Abstract
Diabetic heart disease is highly prevalent and is associated with the early development of impaired diastolic relaxation. The mechanisms of diabetic heart disease are poorly understood, and it is a condition for which there are no targeted therapies. Recently, disrupted glycogen autophagy (glycophagy) and glycogen accumulation have been identified in the diabetic heart. Glycophagy involves glycogen receptor binding and linking with an ATG8 protein to locate and degrade glycogen within an intracellular phagolysosome. Here we show that glycogen receptor protein starch binding domain protein 1 (STBD1) is mobilized early in the cardiac glycogen response to metabolic challenge in vivo, and that deficiency of a specific ATG8 family protein, γ-aminobutyric acid type A receptor-associated protein-like 1 (GABARAPL1), is associated with diastolic dysfunction in diabetes. Gabarapl1 gene delivery treatment remediated cardiomyocyte and cardiac diastolic dysfunction in type 2 diabetic mice and the diastolic performance of ‘diabetic’ human induced pluripotent stem cell-derived cardiac organoids. We identify glycophagy dysregulation as a mechanism and potential treatment target for diabetic heart disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Source data are provided with this paper.
References
Hoek, A. G. et al. Epidemiology of heart failure in diabetes: a disease in disguise. Diabetologia 67, 574–601 (2024).
Varma, U., Koutsifeli, P., Benson, V. L., Mellor, K. M. & Delbridge, L. M. D. Molecular mechanisms of cardiac pathology in diabetes—experimental insights. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 1949–1959 (2018).
Heather, L. C., Gopal, K., Srnic, N. & Ussher, J. R. Redefining diabetic cardiomyopathy: perturbations in substrate metabolism at the heart of its pathology. Diabetes 73, 659–670 (2024).
Mellor, K. M., Bell, J. R., Young, M. J., Ritchie, R. H. & Delbridge, L. M. D. Myocardial autophagy activation and suppressed survival signaling is associated with insulin resistance in fructose-fed mice. J. Mol. Cell Cardiol. 50, 1035–1043 (2011).
Mellor, K. M., Reichelt, M. E. & Delbridge, L. M. D. Autophagy anomalies in the diabetic myocardium. Autophagy 7, 1263–1267 (2011).
Delbridge, L. M. D., Mellor, K. M., Taylor, D. J. & Gottlieb, R. A. Myocardial stress and autophagy: mechanisms and potential therapies. Nat. Rev. Cardiol. 14, 412–425 (2017).
Koutsifeli, P. et al. Glycogen-autophagy: molecular machinery and cellular mechanisms of glycophagy. J. Biol. Chem. 298, 102093 (2022).
Jiang, S., Wells, C. D. & Roach, P. J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 413, 420–425 (2011).
Mellor, K. M., Varma, U., Stapleton, D. I. & Delbridge, L. M. D. Cardiomyocyte glycophagy is regulated by insulin and exposure to high extracellular glucose. Am. J. Physiol. Heart Circl. Physiol. 306, H1240–H1245 (2014).
Faulkes, C. G. et al. Naked mole-rats have distinctive cardiometabolic and genetic adaptations to their underground low-oxygen lifestyles. Nat. Commun. 15, 2204 (2024).
Pierce, G. N. & Philipson, K. D. Binding of glycolytic enzymes to cardiac sarcolemmal and sarcoplasmic reticular membranes. J. Biol. Chem. 260, 6862–6870 (1985).
Xu, K. Y., Zweier, J. L. & Becker, L. C. Functional coupling between glycolysis and sarcoplasmic reticulum Ca2+ transport. Circ. Res. 77, 88–97 (1995).
Mellor, K. M. et al. Myocardial glycophagy flux dysregulation and glycogen accumulation characterize diabetic cardiomyopathy. J. Mol. Cell. Cardiol. 189, 83–89 (2024).
Koutsifeli, P. et al. Methods for detection of cardiac glycogen-autophagy. Autophagy Rep. 3, 2405331 (2024).
Ke, J., Pan, J., Lin, H. & Gu, J. Diabetic cardiomyopathy: a brief summary on lipid toxicity. ESC Heart Fail. 10, 776–790 (2023).
Jiang, S. et al. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J. Biol. Chem. 285, 34960–34971 (2010).
