide variants and insertions/deletions from tumor transcriptomes.
Methods
VarRNA distinguishes transcriptome variants as germline, somatic, or artifact using a combination of two XGBoost machine learning models. These models were trained and validated using a cohort of pediatric cancer samples with paired tumor and normal DNA exome sequencing data serving as ground truth. We performed additional validation on RNA-Seq data from two distinct cancer datasets, demonstrating that VarRNA outperforms existing RNA variant calling methods.
Results
VarRNA identifies 50% of the variants detected by exome sequencing and detects unique RNA variants absent in paired tumor and normal DNA exome data. Some variants classified by VarRNA exhibit variant allele frequencies distinct from the corresponding DNA exome data. Strikingly, this phenomenon is prevalent in cancer-driving genes, where VarRNA analysis of the RNA-Seq data reveals the variant allele expression as much higher than expected based on the exome sequencing data.
Conclusions
These findings highlight the potential of RNA-Seq not only to uncover clinically relevant genetic variants but also to offer a deeper understanding of disease-specific expression dynamics that influence cancer pathogenesis, with implications for prognosis and therapeutic strategies.
Plain language summary
When a cell’s genetic material (DNA) is mutated, it’s growth may become uncontrolled, and cancer develops. Decoding the mutations in the DNA of cancer cells can tell doctors how best to treat a cancer, but if a cancer sample is too small to provide enough DNA, this information is lost. RNA, which is copied from DNA, also can be used to decode cancer mutations, allows use of very small samples, and provides unique information about which mutations may be driving cancer cell growth. We used cancer and matched normal DNA data to train a computing method called VarRNA that determines, using only RNA data from a cancer sample, whether each mutation is unique to the cancer cells or is inherited.
Similar content being viewed by others
Introduction
Genetic variants are critical in the initiation and progression of many diseases, including cancer. Next-generation sequencing (NGS) has radically improved our ability to detect these variants, leading to a better understanding of their roles in disease. RNA sequencing (RNA-Seq) is a powerful NGS assay for cancer research. Over the past decade, numerous analytical methods have been developed to elucidate the information contained within RNA-Seq data, such as gene expression analysis1, gene fusion identification2, and allele-specific expression analysis (ASE)3,4. Gene expression analysis is used to compare differential expression profiles between samples, such as primary and recurrent tumors, to uncover variations in tumor biology in the setting of relapse5 or across groups of cancers with similar histology to pinpoint therapeutic targets6. ASE, which quantifies the difference in expression of two alleles, can reveal mechanisms that promote tumor progression7.
Reliably identifying variants in sequencing data from cancer and matched normal samples is the first step in studying the relationship between genotype and cancer pathogenesis. These variants are typically identified in NGS analysis of tumor DNA (DNA-Seq) through targeted approaches, such as exome sequencing (ES). However, variants can also be called from RNA-Seq data, providing unique insights into ASE by revealing whether a variant observed in DNA is expressed in the tumor, or confirming that a nonsense mutant allele undergoes nonsense-mediated decay, for example4,8. RNA-Seq can present additional advantages over DNA-Seq for variant detection. For example, high expression levels yield increased sequencing depth and potentiate higher variant calling accuracy in those genes. Post-transcriptional modifications, such as RNA editing, can also be identified through variant calling in RNA-Seq. These modifications are important to identify because they can increase protein diversity9 and regulate gene expression10. Variants detected with RNA-Seq are directly tied to the transcriptome, providing immediate insights into potential functional impacts by determining whether variants are associated with upregulated or downregulated genes or pathways.
Outside of the cancer setting, several computational approaches have been published that detect genetic variants using RNA-Seq data. These methods generally use tools initially developed for variant calling in DNA (e.g., GATK HaplotypeCaller or Mutect211), and apply additional methods for sequencing alignment and variant filtering to reduce the higher number of false positive calls produced from using RNA-Seq data as input. SNPiR identifies single nucleotide variants (SNVs), specifically focusing on reducing false positive variant calls that can arise from common issues, such as mapping errors around splice sites or at repetitive loci12. RVBoost implements a machine learning model trained on common variants to prioritize true RNA variant calls over false positives13. DeepVariant RNA-Seq does not use existing variant calling methods, but instead scans the input RNA-Seq BAM file to identify evidence of variants and then uses a convolutional neural net to classify the genotype of each candidate position14.
A significant limitation of these methods is that they were developed to identify germline variants from normal (e.g., non-disease involved) tissue samples. Discerning somatic variants in cancer samples is, by contrast, a much more complex problem. Identifying somatic variants from bulk tissue extracts is primarily challenged by tumor content and tumor heterogeneity in the sample under study, which results in a range of variant allele frequencies (VAFs) for detected variants. VAFs of somatic variants typically range from 5 to 100%, while some assays allow for measurement of VAFs below 5%. This variability makes it more difficult to identify variants that do not follow the expected distribution of variants seen in a germline genome, where the allele frequency should be 0% (reference), 50% (heterozygous), or 100% (homozygous or hemizygous). To overcome this challenge, somatic variant calling in DNA-Seq data typically relies upon a matched normal comparator sample to differentiate germline vs. somatic variants. This approach is not plausible with RNA-Seq data, which is generated from cancer tissue alone, because matched adjacent normal tissue (the optimal comparator) is not routinely obtained surgically. No existing approach has been described to classify germline and somatic SNV and insertions or deletion (indel) calls from cancer tissue-derived RNA-Seq data alone.
We present VarRNA15, a computational classification approach that identifies SNV and indel variant calls from tumor RNA-Seq data and classifies them as artifact, germline, or somatic. VarRNA was developed using two XGBoost16 machine learning models: one to classify variants as true variants or artifacts, and a second that classifies true variants as either germline or somatic. The resulting variant calls are highly accurate compared to the ground truth data, and outperform existing RNA-Seq variant calling methods. In our application of VarRNA to RNA-Seq data from a pediatric cancer cohort, we demonstrated its capability to identify about 50% of the variants detected by ES, while uniquely detecting ASE of mutant alleles in oncogenes and additional variants not found by ES, underlining the value of variant identification from RNA-Seq to uncover RNA editing and allele-specific expression dynamics. These insights enhance our understanding of cancer pathogenesis, with potential implications for prognosis and treatment strategies, promising to elevate the clinical relevance of RNA-Seq variant analysis in precision oncology.
Methods
Ethical compliance
Ethical approval was not sought or required for this study, as it exclusively employed publicly available datasets and thus did not involve direct human subjects research. In conducting this analysis, we strictly adhered to the dataset’s terms of use, access, and distribution as outlined by their respective sources. We ensured that our research methods and objectives were aligned with the ethical guidelines for research and data use, including respecting privacy, intellectual property rights, and data integrity.
