Abstract
Volatile organic compounds (VOCs) are a biologically important subset of an organism’s metabolome, yet in vitro techniques for the analysis of these small molecules vary substantially in practice, restricting the interpretation of study findings. Here, we present an engineered culture tool, termed the “Biodome”, designed to enhance endogenous analyte recovery by integrating dynamic headspace sampling methodology for the recovery of VOCs from biological cultures. We validate the functionality of the device for in vitro volatile metabolomics utilizing computational modeling and fluorescent imaging of mammalian cell culture. Leveraging comprehensive two-dimensional gas chromatography coupled with a time-of-flight mass spectrometer and the enhanced sampling capabilities afforded by our tool, we identify 14 statistically significant VOCs not found in the media or exogenously derived from the sampling method, four of which have not been previously reported in vitro. To demonstrate applicability beyond mammalian cell culture, we assess the production of VOCs throughout the log and stationary phases of growth in ampicillin-resistant DH5α Escherichia coli. We identified 19 compounds with results supporting endogenous production, two of which had not previously associated with E. coli, 3-Octanone and 3-Tridecanone. Our findings emphasize the improved capabilities of the Biodome for in vitro volatile metabolomics.
Similar content being viewed by others
Introduction
Volatile metabolites, part of the broad class of compounds known as volatile organic compounds, are products of cellular metabolism with low molecular weight and a sufficient vapor pressure to exist as a gas at standard temperature and pressure. VOCs have long been recognized to confer information about biological processes in health and disease1,2,3, yet reliable methods for the study of these molecules in vitro have lagged behind. Works overcoming the challenges associated with in vitro sampling methodology have provided evidence for the extensive roles of volatile metabolites in biology, including but not limited to, serving as putative disease biomarkers4,5,6, extracellular signaling molecules in archaea, fungi, bacteria, protists, plants, and animals7,8,9,10,11,12,13,14,15,16, and drivers of inter– and intra–kingdom phenotype and function7,9,10,11,17,18,19,20. Despite the insights gained, nearly all approaches suffer from one or more of the following major limitations: i) detection of exogenous VOCs from plastics or the sampling environment18,21,22, ii) low analyte sensitivity and poor signal-to-noise relative to VOCs originating from non-endogenous sources, as a consequence of indirect18,23 and passive (equilibrium-based) sampling techniques21,24,25 and/or iii) disruption of culture viability26,27,28, prohibiting biological response to environmental stimuli or perturbation. Thus, in vitro VOC studies would largely benefit from a standardized method and tool to reduce signal from contaminants, enhance endogenous signal recovery, and maintain viability for time-dependent applications.
Previous groups have recognized these limitations and worked to develop a broad family of approaches known as purge-and-trap or dynamic headspace sampling (DHS)4,29,30,31,32,33. These approaches involve the continuous sampling or removal of the headspace, often trapping the VOCs on sorbent material fixed to solid support33,34,35,36, or directly analyzing the extracted gas by coupling the sampling methodology to a mass spectrometer37,38,39 or sensor array24,40. DHS has been shown to yield an increase in signal when considering the volatilome emitted from fruit extract41,42 and other complex matrices43. Despite the advantages of DHS methodology, engineered tools and systems designed to facilitate its use in an in vitro biological setting have been limited and a standardized approach does not yet exist4,18,29,30,31,44. Challenges contributing to this gap include organism viability and material compatibility, exogenous contaminants, imaging capabilities, liquid-to-headspace ratio and signal sensitivity, gas flow characteristics and reproducibility, reusability, throughput, and adaptability for automation.
Here, we worked to address many of these gaps surrounding the analysis of VOCs from in vitro cultures through the design, development, and application of a custom borosilicate glass culture vessel (the “Biodome”) and demonstrate the advantages of our tool for biological analysis through the unbiased characterization of mammalian cell and bacterial volatilomes. Similar to a recently published approach32, this work leverages the enhanced sensitivity, resolution, and separation capabilities of thermal desorption coupled to comprehensive two-dimensional gas chromatography with time–of–flight mass spectrometry (GC×GC-TOFMS) for metabolomic analysis45. We pair this instrumentation with an offline, open flow system designed to trap culture-derived VOCs using a tri-phase thermal desorption tube. The modular aspects of our system support offline culturing in a standard incubator and easy replacement of the input gas for adaptation to oxygen-sensitive organisms. The borosilicate glass culture vessel enables sterilization by autoclaving while also allowing for imaging directly through the device.
Given the intended biological applications, we first use fluorescent imaging to demonstrate that SK-OV-3 mammalian cell growth is roughly equivalent to a standard T-75 flask. We then work to extensively characterize the exogenous VOCs originating from the flow system and culture media over 96 h of continuous sampling to identify 31 VOCs observed in ≥50% of SK-OV-3 ovarian adenocarcinoma volatilome samples and in fewer than 50% of all controls, four of which have not been previously reported in vitro. Mammalian volatilomes typically show hydrocarbons and esters as major components, both in biofluid and cell lines46,47. Therefore, to validate endogenous origin using an independent approach, we apply RNA interference (RNAi) to globally evaluate the role of fatty acid metabolism in the production of VOCs in vitro, transiently inhibiting expression of mitochondrial lipid transporter protein carnitine palmitoyltransferase 2 (encoded by the gene CPT2). In doing so, we identify at least four VOCs with evidence supporting a weak association with lipid metabolism. We extend the functionality of the tool by characterizing the Escherichia coli (E. coli) volatilome across the log and stationary phases of growth and identify seven previously reported and two low abundance VOCs not formerly observed in E. coli volatilome. Finally, through direct comparison to more traditional equilibrium-based SPME methodology, we find reduced contaminant signal compared to a T-25 flask in the SK-OV-3 volatilome, and higher signal recovery of E. coli specific VOCs throughout the log and stationary phases of growth. Through this work, we demonstrate that our tool enables the reproducible sampling of complex biological matrices and provides sufficient analyte recovery to allow for the identification of endogenous volatile metabolites and low abundance VOCs not present in the culture media or flow system. Thus, the Biodome tool should serve as a promising platform for the growing field of volatile metabolomics.
Results
Design and Development
The biological VOC sampling system is comprised of a circular glass culture vessel, compressed gas cylinder and dual stage regulator, NalgeneTM 890 FEP tubing (Catalog # 8050-0310, Thermo Scientific, Waltham, MA, USA), oxygen-compatible PTFE tape (Catalog # 22485, Restek, Bellefonte, PA, USA), hydrocarbon trap (Catalog # 22013, Restek), flow meter (Catalog # 40402-0010, Riteflow, Bel-Art, Wayne, NJ, USA), sterile filter (0.2 µm pore size, Pall Corporation, Port Washington, NY, USA), bead bath (LabArmor, Plano, TX, USA), Carbopack C, Carbopack B, and Carbosieve SIII sorbent thermal desorption tube (TDT; Gerstel, Linthicum, MD, USA), and inexpensive laboratory-made TDT adapter – incorporating a modular design to facilitate adoption for a variety of biological organisms (Fig. 1a – d). For example, the bead bath temperature can be raised to promote growth of thermophiles or replaced with an ice bath to allow study of psychrophiles. Similarly, the inflow gas can be replaced to support the analysis of oxygen-sensitive microorganisms. The system is designed to enable the continuous sampling of the headspace VOCs in the Biodome by flowing the desired compressed gas through the culture vessel, effectively increasing the cumulative VOC signal as compared to static equilibrium-based systems (i.e. solid phase microextraction (SPME) in a culture vessel or closed vial), as a consequence of Henry’s Law.