Chandramouli, C. et al. Myocardial glycogen dynamics: new perspectives on disease mechanisms. Clin. Exp. Pharmacol. Physiol. 42, 415–425 (2015).
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Zhang, Y. et al. Decoding the molecular mechanism of selective autophagy of glycogen mediated by autophagy receptor STBD1. Proc. Natl Acad. Sci. USA 121, e2402817121 (2024).
Delbridge, L. M. et al. Glycophagy—the physiological perspective on a newly characterized glycogen-selective autophagy. Curr. Opin. Physiol. 30, 100598 (2022).
Daniels, L. J. et al. Myocardial energy stress, autophagy induction, and cardiomyocyte functional responses. Antioxid. Redox Signal 31, 472–486 (2019).
Heather, L. C. et al. Guidelines on models of diabetic heart disease. Am. J. Physiol. Heart Circ. Physiol. 323, H176–H200 (2022).
Zoppini, G. et al. The E/e′ ratio difference between subjects with type 2 diabetes and controls. A meta-analysis of clinical studies. PLoS ONE 13, e0209794 (2018).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 17, 1–382 (2021).
Calmettes, G., John, S. A., Weiss, J. N. & Ribalet, B. Hexokinase–mitochondrial interactions regulate glucose metabolism differentially in adult and neonatal cardiac myocytes. J. Gen. Physiol. 142, 425–436 (2013).
Janssens, J. V. et al. Mechanical loading reveals an intrinsic cardiomyocyte stiffness contribution to diastolic dysfunction in murine cardiometabolic disease. Journal of Physiology (in the press).
Mills, R. J. et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl Acad. Sci. USA 114, E8372–E8381 (2017).
Voges, H. K. et al. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development 144, 1118–1127 (2017).
Zlobine, I., Gopal, K. & Ussher, J. R. Lipotoxicity in obesity and diabetes-related cardiac dysfunction. Biochim. Biophys. Acta 1861, 1555–1568 (2016).
Le Grand, J. N. et al. GABARAPL1 antibodies: target one protein, get one free!. Autophagy 7, 1302–1307 (2011).
Meddeb, M. et al. Myocardial ultrastructure of human heart failure with preserved ejection fraction. Nat. Cardiovasc. Res. 3, 907–914 (2024).
Reichelt, M. E., Mellor, K. M., Curl, C. L., Stapleton, D. & Delbridge, L. M. D. Myocardial glycophagy—a specific glycogen handling response to metabolic stress is accentuated in the female heart. J. Mol. Cell Cardiol. 65, 67–75 (2013).
Vernier-Magnin, S. et al. Analysis of the guinea-pig estrogen-regulated gec1/GABARAPL1 gene promoter and identification of a functional ERE in the first exon. Biochim. Biophys. Acta 1731, 23–31 (2005).
Kim, Y. et al. Gene therapy in cardiovascular disease: recent advances and future directions in science: a science advisory from the American Heart Association. Circulation 150, e471–e480 (2024).
Takeshige, K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 (1992).
Abeliovich, H. et al. Where is the field of autophagy research heading? Autophagy 19, 1049–1054 (2023).
Daniels, L. J. et al. Myocardial deformation imaging by 2D speckle tracking echocardiography for assessment of diastolic dysfunction in murine cardiopathology. Sci. Rep. 13, 12344 (2023).
Chandramouli, C. et al. Diastolic dysfunction is more apparent in STZ-induced diabetic female mice, despite less pronounced hyperglycemia. Sci. Rep. 8, 2346 (2018).
Mellor, K. M., Wendt, I. R., Ritchie, R. H. & Delbridge, L. M. D. Fructose diet treatment in mice induces fundamental disturbance of cardiomyocyte Ca2+ handling and myofilament responsiveness. Am. J. Physiol. Heart Circ. Physiol. 302, H964–H972 (2012).
Stapleton, D. et al. Analysis of hepatic glycogen-associated proteins. Proteomics 10, 2320–2329 (2010).
Parker, S. J., Venkatraman, V. & Van Eyk, J. E. Effect of peptide assay library size and composition in targeted data-independent acquisition-MS analyses. Proteomics 16, 2221–2237 (2016).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Kuleshov, M. V. et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016).