DNA sequencing, alignment, and variant calling
Sequencing reads from both Nationwide Children’s Hospital (NCH), ZERO Childhood Cancer Program (ZCC), and glioblastoma (GBM) datasets were processed with the same alignment and variant calling steps, using our Churchill secondary analysis pipeline17. The FASTQ files for both tumor and normal samples were aligned to the human reference genome assembly GRCh38 using BWA-MEM. SNV and indel calling were performed with GATK (v4.1.9). GATK HaplotypeCaller was run on each normal sample using the GVCF workflow to generate a per-sample intermediate GVCF. The GATK GenotypeGVCFs function was used to obtain genotypes for each germline sample in VCF format. We then performed variant recalibration and filtering in the two-stage process using the GATK functions VariantRecalibration and ApplyVQSR. GATK Mutect2 was run using both tumor and normal samples as input. The GATK function FilterMutectCalls was used to label false positives with a list of failed filters and true positives with ‘PASS.’’ Only ‘PASS’’ somatic variants were used for further analysis.
VarRNA: RNA-Seq alignment and variant calling
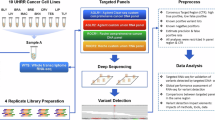
We developed a computational approach, VarRNA, to process RNA-Seq data to call and classify germline and somatic SNVs and indels (Fig. 1). These steps included 1) alignment of raw sequence reads in FASTQ format, 2) postprocessing of aligned reads, 3) variant calling, 4) variant annotation, and 5) variant filtering and application of the VarRNA model predictions. The pipeline is available at https://github.com/nch-igm/VarRNA, where all methods described below are fully documented and implemented with the accompanying scripts. VarRNA was primarily developed in Python and uses Snakemake18 to enhance workflow scalability. It is designed for efficient deployment on high-performance computing systems by facilitating job scheduling.
VarRNA follows a modified version of the GATK best practices for short variant discovery from RNA-Seq data (https://gatk.broadinstitute.org/hc/en-us/articles/360035531192-RNAseq-short-variant-discovery-SNPs-Indels). RNA-Seq reads in FASTQ format were aligned to GRCH38.p13 with STAR19 v2.7.10 two-pass alignment. The total reads for all samples ranged from 70 to 400 million, and the percentage of reads aligned to the reference ranged from 70 to 96%. The percentage of reads mapped to the ribosome ranged from <1 to 10%, indicative of effective ribosomal RNA depletion. GATK v4.2.6.1 was used for the following steps: add read groups, split reads with N in the cigar string, which signifies spliced alignments where the read jumps from one exon to the other, base quality score recalibration with known sites from dbSNP20 (build 151), and variant calling with GATK HaplotypeCaller. Variants were called with “do not-use-soft-clipped-bases,” “standard-min-confidence-threshold-for-calling” set to 20, and “max-reads-per-alignment-start” set to 0 to disable down-sampling reads.
Variant filtering
Variants called in the DNA-Seq and RNA-Seq data were filtered using the same criteria. Two region-based filters were implemented: the removal of repetitive regions and the removal of non-exonic regions. Repetitive regions defined by RepeatMasker21 were downloaded from UCSC, and the chromosome, start, stop, and repeat type fields were extracted and written to a BED file. Variant calls were restricted to exonic regions using a targeted BED file of the exome capture reagent used for the ES samples (IDT xGen Exome Hyb Panel v2 targets hg38 BED file). Variants were removed if they did not meet the minimum criteria of DP > = 10, QUAL > = 100, quality by depth (QD) > = 2, and VAF > = 0.1. Additionally, we restricted variants to biallelic only. These steps were implemented using BEDtools22 and BCFtools23 (Supplementary Equation (1)).
Ground truth label filtering
To train the model for predicting whether variants in the RNA-Seq data were germline, somatic, or artifact, in addition to the filtering described above, more stringent filtering was applied to ensure that our ground truth labels coming from the DNA-Seq were correct. In creating the model, it was also important to ensure that there was adequate coverage of each base in the tumor sample DNA-Seq, the normal sample DNA-Seq, and the tumor sample RNA-Seq. For example, when there is evidence for the variant from RNA but not from DNA (i.e., insufficient sequencing coverage or an exonic region not well targeted by the ES capture kit), these regions were removed from the model training data. To do this, BAM files from the normal DNA, tumor DNA, and tumor RNA were used to generate three separate coverage files, where the number of reads at each genomic position was recorded using BEDTools. Regions with coverage equal to or greater than a minimum threshold of ten reads were used to create a custom bed file for each sequencing dataset (normal DNA, tumor DNA, tumor RNA). (Supplementary Equation (2)). A single BED file for filtering was created by intersecting the three coverage files and a BED file defining the exome regions. (Supplementary Equation (3)). This file was used to restrict variant calls across the three datasets per sample to those that fall within the regions with sufficient coverage.
RNA editing sites were removed to avoid mislabeling any RNA variant calls resulting from RNA editing as artifacts since they are not present in the DNA. This was done using the REDIportal24 database of known RNA editing sites, which were downloaded from their website (http://srv00.recas.ba.infn.it/webshare/ATLAS/donwload/TABLE1_hg38.txt.gz). These sites were extracted to a VCF file and intersected with the query VCF (Supplementary Equation (4)).
Transcript quantification
RNA-Seq transcripts were quantified with salmon52 v1.9.025 to derive transcripts per million (TPM) values for each transcript. To get the corresponding HUGO gene name and other information, the Ensembl transcript names in the ‘Name’’ column of the Salmon quantification output were translated to Ensembl gene name, HUGO gene name, and biotype (e.g., ‘protein coding,’’ ‘‘processed transcript,’’ ‘‘lncRNA’’). This translation was achieved by cross-referencing sequence names (which include all these identifiers) from the gentrome.fa.gz file. This file was generated using the generateDecoyTranscriptome.sh script found in the Salmon documentation (https://github.com/COMBINE-lab/SalmonTools/blob/master/scripts/generateDecoyTranscriptome.sh).
Variant annotation
ANNOVAR26 was used to annotate RNA-Seq variant calls with information, such as gene name and functional consequence (refGene27), population allele frequencies (gnomad30_genome28), disease-specific variants (clinvar_2022123129), functional predictions (dbnsfp42a30), and somatic variants in cancer (cosmic7031). The identifiers in parentheses correspond to existing ANNOVAR tables accessible for download using their tool.
Results from the Salmon quantification were also added to the VCFs using the following method. HUGO gene name and biotype were extracted from the ‘Name’’ column, and all other columns from the quantification output (length, effective length, TPM, and number of reads) were extracted and written to a tab-delimited annotation file. Chromosome start and stop positions were added to the annotation file for each gene. Bcftools23 annotate was used to annotate the VCF with results from Salmon.