a The borosilicate glass culture vessel, termed the Biodome, containing 5mL RPMI 1640 media. T-25 gas permeable flask caps were allowed to rest over the inlet and outlet of the vessel while in an incubator. b Placement of the Biodome during VOC sampling shows the glass vessel is fully submerged in beads at 37 °C. Also shown is the interface between the Biodome, thermal desorption tube (TDT), and TDT adapter. A sterile rubber stopper and PTFE tape are used to create an airtight seal at the interface between the TDT and the adapter. c 3D rendering showing the major components of the gas flow system including the hydrocarbon trap, flow meter, sterile filter, tubing, Biodome, TDT adapter, and TDT sampling tube. The compressed gas cylinder is not shown. d Schematic showing relevant dimensions (in mm; OD) for the Biodome culture vessel. e Representative two-dimensional gas chromatography chromatogram spanning 400–2200 seconds in the first dimension and 0.5-2 seconds in the second dimension, following 24 hours of sampling SK-OV-3 ovarian adenocarcinoma cells. Fluid modeling in ANSYSTM demonstrates f laminar flow conditions are maintained at flow rates ≤20 mL/min (TDT adapter insert included) and g gauge pressure and h velocity changes due to the inflowing gas are negligible at roughly half the Biodome height, limiting turbulent interactions with the surface of the cell culture media. i Live/dead assay images in the Biodome show viability across 0.5–4 days. A T-75 flask at t = 0 (24 hours post seeding) is shown for reference. j Independent live-dead assays were performed using NucBlueTM (Hoechst 33342) and NucGreenTM ReadyProbesTM at 24-hour intervals, with live percentages estimated from 10 independent regions of the T-75 or Biodome culture vessels. Error bars represent standard error of the mean. No statistically significant differences in cell viability are observed using a two-sided t-test, demonstrating equivalence as compared to standard culture vessels. Scale bar = 100 µm. k Custom imaging tray was designed in SolidWorks® 2019 and 3D printed to allow imaging using a Leica DMi8. l-n Typical SK-OV-3 morphology was isualized using fluorescent imaging and a live actin tracking stain (green color), Hoechst 33342 (blue color; live stain), and NucGreen (red color; dead stain), and no differences are observed using different culture vessels and growth environments. All brightfield and fluorescent images were taken using cells at passage <10. Scale bar = 50 µm.
The Biodome culture vessel is glass blown from borosilicate glass and features a 6.35 mm OD (1/4 inch) inlet and 18 mm OD outlet (Fig. 1a, d), with heights equal to 10 mm. The internal chamber height also measures 10 mm, selected to enhance VOC signal recovery and reproducibility by optimizing surface area to volume ratio while limiting turbulent interactions between the inflowing gas and the surface of the liquid culture media. The thickness of the glass is roughly 1–2 mm and the diameter of the biodome is 70 mm, giving an approximate surface area of 38.5 cm2. Vented caps from standard T-25 flasks were found to maintain a favorable environment for growth when allowed to rest over the inlet and outlet (Fig. 1a). Worth noting, no surface coating was required to promote cell adhesion and growth, increasing throughput, and reducing user burden.
The TDT adapter (Fig. 1b, c, Fig. S1) is comprised of a size 3 silicon rubber stopper adjusted to a ~15 mm diameter using a sterile razor (VWR, Radnor, PA, USA) with 6 mm hole introduced by a biopsy punch (Integra LifeSciences, Princeton, NJ, USA), Nalgene tubing (6.35 mm ID, 7.9375 mm OD, see above), one 6.35 mm (1/4 inch) and one 7.9375 mm (5/16 inch) 316 stainless steel compression nut with metal ferrule (Swagelok Southwest Co., Phoenix, AZ, USA), and a 316 stainless steel 6.35 mm male to 7.9375 mm male adapter (Swagelok). For assembly of the adapter, see Methods. Although the initial design was intended for TDT sampling by two dimensional GC×GC-TOFMS (Fig. 1e), given the popularity of SPME for VOC analysis, we further modified the adapter in an inexpensive manner to enable this method by adding a 1 mL disposable syringe body (Electron Microscopy Sciences, Hatfield, PA), with the plunger removed, in line with the 6.35 mm (1/4 inch) nut (replacing the TDT, Fig. S1) and completely sealing the interface with oxygen-compatible PTFE.
To assess the gas flow characteristics through the headspace of the culture vessel, fluid modeling was performed in ANSYS Fluent® 2019 R3. Results show that laminar flow is maintained for gas flow rates ≤20 mL/min, using the defined geometry (Fig. 1d, f – h, Supplementary Video 1). A flow rate of 11.7 mL/min was maintained for all testing, corresponding to a reading of 30 on the flow meter, unless otherwise noted. To enhance throughput, a multiplexed part was designed in SOLIDWORKS® 2019 and flow characteristics were assessed using ANSYS Fluent (Fig. S1). Results show the part provides consistent flow rates at each of the three outlets, equal to 1/3 the total flow rate, while maintaining a single inlet (Fig. S1, Supplementary Video 2). Given an internal chamber volume of approximately 200 cm3, the internal chamber can be flushed of environmental gas using a purge flow rate of 20 mL/min and a duration of 10 min.
Cell viability was considered in SK-OV-3 ovarian adenocarcinoma cells (HTB-77, ATCC, Manassas, VA) using live/dead fluorescence assays. 0.5 × 106 cells were seeded into the Biodome containing a total of 5 mL of RPMI1640 culture media containing 10% FBS and 1% Pen/Strep and given 24 h to allow cell attachment. Brightfield images show time-dependent increases in Biodome confluency and high viability (Fig. 1i). A standard T-75 flask (VWR) 24 h after seeding is shown for reference, equivalent to t = 0 in the Biodome (Fig. 1i). Results from independent live/dead fluorescence assays (NucBlue and NucGreen ReadyProbes, Invitrogen, Carlsbad, CA) show no significant differences in SK-OV-3 cell viability in the Biodome, as compared to a standard T-75 flask in a humidified incubator, for a minimum duration of 4 days (Fig. 1j). Growth rates are comparable in the Biodome to that seen in plastic culture flasks, as cells reach 70-90% confluency in roughly four days when seeded at 500,000 cells total. We further considered cell viability through morphological fluorescence staining using a live actin tracking stain (CellMask, Invitrogen). To enable imaging on a Leica DMi8 microscrope, we designed a custom imaging tray in SOLIDWORKS® 2019 (Fig. 1k). Following seeding in the appropriate culture vessel, SK-OV-3 cells were allowed to proliferate for 48 h before staining, and our results show that cell morphology in the Biodome is comparable to a T-75 flask grown in a standard incubator (Fig. 1l – n). Taken together, our findings demonstrate that the Biodome tool maintains favorable growth conditions for at least 4 continuous days, without the need for surface treatment.