Zybailov, B. et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 5, 2339–2347 (2006).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128 (2013).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Acknowledgements
We acknowledge D. Stapleton for providing the glycogen phosphorylase antibodies. This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRCA; 1027865, 628643, 1082215, 1037320, 1067869, 1157320), the Diabetes Australia Research Trust, Stem Cells Australia, the National Heart Foundation of Australia (NHFA), the New Zealand Marsden Fund (14-UOA-160, 19-UOA-268), the Health Research Council of New Zealand (19/190) and the University of Auckland Faculty Research Development Fund, and the National Institutes of Health, USA (R01 HL155346-01 and R01 HL144509-01). Fellowship support is acknowledged from NHMRCA (J.E.H., R.G.P., E.R.P.) and Heart Foundation of Australia (E.R.P., J.E.H., K.L.W.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
L.M.D.D. and K.M.M. conceived and designed the experiments. K.M.M., U.V., P.K., C.L.C., J.V.J., L.J.D., G.B.B., A.J.A.R., M.A., X.L., S.L.J., D.J.T., K.R., R.J.M. and R.G.P. performed the experiments. K.M.M., U.V., P.K., C.L.C., J.V.J., L.J.D., G.B.B., A.J.A.R., M.A., X.L., S.L.J., D.J.T., K.R., K.L.W., R.J.M., J.R.B., E.R.P. and J.E.H. analyzed the data. L.M.D.D., K.M.M., E.R.P., J.E.H., J.E.V., X.H., R.P.X., R.K., T.J.O. and R.A.G. contributed materials and analysis tools. L.M.D.D. and K.M.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Juan Bernal, Gary Lopaschuk, Manuel Mayr, Jane Reznick and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cardiac function and glycogen handling in diabetic hearts.
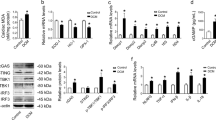
a, Ratio of phosphorylated glycogen synthase (pGS Ser641) to total glycogen synthase (GS) is unchanged in T2D mouse hearts (n = 8 animals/group, molecular weight indicated in kilodaltons, kDa, analyzed by unpaired 2-sided T-test). b, Ratio of phosphorylated glycogen phosphorylase (pGP Ser14) to total glycogen phosphorylase (GP) is unchanged in in T2D mouse hearts (Ctrl: n = 8, T2D: n = 10 animals/group, analyzed by unpaired 2-sided T-test). c, Exemplar M-mode and flow Doppler echocardiography traces. d, Echocardiography M-mode-derived ejection fraction and fractional shortening are unchanged with diabetes in type 2 diabetic (T2D, 14 week high fat sugar diet) vs control (Ctrl) mice (%EF n = 10 animals/group. %FS Ctrl: n = 10, T2D: n = 9 animals/group, analyzed by unpaired 2-sided T-test). e, Glycogen content normalized to protein in adult rat cardiomyocyte lysate from Ctrl and streptozotocin (STZ)-induced diabetic rats (8 weeks post-STZ, Ctrl: n = 10, STZ: n = 9 animals/group, analyzed by unpaired 2-sided T-test, p = 0.0037). f, Ca2+ time constant of decay (Tau) in isolated adult cardiomyocytes from control and STZ rats (8 weeks post-STZ, Ctrl: n = 10, STZ: n = 9 animals/group, analyzed by unpaired 2-sided T-test, p = 0.0022). g, Correlation of cardiomyocyte glycogen content and Ca2+ time constant of decay (Tau) in control (blue) and STZ (pink) rat cardiomyocytes (unloaded cells; r, Pearson correlation coefficient). h, Gabarapl1 mRNA is lower in atrial appendage samples from patients with T2D (n = 12 patients/group, analyzed by unpaired 2-sided T-test, p = 0.043). i, GABARAPL1 protein content in fractionated atrial appendage samples from patients with T2D (Homog: Ctrl n = 9, T2D n = 4; Memb, Cyt, Memb:Cyt ratio: n = 5 patients/group, analyzed by unpaired 2-sided T-test). j, STBD1 protein content is increased in atrial appendage samples from patients with T2D (Ctrl: n = 9, T2D: n = 4 patients/group, analzyed by unpaired 2-sided T-test, p = 0.0418). Data presented as truncated violin plots with median and upper & lower quartiles indicated, or bar graphs mean +/- sem. *p < 0.05. Related to Fig. 2.