Classifier development: dataset/labeling
The classifier development data consisted of pediatric cancer patients from NCH with RNA-Seq of the tumor and DNA-Seq of the tumor and normal DNA. The DNA-Seq variant calls were used as ground truth to label the variant calls from RNA-Seq. After the machine-learning-specific filtering, RNA-Seq variants were labeled as ‘Germline’’ if they were present in the DNA-Seq Haplotype Caller VCF, ‘Somatic’’ if they were present in the DNA-Seq Mutect2 VCF, and ‘Artifact’’ if they were not called by either, i.e., variants that were entirely unique to the RNA-Seq data.
The variants were divided on a per-sample basis to create a training set used for cross-validation to tune hyperparameters (25 subjects, 8612 variants) and a holdout benchmarking set to assess model performance (26 subjects, 8931 variants) (Supplementary Fig. 1). This is done so that variants from one sample are not used for both training and testing.
Classifier development: feature engineering
A variety of features were used, including quality scores, sequence context, variant effect predictions, clinical significance, and population frequencies. Sequence context features were calculated as follows. Nucleotide context (reference and alternate nucleotide) was used to indicate whether the variant represents a transition or a transversion event. GC content was calculated by counting the number of G and C nucleotides six base pairs upstream and downstream of the variant. Linguistic complexity, or vocabulary richness, of the six-base-pair window was calculated using an approximation previously described32. Whether the variant is an SNV or an indel was added as a feature, as separate models for these variant types were not created.
Categorical features (e.g., functional consequence, ClinVar significance) were transformed into numerical values using one-hot encoding. Features that were estimated to have low model impact or that were highly correlated with other features were removed. To define low impact, we first transformed each feature with a min–max scaler such that it was within the range from zero to one. Then, we computed the standard deviation across each transformed feature. We also performed an analysis of variance to obtain an F-statistic for each transformed feature, where the groups were the three model labels: Germline, Somatic, or Artifact. A total of 124 features were tested for association with the labels, based on 308,374 observations. Features projected to have low impact were those that had both a standard deviation less than 0.1 and an F-statistic less than 100, which corresponds to the bottom 25th percentile (mean F-statistic is 4142) with an associated p value less than 0.05. We calculated the effect size using Eta squared. The mean Eta squared value for the features with low impact was 0.0002, while the mean Eta squared value for the remaining features was 0.274. Correlation between each pair of remaining features was calculated, and if a feature pair had a greater than 0.95 correlation, one feature from the pair was removed. A final set of 64 features was selected for model training (Supplementary Data 4).
The gnomAD annotation was used to filter out variants with gnomAD raw allele frequency greater than 0.001, as these variants were contributing to extreme class imbalance. The ratio of variants in the germline: artifact: somatic labels was about 1000:50:1, and removing variants with high gnomAD allele frequency, which were mostly Germline variants, reduced the imbalance to a ratio of about 20:15:1. The breakdown of labels in the training data was 4761 Germline variants, 3601 Artifact variants, and 250 Somatic variants.
Classifier development: model training
A gradient boosting framework implemented by the XGBoost library was used for model training because of its computational efficiency, feature importance capabilities, and ability to handle missing values easily. Scikit-learn33 was used to create a pipeline to tune hyperparameters. Two separate binary models were trained: one for classifying variants as true variants or artifacts, and one for classifying true variants as germline or somatic.
A min–max scaler was used to normalize the feature values between 0 and 1 to help prevent the model from being biased toward features with larger magnitudes. A randomized search of the hyperparameter space (1000 combinations for True vs. Artifact and 2000 combinations for germline vs. somatic) was performed to reduce training time. Stratified k-fold cross-validation was used to fit the models across the hyperparameter space. The model performance was reported on the benchmarking data. The Germline vs. Somatic classifier was trained and evaluated on true variants only. Also, this model includes an additional hyperparameter to handle class imbalance; random under-sampling of the germline class, implemented with imblearn34.
Statistics and reproducibility
Statistical methods are described in the context of the aforementioned analytical methods. For reproducibility, all code is stably archived and open sourced.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Composition and utilization of cancer cohorts for machine learning and variant analysis
We analyzed data from 95 subjects with cancer, including 77 patients from our NCH in-house cancer cohort35,36,37,38, nine subjects from the ZCC39, and nine subjects from an adult cohort GBM. The data comprise patients with both pediatric (NCH and ZCC) and adult cancer (GBM) across 40 cancer subtypes (Supplementary Data 1). Each patient in the NCH and GBM datasets had RNA-Seq of the tumor and corresponding ES of the tumor and matched normal DNAs (peripheral blood mononuclear cells or buccal epithelial cells). The ZCC dataset contained RNA-Seq of the tumor and matched genome sequencing (GS) of DNA from tumors and matched normal tissues.
From these patients, a subset of 8612 variants (identified across 25 subjects) was used for training our XGBoost machine learning models, and 8931 variants (identified across 26 subjects) were used for benchmarking our models against existing methods. The remaining data (identified across 44 subjects) were held out for additional variant analyses (Supplementary Fig. 1). We processed all RNA data with the VarRNA pipeline (Fig. 1) (See “Methods”; RNA-Seq alignment and variant calling).
Evaluation of variant classification models
VarRNA’s two models, true variant vs. artifact and germline vs. somatic variant, were evaluated using the benchmarking data across all three datasets (8931 variants). The true variant vs. artifact model showed high accuracy, demonstrating high precision (92.3%) and recall (95.1%) for true variants (combined germline and somatic), and artifact precision and recall of 93.6 and 90.0%, respectively (Table 1). This high recall rate for true variants is crucial in a clinical setting, indicating that VarRNA has a low probability of incorrectly classifying true variants as artifacts. This capability allows VarRNA to retain important variant information while significantly reducing the burden of artifactual variants.
The germline vs. somatic variant model was evaluated on the subset of the hold-out test set consisting of true variants (5276 variants). This subset comprised 4908 germline variants and 368 somatic variants. The model demonstrated high precision (97.3%) and recall (89.4%) for germline variants. For somatic variants, the precision and recall were 32.0 and 66.6%, respectively.
Model results were accompanied by prediction probability scores calculated using XGBoost. The germline vs. somatic labels were ranked by model probabilities so that informed decisions on the likelihood of label accuracy could be made (Supplementary Fig. 2). When we restricted the model probability to greater than 60%, the somatic precision increased from 32.0 to 42.2%, and recall increased from 66.6 to 73.6% (Supplementary Table 1). As we further restricted the model’s probability, these metrics continued to improve; however, this performance gain comes at the expense of losing additional germline variant calls. Beyond a 75% probability threshold, there is a noticeable drop in performance. (Supplementary Fig. 3).