Characterizing device performance in mammalian culture
SK-OV-3 cells (human origin) were seeded into the Biodome glass culture vessel and allowed to adhere and proliferate for 24 h. Prior to sampling, culture media was replaced to ensure the accumulation of VOCs were minimized. Based on the previously observed growth characteristics (Fig. 1h–l), cells were subject to VOC collection for 4 continuous days with the TDT replaced every 24 h, although shorter sampling times were also shown to be viable (Fig. S2). RPMI 1640 was maintained as the primary medium, however dialyzed FBS was implemented in place of FBS. This change is supported by a previous work that demonstrated serum-free growth media is favorable for VOC analysis48 and will be further considered in a future work. Chromatograms were aligned using ChromaTOF® (see Methods), and VOCs were reported according to the naming confidence scheme presented by the Metabolomics Standards Initiative49. TDU tubes were stored for a maximum of 8 days at 4 °C to limit degradation50 prior to analysis by GC×GC-TOFMS. All experimental conditions were performed in biologically independent triplicate.
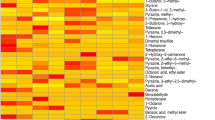
Results show the Biodome system supports the identification of cellular VOCs absent in the flow system and culture vessel, both with and without culture media. Features observed in fewer than 50% of observations within an experimental condition, those observed in all internal blank samples, and analytes eluting with a first-dimension retention time <400 s were removed (see Methods), resulting in a total of 254 unique peaks (Fig. 2a). The filtered volatilome was subsequently subject to principal component analysis (PCA) and clusters corresponding to the control and SK-OV-3 conditions were observed (Fig. 2b). Noticeable batch effects were seen in the first replicate of the SK-OV-3 and empty tool volatilomes and are attributed to instrument maintenance and user variability (Fig. 2b). Roughly 40% (102/254) of the unique features were observed in ≥50% of the empty Biodome controls and fewer than half of SK-OV-3 samples, suggesting these are inherent contaminants associated with the use of the in vitro tool or analytical system (Fig. 2a, Supplementary Dataset 1). An alkane standard (Fig. S3) was run to allow identification of VOC retention indices (RI) and used to support name assignment where possible (see Methods). A further 45% (115/254) of the unique features were observed in ≥50% of the experimental conditions containing culture media but not the empty Biodome control, suggesting media origin (Fig. 2a, Supplementary Dataset 2). Interestingly, 6 low abundance VOCs (2.4%) with a mean signal-to-noise ratio <200:1 were observed in the media volatilome but not the SK-OV-3 or empty volatilomes (allowing for one spurious peak in both), which may indicate SK-OV-3 metabolic biotransformation (Supplementary Dataset 3). Lastly, a total of 31 VOCs (12.2%) were observed in ≥50% of the experimental runs containing SK-OV-3 cells and fewer than 50% of controls (Table 1; Fig. 2c, d; Fig. S4). Four of these VOCs were significantly more abundant in cellular volatilomes after application of a students’ t-test with a Benjamini-Hochberg p-value correction51, with a mean signal-to-noise ratio <200:1 and have not previously detected in mammalian cells in vitro (bolded names), emphasizing the improved signal recovery afforded by the Biodome tool (Table 1; Fig. 2c, d; Fig. S4).
a Bar chart showing the number of VOCs detected after filtering inconsistently observed peaks, categorized according to their proposed origin. b Principal component analysis plot shows distinct clusters when considering the first two principal components, using the 254 features comprising the filtered volatilome. Batch effects are primarily attributed to i) instrument maintenance and ii) user variability that occurred between the first replicate and the remaining runs. Representative c boxplots and d line plots show three low-abundance VOCs reproducibly detected in the SK-OV-3 ovarian adenocarcinoma in vitro volatilome. Abundance values were calculated by integrating the peak area using the unique mass and log10 transformed. The adjusted p value was calculated using a two-tailed student’s t-test and corrected using the Benjamini-Hochberg procedure51. Day 1 represents the cumulative volatilome between 0 and 24 hours, and so forth. Each time point was measured in biologically independent triplicate, with line plot error bars representing ± standard error of the mean. For boxplots, the median is indicated by the solid black line, and first and third quartiles are captured by the bounds of the box. Boxplot whiskers are defined as the first and third quartiles –/+ interquartile range times 1.5, respectively. Minimum and maximum values are captured by the lowermost and uppermost points, respectively, or whisker bound if no outliers are shown. The orange color represents SK-OV-3 observations, light blue captures the RPMI1640 media control, and purple indicates the empty flow system control.
Cell-specific VOCs spanned multiple chemical classes and includes the detection of two hydrocarbons and one ester that fell below the naming threshold. Despite the lower naming confidence, mass spectral data could be used to support their identification as a cyclic hydrocarbon eluting at 1213 and 0.69 s in the first- and second-dimension columns (average), respectively, an alkane at 2102 and 0.71 s, and an ester eluting at 2535 and 1.20 s (Table 1, Fig. S5). Of the VOCs observed primarily in the SK-OV-3 volatilome, roughly 80% (25/31) had a mean signal-to-noise ratio <200:1, emphasizing the capability of our approach to recover low abundance volatiles that may be missed using equilibrium-based sampling approaches.
To determine if the noise due to plastic contaminants is reduced in the Biodome compared to more standard in situ SPME techniques (in a culture flask), we analyzed 11 of the most abundant contaminants reported by Schallschmidt et al.21 that were present in the volatilome from either culture technique (2-butanone, acetic acid, benzene, cyclohexanone, di-tert-butylphenol (2,4-di-tert-butylphenol), decane, dodecane, ethylbenzene, m-di-tert-butylbenzene (1,3-bis(1,1-dimethylethyl)-benzene), octane, and styrene). The aggregated signal abundance was compared between the two methods, with missing values imputed to 0 (Supplementary Dataset 4). Cell seeding densities and media volume were scaled to be equivalent to that of the Biodome tool with triplicate measures performed at the time point of 0-24 h. Demonstrating that the Biodome effectively addresses one major existing limitation associated with the study of cellular volatilomes, DHS in the glass Biodome led to the recovery of a fraction of the total contaminant signal ( ~18%) as compared to the in situ SPME methodology. It is important to note that sorbent material differences were present between the two approaches. Given that SPME fibers comprised of TDT carbopack and carbosieve materials were not commercially available, we elected to utilize the multi-phase SPME fiber consisting of divinylbenzene, carboxen, and PDMS (Sigma-Aldrich, St. Louis, MO, USA).