Extended Data Fig. 2 Crispr-Cas9 Gabarapl1-KO mouse validation.
a, Gabarapl1 sequence details for the 8 founder Crispr-Cas9 mouse lines confirming excision of exons 2-4 from the Gabarapl1 gene (Next Generation sequencing). Text in red highlights mutations differing from the predicted sequence. b, Validation of Gabarapl1 knockdown using DNA electrophoresis to identify the presence of the Gabarapl1-KO allele in the heterozygote Gabarapl1-KO (tail sample). c, Immunoblot to confirm the absence of the GABARAPL1 band in the homozygote knockout mouse (membrane-enriched fraction of heart homogenate). d, Heterozygote Gabarapl1-KO mice exhibit ~50% knockdown of the Gabarapl1 gene (qPCR) in the heart, and absence of the Gabarapl1 gene is confirmed in the homozygote Gabarapl1-KO mouse heart (male mice, 30 weeks old, WT n = 4, KO/WT n = 5, KO/KO n = 5 animals/group, analyzed by 1-way ANOVA with Bonferroni post-hoc, p < 0.0001). Data presented as mean ± s.e.m. *p < 0.05. Related to Fig. 3.
Extended Data Fig. 3 AAV9-Gabarapl1 gene delivery in vitro and in vivo, cardiac stiffness protocol and iPS cardiomyocyte glycogen.
a, Confirmation of Gabarapl1 mRNA overexpression with AAV-Gabarapl1 in NRVMs (n = 3 independent culture wells/group, analyzed by unpaired 2-sided T-test, p < 0.0001). b, Immunoblot of GABARAPL1 protein expression in mouse hearts 4 weeks post-injection (i.v.) of AAV-Null or AAV-Gab (1010, 1011 or 1012 gc/mouse) with homozygote Gabarapl1-KO mouse heart as negative control. c, AAV vector burden in mouse heart 12 weeks post-injection (i.v.) of 1012 gc/mouse AAV9-cTnT-Null (AAV-Null) or AAV9-cTnT-Gabarapl1 (AAV-Gab) as measured by digital-droplet PCR detection of the WPRE viral element (Ctrl-Null n = 9, T2D-Null n = 9, Ctrl-Gab n = 12, T2D-Gab n = 13 animals/group, analyzed by 2-way ANOVA with Bonferroni post-hoc test). d, M-mode and flow Doppler echocardiography exemplar traces in T2D mice treated with AAV-Null or AAV-Gab. e, Ratio of phosphorylated (Thr172) to total AMPK protein expression is decreased with T2D in mice with Gabarapl1 gene delivery (AAV-Gab, Ctrl-Null n = 9, T2D-Null n = 11, Ctrl-Gab n = 11, T2D-Gab n = 13 animals/group, analyzed by 2-way ANOVA with Bonferroni post-hoc, Ctrl-Gab vs T2D-Gab p = 0.0107). f, Myocardial sections from T2D mice with Gabarapl1 gene delivery (T2D-Gab) stained with Oil Red O for visualization of cellular neutral lipids (scale bar, 100 µm). g, Exemplar images of a non-stretched and stretched cardiomyocyte attached between two glass rods. Cardiomyocyte stretch protocol. h, Glycogen is increased in human iPS cardiomyocytes in response to high glucose exposure (n = 6 wells/group, analyzed by unpaired 2-sided T-test, p = 0.0021). Data presented as mean ± s.e.m. *p < 0.05. Related to Figs. 4, 5 & 6.
Supplementary information
Source data
Source Data Figs. 1–6
Statistical source data and unprocessed blots.
Source Data Extended Data Figs. 1–3 and Source Data Extended Data Tables 2, 5 and 6
Statistical source data and unprocessed blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mellor, K.M., Varma, U., Koutsifeli, P. et al. Targeted glycophagy ATG8 therapy reverses diabetic heart disease in mice and in human engineered cardiac tissues. Nat Cardiovasc Res 4, 1487–1500 (2025). https://doi.org/10.1038/s44161-025-00726-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-025-00726-x
This article is cited by
-
Targeting glycogen clearance to treat diabetic cardiomyopathy
Nature Cardiovascular Research (2025)