We explored model performance as a function of tumor purity, defined as the proportion of cancer cells in the tumor tissue utilized for nucleic acid extraction. Tumor purity was calculated using ESTIMATE40, which uses gene expression signatures to infer the fractional proportion of stromal and immune cells. We evaluated the effects of tumor purity on the subset of benchmarking data, consisting of eight NCH samples, that were analyzed with ESTIMATE. We found that macro-averaged precision, recall, and F1 scores for the true variant vs. artifact model remained consistently high across the tumor purity levels observed in our cohort (Supplementary Fig. 4a). The germline vs. somatic model is more sensitive to tumor purity, and model performance tends to increase as tumor purity increases (Supplementary Fig. 4b).
We evaluated variant calls and model performance across different numbers of aligned reads, ranging from 60 to 150 million. As expected, fewer mapped reads resulted in fewer variant calls (Supplementary Table 2). For the True vs. Artifact model, performance slightly declined with fewer mapped reads, though it remained high across all read levels (Supplementary Fig. 5a, b). The germline vs. somatic model showed a more noticeable drop in performance with fewer aligned reads (Supplementary Fig. 5c, d), but no consistent trend was observed linking lower read counts to worse results.
We compared the performance of our models across the three benchmarking datasets, consisting of pediatric and adult brain cancer subjects. For the True vs. Artifact model, performance was consistent between the NCH and GBM datasets, with area under the receiver-operator curve (AUROC) scores of 0.990 and 0.987, and area under the precision-recall curve (AUPRC) scores of 0.993 for both (Supplementary Fig. 6a, b). In contrast, the ZCC dataset showed lower performance, with an AUROC of 0.943 and an AUPRC of 0.886. For the germline vs. somatic model, the NCH dataset exhibited the highest performance, followed by the GBM and ZCC datasets (AUROC–NCH 0.924, GBM 0.873, ZCC 0.864; AUPRC–NCH 0.633, GBM 0.494, ZCC 0.280; Supplementary Fig. 6c, d).
Benchmarking VarRNA
While VarRNA offers comprehensive variant classification capabilities for tumor samples by identifying artifacts and somatic variants, we benchmarked its performance against two specialized tools, each addressing specific aspects of VarRNA’s functionality. GATK Variant Filtration, typically used for detecting germline variants from RNA-Seq data, was employed to assess VarRNA’s artifact identification capability. Conversely, RNAIndel41, designed to call indels from tumor RNA-Seq data, provided a comparative framework for VarRNA’s ability to categorize these variants as artifacts, germline, or somatic. This benchmarking approach highlights VarRNA’s multifaceted utility in a field where specialized tools often focus on narrower tasks.
Two separate comparisons are presented because GATK variant filtration only distinguishes variants as true or artifact, and although RNAIndel classifies all three labels, it provides only results for indels. The VarRNA True Variant vs. Artifact model was validated against GATK variant filtration for both SNVs and indels. The combined VarRNA models, which produce artifact, germline, and somatic labels, were validated against RNAIndel for indels only. A detailed description of the steps to run each model and gather performance metrics is provided (Supplementary Notes). The three benchmarking datasets were used for this comparison (Supplementary Fig. 1).
The VarRNA True vs. Artifact model substantially outperforms GATK Variant Filtration with an AUROC of 0.967 compared to 0.764, and an AUPRC of 0.961 compared to 0.786 (Fig. 2a, b). We compared performance metrics (recall, precision, F1-score, and false positive rate per 1 Mb) across multiple VAF bins (Supplementary Fig. 7a, b). In general, both tools tended to have a better performance as VAF increases. GATK variant filtration had a higher recall across multiple VAF bins, while VarRNA had a higher precision, F1-score, and lower false positive rate. We see higher variability for indels because there are far fewer observations, particularly at lower VAFs.
To compare VarRNA with RNAIndel, the output of both VarRNA models was combined by multiplying the Germline vs. Somatic model probabilities by the True Variant probability so that the artifact, germline, and somatic probabilities sum to 1. The observations were subset so that only indels were included to evaluate performance, reducing the number of variants from 8931 to 2317. Additionally, we only compared the subset of variants that were called by both VarRNA and RNAIndel, reducing this number to 1507. Despite this RNAIndel limitation, we could determine that VarRNA and RNAIndel performed comparably across indel variant calls, with similar areas under the ROC and PR curves (Fig. 2c, d). VarRNA shows a slightly increased precision and F1-score and a slightly decreased false positive rate across most VAFs (Supplementary Fig. 7c). We did not observe a trend of increased recall, precision, or F1-score as VAF increased for indels.
Feature interpretation
We employed Shapley additive explanations (SHAP)42 values to rank the importance of features used in training the VarRNA model. SHAP is a powerful method that demystifies the black-box nature of machine learning models, enabling us to identify which features were most informative for model classification. SHAP also permits an understanding of the rationale behind our model’s predictions to rank the importance of features used to train the VarRNA model. We calculated the mean of the absolute value of each feature across all training observations to summarize the ranked order of how informative each feature was (True Variant vs. Artifact: Supplementary Data 2; germline vs. somatic: Supplementary Data 3). We additionally described their orientation, defined by the feature correlation with the model output.
We summarized the top 20 most informative features for the True Variant vs. Artifact model, ordered by the mean of the absolute SHAP value of each feature (Fig. 3a). High VAF and high gnomAD allele frequency generally increased the True Variant prediction. We observed that a large median fragment length of reads supporting the alternative allele (MFRL_ALT), consistent with findings in DNA from a previous study that showed longer fragment length tended to yield high error rates43, and high strand odds ratio, which is a quality metric used to identify potentially false-positive variants, both tended to increase the Artifact prediction. For the Germline vs. Somatic model, a high gnomAD allele frequency (INFO/AF_raw) increased the Germline prediction (Fig. 3b). Another important set of features is ClinPred Pred: D (deleterious) and ClinPred Pred: T (tolerated), which are based on the output of the ClinPred model, aiming to predict the impact of nonsynonymous variants using an ensemble of existing pathogenicity classifiers. These features indicate whether the variant is deleterious (e.g., has a damaging effect on the protein), or tolerated (e.g., likely has a minimal impact on protein function). A deleterious outcome increases the Somatic prediction, and a tolerated outcome increases the Germline prediction. We see a similar trend in the 30-way mammal PhyloP score, where higher values increase the somatic prediction. PhyloP scores quantify the conservation rate at a particular region, and positive scores indicate higher conservation rates. Both features suggest that variants occurring at more conserved loci are more likely to be somatic variants.