Gene knockdown supports endogenous production of cellular VOCs
Most mammalian VOCs have an unknown or poorly understood biosynthesis pathway, with traditional approaches reaching limited insight, restricted by the pitfalls described above. Highlighting many of the improvements afforded by the Biodome tool, including temporal viability, reduced contaminant signal, and improved analyte recovery, we investigated the relationship between the biosynthetic origin of cellular VOCs and fatty acid metabolism via CPT2 transcriptional levels in SK-OV-3 cells. The CPT2 expression levels were altered by treating the SK-OV-3 cells with pooled siRNA oligonucleotides mediating gene knockdown (see Methods). The expression levels of CPT2 relative to the housekeeping gene GAPDH was determined using RT-qPCR (see Methods). Results show CPT2 expression was significantly reduced in siRNA-treated SK-OV-3 cells to 35.7 ± 13.6% (mean ± S.E.M; p = 6.76E–9) at t = 0 hrs, indicating successful transfection and gene knockdown (Fig. 3a). The expression levels of CPT2 in the transfected SK-OV-3 cells were also monitored transiently in a 6-well plate format over a 96-hour timeframe. As depicted in Fig. 3b, the CPT2 expression level during the initial 24 h showed a significant decrease to 9.83 ± 4.20% (p < 0.05). Subsequently, after 48 h, the expression levels increased to 53.0 ± 7.82% (p < 0.05). Following 72 h, the expression levels decreased to 36.8 ± 6.49% (p < 0.05) and were recorded as 56.8 ± 10.5% (p < 0.05) after 96 h.
a Expression of CPT2 quantified by RT-qPCR at the start of volatile organic compound (VOC) collection (time = 0) using biologically independent triplicate in a 6-well plate to estimate Biodome CPT2 expression levels. P-values were determined using a one-tailed t-test. b The expression of CPT2 was independently mapped across the four days of VOC sampling following siRNA-mediated knockdown in SK-OV-3 cells at equal passage number using biologically independent duplicate or triplicate as indicated with individual points. c Four VOCs not found in controls showing decreased abundance following the inhibition of β-oxidation via CPT2 knockdown, across 4 days of measurement. P values were determined using a one-tailed t-test and adjusted using the Benjamini-Hochberg procedure51. * denotes an adjusted p values < 0.05. Each time point was measured in biologically independent triplicate. Error bars represent the standard error of the mean. d Boxplots showing significant or trending toward significant decreases in endogenous VOC abundance following integration by unique mass and log10 transformation. P-values, same as c. The median is indicated by the solid black line, and first and third quartiles are captured by the bounds of the box. Boxplot whiskers are defined as the first and third quartiles –/+ interquartile range times 1.5, respectively. Minimum and maximum values are captured by the lowermost and uppermost points, respectively, or whisker bound if no outliers are shown. The gray color represents the non-targeting siRNA pool while red indicates the CPT2 targeted siRNA pool.
The knockdown of fatty acid metabolism via CPT2 RNA inhibition in transfected SK-OV-3 cells was carried out with the goal of perturbing endogenous hydrocarbon (alkane, alkene) and ester production, although ketones and aldehydes are also known to arise as a product of fatty acid metabolism52,53. Nearly all VOCs were found to have decreased median abundances when compared to the unperturbed SK-OV-3 volatilome across the 96-hour timeframe (Fig. 3c, d; Fig. S6; Supplementary Dataset 5). In the case of 2-Ethyl-1-hexanol, the average decrease in raw peak abundance across all samples was found to be 70.0%, with a roughly 87% decrease observed during the first day of sampling, although average expression increased on the second and fourth days, which may reflect the complex relationship between alcohols and fatty acid metabolism54 (Fig. 3c, d). Furfural was observed to have an average raw abundance decrease of 56.5% across all samples (adjusted p = 0.053), with a similar decrease observed during the first two days, but followed by lower abundance on days 3, and 4 – weakly consistent with the transient recovery of CPT2 expression (Fig. 3c, d). The average raw abundance of pyrrole after 24 h was roughly equivalent to the control following knockdown of CPT2 (1% increase), with a mean signal reduction of 8%, indicating pyrrole production is not related to fatty acid metabolism and consistent with other studies that show amino acids as precursors55 (Fig. 3c, d). The unknown analyte was seen to have an average raw abundance decrease of 66.3% across all samples, with transformed abundances mirroring the trend observed in furfural. Collectively, results show some possible candidates for endogenous origin associated with fatty acid metabolism (Fig. 3c, d; Fig. S6), although VOC abundance generally does not correlate well with the transient recovery of CPT2 expression and additional work is needed to confirm the proposed link.
Bacterial adaptation
To extend the functionality of our tool, we analyzed VOCs originating from Escherichia coli, strain DH5α, across 3 days, during the log and stationary phases of growth. E. coli was transformed with plasmid containing ampicillin resistance gene and seeded into the Biodome containing LB Broth and 100 µg/mL ampicillin. Turbid broth, indicating an increase in cell density, was observed at the conclusion of the sampling period for the run containing E. coli but not in the broth control (Fig. S7a). Following in vitro VOC sampling and analysis by GC×GC-TOFMS, chromatographic peaks were aligned in ChromaTOF®, yielding 206 unique features after the removal of known contaminants and compounds observed in the instrument blanks. Most of the recovered volatiles were variably present in the broth and bacterial conditions (85.4%) (Supplementary Dataset 6). For statistical analysis, compounds not detected in at least 70% of all samples were filtered. VOCs specific to the E. coli or broth volatilome were also retained—allowing up to three spurious observations in the alternative condition, resulting in a total of 30 features (Fig. 4a, Table 2). Using the filtered bacterial and broth volatilomes, we performed PCA and observe separation of the experimental groups when considering the first two components (Fig. 4b). After adjusting for multiple hypothesis testing using the Benjamini-Hochberg procedure51, 12 VOCs were found to be significantly elevated in the condition containing ampicillin-resistant E. coli (Fig. 4c, d, Fig. S7; Table 2). Results show the Biodome has sufficient sensitivity to repeatedly identify low abundance bacterial-derived VOCs, even when initially present in the culture media, including indole (FDR p value = 0.002) and 2-decanone (FDR p value = 0.004) (Fig. 4c, d, Fig. S7). Finally, of the 18 VOCs reproducibly observed in the E. coli volatilome, roughly 45% (8/18) had a mean signal-to-noise ratio <200:1, across the log and stationary phases of growth, highlighting the analyte recovery advantages of our tool for untargeted volatile metabolomics (Table 2).
a Venn diagram shows the number of unique and common volatile organic compounds (VOCs) detected in the bacterial and broth conditions. Pie charts show the breakdown of VOC functional groups specific to each group. b Principal component analysis using VOCs presented in Table 2 shows the E. coli volatilome is distinct from the broth control. c Line plots for four high confidence VOCs showing log10 transformed abundance across three days of sampling. Each time point was measured in biologically independent triplicate, with error bars representing ± standard error of the mean. Day one represents the cumulative volatilome between 0-24 hours of growth, day 2 between 24-48 hours, and the final day capturing 48–72 hours. d Boxplots for the same four VOCs showing expression of these metabolites are significantly elevated in the E. coli experimental condition, after correcting for multiple hypothesis testing using the Benjamini-Hochberg p-value adjustment procedure51. The median is indicated by the solid black line, and first and third quartiles are captured by the bounds of the box. Boxplot whiskers are defined as the first and third quartiles –/+ interquartile range times 1.5, respectively. Minimum and maximum values are captured by the lowermost and uppermost points, respectively, or whisker bound if no outliers are shown. The light red color represents the E. coli observations while the light blue indicates the LB broth control.
Many of the high confidence VOCs observed in the E. coli volatilome have been previously linked to this microorganism, however, we first report the production of 3- Octanone and 3-Tridecanone during the log and stationary phases of growth (Table 2; Fig. S7). Both VOCs had a mean signal-to-noise ratio <200:1. We also briefly consider VOCs distinguishing the log phase from the stationary phase of growth and note distinct production of Bicyclo[3.2.0]hepta-2,6-diene, Dimethyl trisulfide, and an unknown hydrocarbon with average first- and second-dimension retention times of 938 s and 0.64 s respectively, corresponding to a retention index of 840 (Fig. S7). The observation of significantly elevated 3,5-dihydroxybenzamide in the broth controls may suggest consumption by the bacteria during log and stationary phases of growth, provided structural similarity with endogenously produced enterobactin, although additional work is needed to validate the proposed consumption (Fig. 4c, d).