Top 20 most informative features for the True vs. Artifact (a) and Germline vs. Somatic (b) models as calculated by the sum of the absolute SHAP values using the training data. The trend of feature value influence on the model predictions is indicated in red and blue, with the color based on correlation between each feature and the corresponding SHAP values. In a, a positive SHAP value is associated with True Variant prediction and in b, a positive SHAP value is associated with Somatic prediction. (SHAP: SHapley Additive exPlanations).
We also explored why the germline vs. somatic model was miscalling some somatic variants as germline by evaluating the most impactful features (calculated by SHAP) for germline prediction of somatic variants. The most frequently impactful features are QD and gnomAD allele frequency. When the QD is lower, the model tends to predict a variant as somatic, reflecting the tendency in our training data for somatic variants to exhibit lower quality than germline variants. Additionally, the presence of a non-zero allele frequency in gnomAD drives the model toward a germline classification, which aligns with the expectation that germline variants are more commonly found in healthy population databases.
Comparison of variant calls from RNA-Seq or DNA-Seq
The 44 NCH samples were used to compare variant calls between RNA-Seq and DNA-Seq (ES) (Supplementary Fig. 1). We included all variants called, rather than filtering by gnomAD AF < .001 as done for model training and benchmarking (see “Methods”), to compare the total breadth of variants. About half (47.2%) of the variants called in the DNA-Seq data are also called by VarRNA analysis of the RNA-seq data (Fig. 4a). When accounting for expression level, only 38,227 of the 540,243 (7.1%) germline variants called from the DNA-Seq data had at least ten-fold coverage in the RNA-Seq data, and only 6187 of these variants (1.1%) had at least ten reads covering the alternative allele. Hence, 98.9% of the germline variants called only from the DNA-Seq data have an insufficient RNA expression level to permit calling of the variant allele from RNA-Seq data (i.e., if the gene containing the variant observed in the DNA is not expressed, then it will not be possible to call that variant from RNA-Seq data). Similarly, for somatic variants, 96.9% of variants called only from Mutect2 analysis of the tumor DNA-Seq data lacked sufficient RNA-seq data coverage of the variant allele to permit variant detection. These results highlight that many DNA variants are not expressed, and as such, variant calling using RNA-Seq can focus downstream interpretation on genes that are being expressed in the tumor at the time of biopsy or surgical resection.
a Venn diagram of the overlap of variants identified from ES Germline (green), ES Somatic (blue), and RNA-Seq (red) data. Each number in (or pointing to) the Venn diagram corresponds to the number of variants in that section. The union of all the variants is listed as Total variants (1,044,472). The number in parentheses next to each label is the number of variants in each group. b Distribution of the variant locations called in the RNA-Seq but missed in ES. c Distribution of the variant calls missed in ES, based on ES coverage and variant evidence. (ES: Exome Sequencing).
When we evaluated RNA-Seq variants called by VarRNA, most (97.5%) were also present in DNA-Seq data. Of the subset of variants only called in RNA-Seq (12,486; 2.5% of RNA variants), most (99.1%) were in exonic regions; 0.9% were in intergenic, 5’ or 3’ untranslated regions, or at splice sites (Fig. 4b). We reviewed the ES BAM coverage in a subset of 36 of these patients (10,436 variants in exonic regions) to determine what evidence existed for the variant in the DNA data. Of these 10,436 variants, 4869 (46.7%) had insufficient coverage to identify variants, as defined by a depth of fewer than 10 reads over the combined reference and variant alleles. 1295 variants (12.4%) were identified in the DNA BAM with a coverage depth over the variant allele greater than ten reads and a VAF greater than 0.1, yet were filtered out of the results, likely due to low quality. 296 variants (2.8%) showed evidence of a variant in the DNA with a VAF greater than 0.1 but with a coverage depth of fewer than ten reads over the variant allele. Finally, 3976 variants (38.1%) had sufficient coverage in the DNA, but with no evidence of a variant (Fig. 4c). Therefore, we estimate that 62% of these variants uniquely identified by VarRNA in the RNA-Seq data are likely real variants that were missed by DNA-Seq. They either had no DNA-Seq coverage (i.e., regions not covered by the ES hybrid capture probe reagent) or there was evidence of the variant in the DNA but of insufficient depth to call it. The remaining 38% of variants unique to the RNA-Seq data may result from RNA editing or post-transcriptional modifications. These also may be due to RNA strand scission or to sequencing artifacts, but the high performance of the true vs. artifact model performance would argue against the latter possibility.
Allele-specific expression in cancer-driving genes
We combined our VarRNA results and annotations from OncoKB, a curated knowledgebase providing evidence-based information on somatic mutations in patient tumors44, to further characterize variants in genes found to be either oncogenes or tumor suppressors. For variants in cancer-driving genes described by OncoKB and found in both RNA and DNA data, 1125 out of 16,626 variants show ASE of the variant allele, quantified as a VAF in the RNA greater than 0.65 and a VAF in the DNA less than 0.55. Of these variants, 361 were observed to exhibit extreme absence of transcription from the wildtype allele (0–10% VAF) and corresponding allele-specific expression of the variant allele (90–100% VAF) Table 2a, b outlines four such examples of allele-specific expression of pathogenic variants in known cancer genes with clinical significance established by the AMP/ASCO/CAP tiering system45 and oncogenicity classification by ClinGen/CGC/VICC27 that were identified by VarRNA.
Complex variants are classified by VarRNA
We utilized VarRNA to evaluate RNA-Seq data that was produced by an outside commercial laboratory using tumor tissue obtained from an individual with a high-grade glioma. Due to insufficient tumor cell content and limited tissue obtained at the time of biopsy, only RNA-Seq was performed. VarRNA classified two variants of clinical relevance, both of which were supported by results from an outside academic institution that performed a cell-free DNA assay on cerebrospinal fluid from the same patient. A missense somatic variant (NM_023110.3(FGFR1): c.1638 C > A: p.N546K) was classified in a recurrent hotspot in the kinase domain of the FGFR1 gene. Additionally, a somatic in-frame six base pair deletion (NM_181523.3(PIK3R1):c.1362_1367del:p.F456_Q457del) was classified in the PIK3R1 gene. Loss of function somatic variants such as that in PIK3R1 have been described in pediatric high-grade gliomas46,47. This in-frame deletion occurs in a hotspot within the iSH2 domain that is expected to result in loss of function48,49, and likely results in aberrant signaling through the PI3K pathway to drive oncogenesis. These findings emphasize the ability of VarRNA to classify complex variants, including small deletions, and highlight how RNA-Seq may provide a valuable resource for variant identification and classification in cases where DNA-Seq data are unavailable.