Finally, leveraging the Escherichia coli volatilome, we directly compared equilibrium headspace sampling methodology (SPME) to the DHS approach utilized in the Biodome tool (Fig. S8). Triplicate measures were made at each time point after inoculating a baked 20 mL glass vial with ampicillin-resistant Escherichia coli with the same target optical density (0.1) and analytical method, excluding the use of a septumless head inlet (Gerstel) in place of the TDU 2 inlet. To ensure findings were not affected by the liquid to headspace volume ratio, the volume of broth in the 20 mL vial was set at 3 mL to ensure equivalency to the Biodome tool. Results show that the Biodome tool with dynamic headspace sampling outperforms traditional SPME methodology for some VOCs including indole and the previously unreported 3-Octanone (Fig. S8), with total signal recovery 77% greater compared to traditional SPME sampling after filtering contaminant peaks. Inconclusively, others show lower variability and higher abundance using SPME including VOCs in the same chemical class (ketone), which is attributed to sorbent material differences (Fig. S8). SPME fibers comprised of the TDT carbopack and carbosieve materials were not commercially available, therefore we elected to use multi-phase SPME fiber consisting of divinylbenzene, carboxen, and PDMS (Sigma-Aldrich, St. Louis, MO, USA). While these differences limit the interpretation to some extent, we note that this specific sorbent coating is commonly reported in the study of human and microbial volatiles using SPME56,57,58 and is known to be well-suited to capture a wide range of analytes58.
Discussion
In this study, a glass culture vessel was designed and developed to facilitate the in vitro analysis of biological volatilomes and pair the engineered tool with the enhanced analytical sensitivity associated with GC×GC-TOFMS, which is recognized to directly support the improved recovery of endogenous low abundance VOCs independent of the tool. We validate the functionality of our tool by integrating computational modeling and fluorescent imaging to demonstrate desirable sampling and growth conditions. We then extensively characterize the origin of VOCs from the flow system itself, the culture media, and human-derived SK-OV-3 ovarian adenocarcinoma cells. In doing so, we identify at least low abundance three compounds not previously observed in vitro but reported in vivo (furfural, aniline, and naphthalene). With the aim of verifying the endogenous origin of some cell-specific VOCs, we inhibit lipid metabolism leveraging RNAi-mediated knockdown of CPT2 and find four volatile metabolites that show a decrease in abundance across the 4 days of analysis weakly consistent with the transient recovery of CPT2 expression (furfural and unknowns 4, 7, and 8). Finally, to broaden the scope of use for in vitro volatile metabolomics, we characterized the DH5α Escherichia coli volatilome across the log and stationary phases of growth (0-72 h) and identify two low abundance VOCs not previously reported (3-Octanone and 3-Tridecanone).
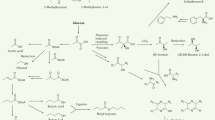
Considering the four mammalian cell VOCs not previously reported46, all were low abundance with a mean signal-to-noise ratio <200:1. Furfural (HMDB0000617)59 has not been previously detected in vitro, however, studies report observing this VOC in urine, feces, breath, and skin of healthy humans47 but has been linked to exogenous exposure59. Clearance is known to be rapid60 and thus sustained detection over 4 days of sampling strictly in the cellular volatilome indicates exogenous sources do not fully explain the observed signal, further supported by the results following RNA inhibition of CPT2. Moreover, furfural has been shown to act as a breath biomarker for gastric cancer61,62 and has been shown to influence DNA replication63, potentially linking this VOC to cancer. Aniline (HMDB0003012)59 is a naturally occurring metabolite that can arise from the condensation of ketones and amines59,64 and has been observed in the breast milk of healthy women47, suggesting sex-dependent differences in natural abundance. Thus, our use of an ovarian adenocarcinoma cell line for this study may partially explain the in vitro recovery of aniline using the Biodome. Aniline been detected exclusively in the breath of lung cancer patients but not healthy controls65, potentially linking increased abundance to cancer. Naphthalene (HMDB0029751)59 has been observed in the feces, breath, skin, milk, and saliva of healthy individuals47 and is generally associated with exogenous sources like environmental exposure59. It is bioactive and known to be metabolized by intracellular enzymes like cytochrome P45066 and detected in the breath of lung cancer patients67 – although biosynthesis routes through polyketide synthases are not known to be present in the human genome68. Lastly, very little has been reported on succinic acid diisopropyl ester as a mammalian metabolite46,47. Ester functional groups may arise from the metabolism of fatty acids, although abundance measures following inhibition of CPT2 suggests this VOC is not directly associated with the process. The expansion and culturing of cells prior to analysis is known to influence the volatile profile21 and potentially providing a source for the observed naphthalene signal and possibly others. In general, our results demonstrate that the Biodome enhances total signal recovery allowing for repeatable detection of low-abundance VOCs, and given the comparison to in situ SPME methodology, reduces signal abundance of common plastic contaminants.
In addition to the mammalian VOCs not previously reported, compounds statistically significantly elevated in the mammalian cell volatilome include pyrrole, n-butyl acetate, acetophenone, dimethyl trisulfide, and phenol. All five metabolites have been previously observed in mammalian cell culture in vitro. The A549 cell line (non-small cell lung cancer) has been reported to release pyrrole46 while acetophenone has been consumed by the Lu7387 lung cancer cell line46 and released from HUVEC primary endothelial cells69. Similarly, n-butyl acetate is reportedly taken up by human fibroblast and lung cancer cell line CALU-146,70 and released by normal (SV-HUC-1) and cancerous (Scaber, J82, 5637) bladder cell lines71. Phenol has been previously reported as a VOC released by healthy human tracheobronchial epithelial cells46,72. Lastly, dimethyl trisulfide has been detected in the headspace of culture flasks containing melanoma cell lines WM983B and WM1158 but not non-cancerous cell lines73, indicating an association with cancer given our findings utilizing the SK-OV-3 cell line. Trending towards significance, 2-Ethyl-1-hexanol (adjusted p = 0.061) is reportedly released by healthy human fibroblasts and lung cancer cell line NCI-H208746,70.
Many VOCs detected from mammalian culture in vitro often feature a relatively long hydrocarbon backbone46 and are generally thought to originate from fatty acid metabolism. To globally evaluate the role of fatty acid metabolism in the production of VOCs in vitro, we elected to transiently inhibit expression of lipid transporter CPT2 using RNA interference. Furfural and three unknown VOCs were primarily observed in cellular volatilomes and demonstrated abundance decreases weakly consistent with the knockdown of CPT2 across 4 days, although other sources of variability cannot be ruled out. Furfural could be associated with the inhibition of fatty acid metabolism through the aldehyde functional group on the 2-position of furan53. However, high abundance on days 1 and 2 followed by an absence on day three entirely and weak recovery on day 4 suggests a complex relationship between its biosynthesis and fatty acid metabolism, with proposed reaction mechanisms involving xylose and O2 protonation74. This approach represents an early application of RNAi to assess pathway origin of VOCs in mammalian cells, however methodological success has recently been demonstrated in plants75,76. Although a specific enzymatic step cannot be assigned to their production, our findings lend some strength to claims for cellular origin and provide a methodological platform for the secondary verification of VOC producing pathways in mammalian volatilomes. Future work will look to apply a similar approach to the less complex bacterial volatilome.
In the case of the E. coli volatilome the significant increase in indole, relative to the broth control, is an indication of the advantages of the Biodome tool for in vitro volatile metabolomics. Conversion of tryptophan to indole by tryptophanase is a well characterized process in E. coli77,78. Tryptophanase catalyzes the production of pyruvate in conditions of excess tryptophan, as expected in starter broth cultures79, generating indole as a byproduct77,78. Here, we find indole is the most abundant peak across the log and stationary phases of growth (0-72 h) in the E. coli volatilome, although one recent study has shown bacterial transformation to induce ampicillin resistance may act as a confounding factor80. Generally, our results agree with many previous studies, which demonstrate indole is a biomarker for E. coli, acting as an extracellular signaling molecule with import and export complexes, and serving to regulate biofilm formation, cell division, and gene expression81,82,83,84,85. Worth discussing, indole was detected with high confidence in the broth controls and had similarly broad peaks. Therefore, to ensure consistency, peaks within the first-dimension retention time window of 2092-2110 s were summed in all chromatograms, likely explaining the high reported signal in broth controls despite clear differences in abundance (Fig. S7). Demonstrating similar agreement, recent studies also report 2-nonanone in the E. coli volatilome, including the DH5α strain we detected it from in this study86,87. Further, biosynthesis of 2-nonanone is known to be naturally derived from the decarboxylation of 10 carbon β-keto acids88 and has been shown to regulate gene expression associated with quorum sensing89. We also observe related 2-ketones: 2-decanone, 2-undecanone, and 2-tridecanone in good agreement with previous studies90,91,92,93. Similarly, both 3-methyl-1-butanol and 2,2,4,6,6-pentamethylheptane have been previously observed in the E. coli volatilome93,94,95. Taken together, our findings demonstrate that volatile profiles acquired using the Biodome tool accurately reflect active metabolic processes in E. coli and agree with previous studies regarding the abundance of these VOCs relative to the remainder of analytes comprising the volatilome.
While we detected the well-established E. coli VOCs, indole and 2-ketones91 (e.g. 2-nonanone) we report the observation of low abundance 3-Octanone and 3-Tridecanone in the DH5α E. Coli volatilome. Interestingly, 2-ketones are known to be derived from fatty acid metabolism through the production of β-keto acyl-CoAs96 and while 3-ketones have not been previously observed in vitro, their detection implies the same precursor molecules may be used but involve a separate mechanism of biosynthesis. Furthermore, 3-octanone has been shown to act as a potent inhibitor of E. coli growth97 and line plots generally show increasing abundance of 3-ketones over the 72 h of sampling (Fig. S7d), possibly suggesting these VOCs may act to regulate growth as nutrient availability grows increasingly scarce, although additional work is needed to confirm their bioactivity and endogenous origin.
As demonstrated, the total signal emitted from cells is far less than that of the media or flow system, lending strength to claims of advantageous sampling in the Biodome associated with the implementation of dynamic headspace sampling. By enhancing the total analyte signal recovered from the biological culture, low abundance analytes have a better ability to generate a sufficient quality spectrum for database searches. While many single advantages of the Biodome tool have been incorporated into other designs, no single device had been developed that leveraged the analyte recovery advantages of dynamic headspace sampling while also addressing common pitfalls associated with contaminant plastic signal or relatively large headspace volumes. Thus, the Biodome tool provides a platform to build upon existing knowledge for in vitro volatilomics by integrating a (i) dynamic headspace methodology and low headspace volume to enhance total signal recovery allowing detection of lower abundance VOCs, and (ii) borosilicate glass culture vessel to reduce exogenous signal due to plastics, support dual imaging function, and allow for sterilization and reuse. Through the direct comparison of E. coli volatilomes by SPME and the Biodome tool we find improved reproducibility for some E. coli specific VOCs, including the volatile biomarker indole, while operating in the laminar flow range. The non-destructive nature of collection allows for analysis of VOCs across time and in response to perturbation, as demonstrated by the application of a standard RNA interference workflow. Given the advantages of the tool established through this work, the engineered Biodome can be readily adapted to meet experimental requirements, including temporal viability and easy modification of the temperature and flow gas composition. Analytical flexibility may be of notable usefulness for the study of anaerobes and extremophiles. Furthermore, off-line sampling enables the convenient storage of captured volatiles, as performed in this work, and the sampling adapter can be readily modified for non-TDU sampling (SPME), expanding tool accessibility. The gas inlet has been specifically designed to enable an airtight seal using standard (US) tubing sizes. The modular aspect of the system enables ease of sample replacement and reuse through autoclave sterilization, while the design of a secondary part to enable multiplexing of Biodomes improves throughput. Beyond the unique elements of the engineered Biodome for in vitro volatilomics, the flow system is generally comprised of low-cost elements and provides important manual control over the flow rate while minimizing exogenous carbon sources from the compressed cylinder and environment. Thus, the Biodome should serve as a powerful tool for the characterization of in vitro VOCs in this growing field of metabolomics.
Methods
Biodome Development and Fabrication
A CAD model of the Biodome culture vessel, with the TDT adapter included, was designed in SOLIDWORKS® 2019. Flow characteristics were then modeled in the CAD part using ANSYS Fluent® 2019 R3. For meshing, the target skewness was adjusted to 0.65, smoothing set to high, a mesh metric set to Orthogonal Quality, and default for all other parameters. The inlet and outlet mesh were then refined using a value of 2. Boundary conditions were then set, using inlet velocities ranging from 1 – 50 mL/min and the pressure outlet condition. The operating pressure was assumed to be 1 atm, the fluid set to air, and acceleration due to gravity was included in the model. The residual threshold was adjusted to 1E-5. Solution parameters were as follows: Pressure-Velocity Coupling Scheme set to SIMPLE, Spatial Discretization Gradient set to Least Squares Cell Based, Pressure set to Second Order, and Momentum set to Second Order Upwind. Solution results were visualized using ANSYS CFD Post GUI.
Fabrication of the glass culture vessel leverages specialized trade techniques, but in brief begins with rotation of Pyrex tubing using a glassblowing lathe. A natural gas or oxygen flame was used to pull the tube down into a flat bottom. The inlet and outlet were then added to onto the flat bottom. Using a custom holder to retain that piece in the lathe, another flat bottom was pulled to create the internal chamber. Additional attention was given to the curvature of the bottom and top to ensure the flattest surface possible for imaging applications.
To assemble the TDT adapter, Nalgene tubing was inserted into the 7.9375 mm nut head (with moderate force) and the other end was inserted into the rubber stopper. A metal ferrule and tubing insert was attached to the free end and the nut-ferrule assembly was then connected to the male-to-male adapter (Fig. S1e). A ferrule was then added to the 6.35 mm stainless steel nut and screwed firmly into the other end of the male-to-male adapter. The opening of the 6.35 mm stainless-steel nut served as the interface for TDT sampling. An airtight seal was ensured by wrapping 7-8 cm of oxygen-compatible PTFE tape around the beveled end of the TDT tube before inserting it into the TDT interface. A visible decrease in the flow rate (>2 mL/min) was always observed when the TDT was inserted into the adapter and the regulator was readjusted to achieve the original flow rate after roughly 5-10 min of equilibration. To adapt the system for SPME sampling methodology, a 1 mL disposable syringe body (Electron Microscopy Sciences, Hatfield, PA), with the plunger removed, in line with the 6.35 mm (1/4 inch) nut (replacing the TDT, Fig. S1f) and completely sealing the interface with oxygen-compatible PTFE. The SPME fiber then rests naturally at the narrow end of the syringe body – without contacting the sides of the syringe. Other SPME adapters were considered, including the use of a GC SPME glass liner insert, however the added volume of the syringe was found to help with the condensation of water during sampling durations >24-48 h (depending on flow rate).