Discussion
We have developed VarRNA, a computational method to classify SNVs and indels identified from tumor RNA-Seq data, which encompasses 1) a pipeline to align RNA-Seq reads and call variants and 2) a machine learning model to predict variants as germline, somatic, or artifact. VarRNA retains important variant information while reducing the burden of artifact variants, exhibiting both high precision and recall identification of true genetic variants from tumor RNA-Seq data. VarRNA can also predict whether the variant identified is a germline or somatic variant. Our results demonstrate that VarRNA can detect somatic variants that would have been missed by performing DNA-Seq alone. Moreover, some variants detected by VarRNA exhibited markedly different VAFs compared to those called in the corresponding DNA exome data. This discrepancy was particularly striking in cancer-relevant genes, where VarRNA analysis revealed that the expression of the variant allele was significantly higher relative to the wild-type allele, with potential clinical relevance.
Variants in MSH6 can result in defective mismatch repair, wherein this activity is reduced, but not absent, when there is one normal copy of the MSH6 gene. As such, one deleterious variant in each MSH6 allele is required to cause a complete loss of function, resulting in a high tumor mutational burden50. In patient IGMCH0061, VarRNA classified a nonsynonymous somatic variant in the MutS_V functional domain of MSH6. The MSH6 protein plays a role in detecting mismatches that arise during DNA replication. Mutations in this domain compromise the mismatch repair system’s ability to accurately identify and correct these errors. The VAF of the MSH6 variant detected in the DNA-Seq is about 50%, indicating the second allele is wildtype. However, the VAF of this variant in the RNA is close to 100%, suggesting that the wild-type protein is not being produced. Indeed, the tumor mutational burden, or the total number of mutations per megabase (Mb) of DNA in the tumor cells is about 45 variants per Mb, indicative of an ultra-hypermutated genome. Thus, the overexpressed variant MSH6 allele indicates predominant expression of an altered and dysfunctional protein product, resulting in significant disruption of DNA repair, which leads to a significant mutational burden and contributes to cancer onset. If DNA-Seq data alone were evaluated, the underlying reason for the high TMB, a highly relevant clinical observation, may have been missed.
The STAG2 gene plays a significant role in several types of cancer, including Ewing sarcoma, due to its encoded protein’s function in the cohesin complex, which regulates chromosomal segregation during cell division51,52. In patient IGMCH0074, a germline frameshift insertion in STAG2 was called with about 50% VAF in the DNA but with nearly exclusive homozygous expression of the variant allele in the RNA. This mutation leads to a premature stop codon seven amino acids downstream of the mutation, which likely confers a loss of function for the only expressed allele of STAG2 in this tumor. Recent studies have provided evidence that a loss of STAG2 function can lead to more aggressive behavior in Ewing sarcoma, which requires corresponding therapeutic adjustment due to poor prognosis53,54. Absent evidence of monoallelic expression of this STAG2 variant, the opportunity to appreciate the prognostic value of the mutation only from DNA-Seq may have been missed.
The clinical implications of our findings regarding the MSH6 and STAG2 mutations reinforce the value of studying RNA from tumor materials to clinically inform cancer therapy. In both cases, understanding these ASE mechanisms could lead to therapeutic approaches that are tailored to the patient’s disease characteristics, possibly improving treatment outcomes. These findings underscore the utility of RNA-Seq not only in detecting variants that may be underrepresented or absent in DNA but also in providing a more nuanced view of disease-specific expression dynamics and potential impact on cancer pathogenesis, thereby influencing both prognosis and therapeutic strategies.
Our model to distinguish true variants from artifacts has a high true variant recall, correctly labeling 95.1% of true variants as true while keeping the averaged F1 score high (92.7%). The F1 score harmonizes recall and precision, ensuring that high performance is not achieved in one metric at the expense of the other, making the F1 score a useful measurement for overall model performance. Indeed, VarRNA retains important variant information while reducing the burden of artifact variants. Our Germline vs. Somatic model also performs well, correctly labeling 89.4% of expressed germline variants and 66.6% of expressed somatic variants. By utilizing the model’s probability scores, somatic recall increased from 66.6 to 73.6%. Due to this increase in performance metrics with minimal loss in correct predictions, we recommend filtering germline vs. somatic results by model probability.
VarRNA outperformed two existing methods to reduce artifact calls in RNA-Seq data or classify variants as germline or somatic in tumor samples. Our True vs. Artifact model was validated against GATK variant filtration for SNVs and indels. Overall, VarRNA outperforms GATK variant filtration with a higher precision and F1-score, and a lower False Positive rate. The GATK variant filtration criteria generally accept more variants that are artifacts, which raises its recall but lowers its precision. Our combined germline, somatic, or artifact results were compared to those from RNAIndel, which provides the same labeling scheme but is limited to only indels called from tumor RNA-Seq. We observed comparable performance to RNAIndel. Overall, VarRNA outperforms both methods and provides more comprehensive results on tumor data than either method on its own.
Our models are built with XGBoost, and the contribution of each feature to the prediction is quantified using SHAP Tree Explainer. This feature of VarRNA is unique amongst the RNA variant classification methods and provides significant value for clinical interpretation of our model predictions. For example, the population frequency of a given variant proved to be highly informative in distinguishing between germline and somatic variants. We found that somatic variants are less commonly found in population databases like gnomAD because they occur in specific tissues or tumors and are not inherited, whereas these databases primarily catalog germline variants present in all cells.
We also observed that the most influential features in predicting somatic variants tend to occur more frequently in conserved genomic loci than do germline variants. Conserved regions have functional importance, including crucial roles in cellular function and development, and therefore, variants in these regions can have significant effects on cell biology that, in cancers, lead to dysregulated cellular proliferation. By contrast, germline variants in these same regions are often subject to evolutionary pressures and may be less likely to be inherited due to their detrimental effects. However, germline variants in cancer predisposition genes contribute to cancer susceptibility, particularly in pediatric disease, and are as such, important to identify55,56.
VarRNA was trained with pediatric cancer samples and applied to both pediatric and adult cancer samples. We observed no substantial decrease in True Variant vs. Artifact model performance between pediatric and adult cancer data. For the germline vs. somatic model, performance was highest in the NCH dataset, followed by the GBM and ZCC datasets. This slight decline in performance is expected, as the NCH dataset likely resembles the training data more closely. Additionally, the ZCC dataset generally contains fewer mapped reads per sample, which may impact the Germline vs. Somatic model’s performance (Supplementary Fig. 5b, d). The ZCC labels were derived from GS, which could introduce differences compared to the ES-derived labels used in the NCH and GBM datasets. Additionally, most of the cancer samples used to train VarRNA were from central nervous system tumors. It is possible that the model may need to be optimized for specific tumor types. This limitation could be overcome by training a model optimized for the user’s specifications. VarRNA is available as open source, with feature annotation and processing provided with detailed code, allowing groups to train their own models efficiently. Given that the germline vs. somatic model performance is reduced at lower tumor purity levels, these results could be mitigated by training a model with samples of the desired tumor purity level to gain higher performance at detecting somatic vs. germline variants.