Cell Culture
SK-OV-3 ovarian adenocarcinoma cells (human origin; HTB-77) were obtained from ATCC. Cells were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS (ATCC) and 1% penicillin (100 I.U. / mL; ATCC) / streptomycin (100 µg/mL; ATCC). Cells were maintained in a humidified incubator at 37 °C and 5% CO2. Cultures were grown in standard T-75 flasks and passaged at 70-85% confluency.
To seed the Biodome for VOC analysis, approximately 500,000 cells – estimated using a hematocytometer – were seeded in the Biodome. Cells were allowed to adhere from within a standard incubator (see above) for 24 h, and the culture media was replaced immediately before the start of analysis. Vented caps from standard T-25 flasks (VWR) were allowed to rest on top of the inlet and outlet of the Biodome – maintaining an enclosed environment and CO2 exchange with the media. The total media volume in the Biodome was maintained at 5 mL for all experiments. To limit the background signal from the culture media and maintain growth rates, dialyzed fetal bovine serum (FBS) was used in place of traditional FBS, ensuring small molecule (<10,000 Da) concentration was reduced. Finally, to improve the consistency of heating and reduce condensation, the Biodome was completely submerged within a bead bath maintained at 37 °C. Gas flow was set 24 h before the start of experimentation to ensure an equilibrium was reached within the two-stage regulator.
Bacterial Culture
DH5α E. coli were transformed with plasmid containing the ampicillin resistance gene (pUC19, Addgene Plasmid #50005) and streaked on LB agar plates spiked with 100 µg/mL ampicillin. Isolated colonies were inoculated in 100 µg/mL ampicillin spiked LB broth and grown for 24 h. OD600 measurements were taken and used to dilute the bacterial culture such that the initial OD600 seeding concentration was approximately 0.1 at the start of VOC collection. Bacteria were seeded into the Biodome containing a total of 5 mL of LB broth spiked with 100 µg/mL ampicillin and TDU tubes were replaced every 24 h. The carrier gas composition was 95% air / 5% CO2, to allow compatibility with cellular analysis, and a flow rate of 18 mL/min was maintained for the duration of sampling. Gas flow was set 24 h before the start of experimentation to ensure an equilibrium was reached within the two-stage regulator.
Fluorescent Imaging
Prior to the start of the live/dead assay, seeded cells were allowed to adhere for 24 h in a humidified incubator at 37 °C and 5% CO2 and culture media was replaced immediately before introduction to the flow system. The Biodome was then incubated in the flow system for durations spanning 0 and 4 days. Following growth in the flow system, live/dead fluorescent probes (Blue/Green ReadyProbes, Invitrogen) were added to the Biodome according to the manufacturer guidelines. In brief, 2 drops of the ready-to-use stain were added per millimeter, totaling 8 drops. The biodome was then placed in a humidified incubator at 37 °C and 5% CO2 for 30 min. After 30 min, the Biodome was imaged using Eclipse Ts2R microscope (Nikon, Minato City, Tokyo, Japan) with a Coolsnap Dyno CCD (Photometrics, Tucson, Arizona, USA) and 10x objective. A FITC filter (EX: 482 nm, EM: 536 nm; Nikon) was used to excite the live stain (Hoechst 33342) while a DAPI filter (EX: 390 nm, EM: 475 nm; Nikon) was used to visualize the dead stain. Fluorescent image processing and cell counting was performed using ImageJ.
To evaluate cell morphology, actin fluorescence staining was performed in live cells using CellMask™ Orange Actin Tracking Stain (Invitrogen). Cells were grown out in the specified incubation system for approximately 3 days until the confluency had reached 70-80%. 1000x stock solution of the actin tracking stain in DMSO was diluted to 1x in the RPMI1640 culture media described previously. Live/dead fluorescent probes were concurrently added to the staining media at a ratio of 2 drops per milliliter and cells were incubated for 30 min in a humidified incubator as previously described. Cells were rinsed twice with a wash buffer comprised of culture media previously equilibrated in a humidified incubator at 5% CO2 and 37 °C for 30 min. Cells were imaged using a Leica DMi8 (Leica, Wetzlar, Germany) microscope with DFC345 FX monochrome camera (Leica) and Texas Red (EX: 560 nm, EM: 645 nm), DAPI, and FITC filters to visualize the actin, dead cell nuclei, and live cell nuclei, respectively. Images were processed and fluorescent channels overlaid using ImageJ. All fluorescent images were captured using cells at passage <10 using a 20x objective.
RNA inhibition
Pooled siRNA oligonucleotides were purchased from Dharmacon (Lafayette, CO, USA) targeting CPT2 [siGENOME SMARTpool Human CPT2; 5’ – GGCAGAAGCUGAUGAGUAG – 3’, 5’ – UGGCAUACCUGACCAGUGA – 3’, 5’ – GGAAAGUGGACUCGGCAGU – 3’, 5’ - CAAGAGACUCAUACGCUUU] and a non-silencing (sham) control [siGENOME Non-Targeting siRNA Pool #1; 5’ – UAGCGACUAAACACAUCAA – 3’, 5’ – UAAGGCUAUGAAGAGAUAC – 3’, 5’ – AUGUAUUGGCCUGUAUUAG – 3’, 5’ – AUGAACGUGAAUUGCUCAA – 3’]. SK-OV-3 cells were grown in a 6-well format and transfected at 80% confluency with complexes consisting of 50 nM pooled RNAi probes and 2 µg/mL LipofectamineTM 2000 (Invitrogen) in Opti-MEM® I Reduced Serum Medium (ThermoFisher, Waltham, MA, USA). In the 6-well format, an extra well was allocated for direct seeding into the Biodome for VOC analysis. After 24 h in a humidified incubator at 37 °C and 5% CO2, the media was aspirated and the transfected cells were trypsinized. Approximately 500,000 cells were then seeded into the Biodome containing 5 mL of RPMI1640 with 10% dialyzed FBS and 1% Pen/Strep. The cells were allowed to adhere for roughly 1 h in an incubator before being transferred to the flow system. For the transient RNAi assay, media was replaced after 24 h, defining the 0-hour time point.
RNA Extraction
Following RNAi, cellular RNA was extracted using PureLink® RNA Mini Kit (Life Technologies, Carlsbad, CA, USA) according to manufacturer guidelines. In brief, fresh lysis buffer containing 1% 2-mercaptoethanol (Sigma Aldrich) was used to lyse the cells directly in the 6-well plate. Lysate was homogenized using 10 repeat passes through an 18-gauge needle (BD, Franklin Lakes, NJ). Binding, washing, and elution of purified RNA was performed according to kit specifications. Purity was assessed using absorbance values from a NanoDropTM One (ThermoFisher Scientific) and all samples were found to have A260/A280 ratios between 1.98 – 2.06. Samples were stored at –80 °C.