There exists a large overlap between variants called in the RNA-Seq and DNA-Seq data, with about half of the variants called in DNA-Seq also being found in RNA-Seq. However, most variants observed in DNA-Seq that were not observed in RNA-Seq occurred in genes that lacked expression in the tissue of study and, as such, had insufficient read coverage to identify the variant from RNA-seq data. This highlights one disadvantage of variant calling from RNA-Seq, in that expression levels dictate whether a variant will be covered by RNA-Seq reads and identified with variant calling tools. Additionally, RNA is generally less stable than DNA due to chemical and structural characteristics57. This results in some degradation during tissue preservation and RNA extraction, which may lead to missed variant calls58. Technical variability in library preparation can also introduce biases; for example, some reverse transcriptases might fail to effectively reverse transcribe regions having secondary structure59.
Conversely, some variants were detected exclusively in the RNA-Seq data. Our evaluation of the RNA- and DNA-Seq data for these variants revealed that most (about 62%) were likely missed in the DNA-Seq due to regions that were not targeted by the ES reagent or had lower sequence coverage from suboptimally performing capture probes. However, it is possible that the somatic variants unique to the RNA-Seq data reflected changes in the underlying biology of the tumor. For example, dysregulation of genes in the tumor sample could result in higher gene expression, leading to increased sequence coverage. This higher coverage enhances the accuracy of variant calling in highly expressed genes. Moreover, unlike DNA-Seq, RNA-Seq has the capability to detect ASE; some variants are identified in the RNA due to preferential expression, leading to higher coverage, whereas the variant may have too low a frequency to be identified in the corresponding DNA. RNA-Seq can also identify post-transcriptional modifications, such as RNA editing, which are not present in DNA. It is also possible that a subset of these variants, only called in the RNA, resulted from technical differences in library preparation and sequencing protocols. While it cannot be ruled out that variants found in the RNA with no evidence of the variant in the corresponding DNA may represent sequencing artifacts, our true variant vs. artifact model performed exceptionally well, making this possibility less likely.
The results in Table 2 highlight several clinically relevant variants with high VAFs in RNA. This naturally raises the question of whether the opposite logic might apply: can clinically relevant variants be identified by their disproportionately low VAFs in RNA data? Initially, this may seem counterintuitive, as variants with decreased presence in a cancer sample are typically less likely to be oncogenic drivers. However, an intriguing possibility arises: focusing on mutations with low RNA VAFs could implicate variants under negative oncogenic selection. Although the overall level of negative selection in cancers is low, with 99% of coding variants being tolerated, there are notable exceptions. For example, essential genes like POLR2A, which can exhibit ploidy 1 in tumors, demonstrate this concept60,61. Identifying such mutations with low VAFs could provide valuable insights and be leveraged in clinical applications.
Variant calling and classification with RNA-Seq, as we have demonstrated through VarRNA, has the potential to revolutionize our understanding of pediatric cancer biology, providing a crucial layer of genetic information not captured from DNA alone. The additional information from tumor-derived RNA greatly enhances the breadth and depth of cancer genomics data interpretation. As a result, we can expand the utility of RNA-Seq data beyond what has already been demonstrated, such as detecting fusion genes and splicing variants, gene expression profiling, cancer classification, and pathway analysis. By incorporating variant calling into this already powerful toolset, VarRNA further broadens the potential applications of RNA-Seq, increasing its impact in both cancer research and clinical settings. This method represents a groundbreaking approach that enhances our understanding of cancer biology by seamlessly integrating expression data with variant identification. It opens new avenues for further research in cancer biology and clinical care, promising significant advancement in the field.
Data availability
NCH ES and RNA-Seq data used in this study are publicly available for general research use and have been deposited in dbGaP, under study accession phs001820.v3.p1. The GBM cohort data are deposited at NCBI’s Sequence Read Archive and are accessible through BioProject ID PRJNA999679 (ES data) and are deposited at NCBI’s Gene Expression Omnibus and are accessible through Gene Expression Omnibus Series accession number GSE238012 (RNAseq data). Additional data used for model validation from the ZCC study were accessed from the European Genome Archive, study accession EGAC00001001683. These data are available for research upon request. Source data for main figures are provided with this manuscript as Supplementary Data 5. All other data are available from the corresponding author on reasonable request.
Code availability
The models developed and the code used to process RNA-Seq data presented in this manuscript are open-source and available for download under the BSD 3-Clause license at https://github.com/nch-igm/VarRNA. The data in this manuscript were processed using VarRNA v1.0.0, corresponding to the 1.0.0 release. The code is also deposited at Zenodo, a DOI-minting repository: https://doi.org/10.5281/zenodo.14699945.
References
Oshlack, A., Robinson, M. D. & Young, M. D. From RNA-seq reads to differential expression results. Genome Biol. 11, 220 (2010).
LaHaye, S. et al. Discovery of clinically relevant fusions in pediatric cancer. BMC Genom. 22, 872 (2021).
Hong, M. et al. RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 13, 1–16 (2020).
Castel, S. E., Levy-Moonshine, A., Mohammadi, P., Banks, E. & Lappalainen, T. Tools and best practices for data processing in allelic expression analysis. Genome Biol. 16, 195 (2015).
Okonechnikov, K. et al. Comparison of transcriptome profiles between medulloblastoma primary and recurrent tumors uncovers novel variance effects in relapses. Acta Neuropathol. Commun. 11, 7 (2023).
Khaled, N. et al. RNA sequencing reveals the differential expression profiles of RNA in metastatic triple negative breast cancer and identifies SHISA3 as an efficient tumor suppressor gene. Am. J. Cancer Res. 11, 4568–4581 (2021).
Sen, A., Huo, Y., Elster, J., Zage, P. E. & McVicker, G. Allele-specific expression reveals genes with recurrent cis-regulatory alterations in high-risk neuroblastoma. Genome Biol. 23, 71 (2022).
Kukurba, K. R. et al. Allelic expression of deleterious protein-coding variants across human tissues. PLOS Genet. 10, e1004304 (2014).
Gabay, O. et al. Landscape of adenosine-to-inosine RNA recoding across human tissues. Nat. Commun. 13, 1184 (2022).
Lo Giudice, C., Tangaro, M. A., Pesole, G. & Picardi, E. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat. Protoc. 15, 1098–1131 (2020).
Poplin, R. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. http://biorxiv.org/lookup/doi/10.1101/201178 (2018).
Piskol, R., Ramaswami, G. & Li, J. B. Reliable identification of genomic variants from RNA-seq data. Am. J. Hum. Genet. 93, 641–651 (2013).
Wang, C. et al. RVboost: RNA-seq variants prioritization using a boosting method. Bioinformatics 30, 3414–3416 (2014).
Cook, D. E. et al. A deep-learning-based RNA-seq germline variant caller. Bioinforma. Adv. 3, vbad062 (2023).
Bollas, A. & Lammi, G. nch-igm/VarRNA: Initial release - v1.0.0. Zenodo https://doi.org/10.5281/zenodo.14699945 (2025).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In: Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794. Available from: http://arxiv.org/abs/1603.02754.
Kelly, B. J. et al. Churchill: an ultra-fast, deterministic, highly scalable and balanced parallelization strategy for the discovery of human genetic variation in clinical and population-scale genomics. Genome Biol. 16, 6 (2015).
Mölder, F. et al. Sustainable data analysis with Snakemake. F1000Res. 10, 33 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Sherry, S. T., Ward, M. & Sirotkin, K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 9, 677–679 (1999).
Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0 [Internet]. 2013. Available from: http://www.repeatmasker.org.
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Picardi, E., D’Erchia, A. M., Lo Giudice, C. & Pesole, G. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 45, D750–D757 (2017).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon: fast and bias-aware quantification of transcript expression using dual-phase inference. Nat. Methods 14, 417–419 (2017).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
O’Leary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Landrum, M. J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Liu, X., Li, C., Mou, C., Dong, Y. & Tu, Y. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 12, 103 (2020).
Sondka, Z. et al. COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res. 52, D1210–D1217 (2024).
Gabrielian, A. & Bolshoy, A. Sequence complexity and DNA curvature. Comp. Chem. 23, 263–274 (1999).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Lemaître, G., Nogueira, F. & Aridas, C. K. Imbalanced-learn: a Python toolbox to tackle the curse of imbalanced datasets in machine learning. J. Mach. Learn. Res. 18, 1–5 (2017).
Liu, H. et al. De novo primary central nervous system pure erythroid leukemia/sarcoma with t(1;16)(p31;q24) NFIA/CBFA2T3 translocation. Haematologica 105, e194–e197 (2020).
Miller, K. E. et al. Genome sequencing identifies somatic BRAF duplication c.1794_1796dupTAC;p.Thr599dup in pediatric patient with low-grade ganglioglioma. Cold Spring Harb. Mol. Case Stud. 4, a002618 (2018).
Richardson, T. E. et al. GOPC-ROS1 fusion due to microdeletion at 6q22 Is an oncogenic driver in a subset of pediatric gliomas and glioneuronal tumors. J. Neuropathol. Exp. Neurol. 78, 1089–1099 (2019).
Darby, C. A. et al. Samovar: single-sample mosaic single-nucleotide variant calling with linked reads. iScience 18, 1–10 (2019).
Wong, M. et al. Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat. Med. 26, 1742–1753 (2020).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Hagiwara, K. et al. RNAIndel: discovering somatic coding indels from tumor RNA-Seq data. Bioinformatics 36, 1382–1390 (2020).
Lundberg, S. & Lee, S. I. A unified approach to interpreting model predictions. Available from: http://arxiv.org/abs/1705.07874 (2017).
Tan, G., Opitz, L., Schlapbach, R. & Rehrauer, H. Long fragments achieve lower base quality in Illumina paired-end sequencing. Sci. Rep. 9, 2856 (2019).
Chakravarty, D. et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017, 00011 (2017).
Li, M. M. et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer. J. Mol. Diagn. 19, 4–23 (2017).
Horak, P. et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): joint recommendations of clinical genome resource (CLINGEN), cancer genomics consortium (CGC), and variant interpretation for cancer consortium (VICC). Genet. Med. 24, 986–998 (2022).
Mackay, A. et al. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 33, 829–842.e5 (2018).
Wu, G. et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat. Genet. 46, 444–450 (2014).
Thorpe, L. M., Yuzugullu, H. & Zhao, J. J. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer 15, 7–24 (2015).
Urick, M. E. et al. PIK3R1 (p85α) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 71, 4061–4067 (2011).
MSH6 gene: MedlinePlus Genetics [Internet]. [cited 2024 Apr 15]. Available from: https://medlineplus.gov/genetics/gene/msh6/.
Tirode, F. et al. Genomic landscape of ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 4, 1342–1353 (2014).
Adane, B. et al. STAG2 loss rewires oncogenic and developmental programs to promote metastasis in ewing sarcoma. Cancer Cell. 39, 827–844 (2021).
Surdez, D. et al. STAG2 mutations alter CTCF-anchored loop extrusion, reduce cis-regulatory interactions and EWSR1-FLI1 activity in ewing sarcoma. Cancer Cell. 39, 810–826 (2021).
von Stedingk, K. et al. Prevalence of germline pathogenic variants in 22 cancer susceptibility genes in Swedish pediatric cancer patients. Sci. Rep. 11, 5307 (2021).
Zhang, J. et al. Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med. 373, 2336–2346 (2015).
Minchin, S. & Lodge, J. Understanding biochemistry: structure and function of nucleic acids. Essays Biochem. 63, 433–456 (2019).
Schuierer, S. et al. A comprehensive assessment of RNA-seq protocols for degraded and low-quantity samples. BMC Genom. 18, 442 (2017).
Verwilt, J., Mestdagh, P. & Vandesompele, J. Artifacts and biases of the reverse transcription reaction in RNA sequencing. RNA 29, 889–897 (2023).
Van den Eynden, J., Basu, S. & Larsson, E. Somatic mutation patterns in hemizygous genomic regions unveil purifying selection during tumor evolution. PLoS Genet. 12, e1006506 (2016).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041 (2017).
Author information
Authors and Affiliations
Contributions
A.B. processed data, trained models, wrote the VarRNA package, and wrote the first draft of the manuscript. J.G. advised on statistical/machine-learning methods. K.S. provided variant interpretation. A.B. and P.W. prepared figures and tables. A.B., J.G., K.S., P.W., and E.M. prepared and/or revised the manuscript. P.W. and E.M. conceptualized and supervised the project.
Corresponding author
Ethics declarations
Competing interests
No competing interests: A.B., J.G., K.S., and P.W. E.M. declares the following competing interests: Qiagen N.V., supervisory board member, honorarium, and stock-based compensation. Singular Genomics Systems, Inc., board of directors, honorarium, and stock-based compensation.
Peer review
Peer review information
Communications Medicine thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bollas, A., Gaither, J., Schieffer, K.M. et al. Variant calling from RNA-Seq data reveals allele-specific differential expression of pathogenic cancer variants. Commun Med 5, 202 (2025). https://doi.org/10.1038/s43856-025-00901-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-00901-y