RT-qPCR
Purified RNA samples were thawed on ice and subject to reverse transcription using the QuantiTect Reverse Transcription Kit (Qiagen, Germantown, MD, USA). In brief, 100 ng of RNA was used as the starting weight. The genomic DNA elimination and reverse transcription reactions were prepared according to kit specifications. Reverse transcription reaction was allowed to proceed for a total of 15 min. Samples were stored for less than two weeks at –20 °C before proceeding to qPCR.
The resulting cDNA was then subject to quantitative PCR to determine expression of CPT2 and housekeeping gene GAPDH. Primers for CPT2 [RefSeq Accession: NM_000098; forward primer 5’ – TGGTTTATCTGCCCGTATGC – 3’ and reverse primer 5’ – TGCTCCAAGTACCATGGC – 3’] and GAPDH [RefSeq Accession: NM_002046; forward primer 5’ – ACATCGCTCAGACACCATG – 3’ and reverse primer 5’ – TGTAGTTGAGGTCAATGAAGGG – 3’] were purchased from IDT (Newark, NJ, USA). The qPCR reaction was prepared in a 96-well format (ThermoFisher Scientific) using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), according to manufacturer guidelines. In brief, a 20 uL reaction size was selected, using 500 nM of the forward and reverse primer, 1 uL of cDNA, 10 uL of 2x iTaq, and remaining volume balanced with nuclease-free H2O. 96-well plates were transferred to a qTOWER 2.0 instrument and expression quantified using the following cycling conditions: 50 °C for 2 min, 95 °C for 5 min, followed by 40 cycles of amplification at 95 °C for 15 s then 60 °C for 1 min. Annealing at 72 °C was allowed to proceed for 10 min, at which point the dissociation characteristics were assessed by ramping the temperature from 65 °C to 95 °C using a step size of 2 °C, equilibration time of 6 s, and heating rate of 0.5 °C/second. Independent biological replicates were analyzed in technical triplicate and averaged.
GC×GC-TOFMS
Thermal desorption tubes (Carbopack C, Carbopack B, and Carbosieve SIII, see Results) containing retained VOCs were analyzed using comprehensive GC×GC-TOFMS (Pegasus 4D®, LECO Corp. St. Joseph, MI), equipped with an autosampler (Multipurpose Sampler RoboticPro®, Gerstel Inc., Linthicum Heights, MD). TDTs were stored for a maximum of 8 days at 4°C to limit degradation49 and analyzed in order of collection. Volatiles were desorbed into the TDU 2 (Gerstel Inc.) inlet for a total of 4 min using variable temperature programming and splitless desorption mode. The TDU 2 inlet was initially held at 50 °C for 30 s after which a ramp rate of 700 °C/min was applied until the final temperature of 300°C was achieved. The inlet temperature was held at 300 °C for 3 min prior to the start of chromatographic analysis. The transfer temperature was maintained at 310 °C. The TDU 2 inlet was paired with a cooled injection system (CIS) to enhance analyte sensitivity. The CIS was initially maintained at –100 °C for the first 30 s of the run. The CIS temperature was then ramped at a rate of 12 °C/sec until the final temperature of 275 °C was achieved. The CIS was then maintained at 275 °C for 3 min.
The instrument was fitted with a two-dimensional column set consisting of a Rxi-624Sil MS (60 m × 250 µm × 1.4 µm; Restek®, Bellefonte, PA) first dimension column and a Stabilwax (1 m × 250 µm × 0.5 µm, Restek) second dimension column joined together by a press-fit connection (length × internal diameter × film thickness, respectively). The first-dimension column was set at an initial temperature of 50 °C and held for 2 min. The temperature was increased at a rate of 5 °C/min until reaching the target temperature of 230 °C, where a final hold time of 5 min was applied. The secondary oven was maintained at a + 5 °C offset relative to the primary oven. A quad-jet modulator was used with a 2 s modulation period and a + 15 °C offset relative to the secondary oven. The transfer line was maintained at 250 °C and the ion source at 200 °C for the duration of the run. Helium (UHP, 99.999%) was used as the carrier gas at a flow rate of 2 mL/min. Mass spectra were acquired at 100 Hz over a mass range of 35-300 Da with an ionization energy of -70eV. Prior to all sample analysis, a PFTBA standard was run to tune the mass spectrometer. Empty vials (blanks) were run at the start of each sample set to monitor the system for contamination. An alkane standard (C8-C20; Sigma Aldrich, St. Louis, MO) was sampled at the conclusion of testing for use in determination of retention indices.
Data Processing
Within individual sample chromatograms, subpeaks in the second dimension were required to meet a signal-to-noise ratio ≥6 to be combined. The signal-to-noise cutoff for peak selection was set at 20:1 for a minimum of two apexing masses. The baseline signal was drawn through the middle of the noise. To align peaks across chromatograms, the Statistical Compare software package in ChromaTOF® Version 4.72.0.0 (LECO Corp.) was utilized. Peaks were allowed to shift by a maximum of 18 s (9 modulation periods) in the first dimension and 0.25 s in the second dimension. The resulting aligned peaks were compared to the National Institute of Standards and Technology (NIST) 2011 Mass Spectral Library and tentative peak names were assigned if the spectral similarity score was ≥600 (60%). A secondary round of peak picking was performed on aligned chromatograms using a signal-to-noise threshold of 5 and a minimum spectral similarity ≥ 60%. Peaks with a spectral similar score that fell below the threshold were labeled ‘Analyte’. The resulting dataset was then manually filtered to remove poorly resolved peaks eluting before 400 s and known contaminants (siloxanes). Split peaks that fell outside the data processing thresholds were manually summed with first- and second-dimension retention times averaged. Quantitative values for signal abundance were obtained by integrating peaks areas using the unique ion mass.
VOC identities were assigned according to the metabolomic reporting standards established previously49. Compounds received an ID confidence level between 1 and 4, with 1 representing the highest confidence supported by two independent and orthogonal data sources49. In brief, compounds with mass spectral match ≥85%, to the NIST 2011 Mass Spectral Library, received an initial confidence level of 3. Compounds below this mass spectral threshold were assigned an ID level of 4 and labeled as unknown. The tentative VOC name and functional group is reported for all compounds with an ID level ≤3. A C8-C20 alkane standard was analyzed (Fig. S3) and used to assign retention indices for all VOCs. An ID level of 2 was the highest classification in this study, verified with a ≥85% mass spectral match and a retention index that is consistent with the mid-polar Rxi-624Sil stationary phase using the mean of published RIs, according to an approach previously established56,98. If mass spectral and polar second dimension chromatographic information supported the assignment of a functional group to a compound with an ID level of 4, then the compound was named by the functional group. The reverse library search was used to identify 3,5-dihydroxybenzamide and 3-octanone in the bacterial volatilome and pyrrole, furfural, aniline, phenol, and 1-dodecanol in the SK-OV-3 volatilome. When compared to the unperturbed SK-OV-3 volatilome, furfural abundance variability in the RNAi volatilome is attributed to alignment post-processing and retention time drift.
Statistics and Reproducibility
To focus on the reproducible aspects of the biological volatilomes, compounds variably present (>20% missing observations) in the experimental and control volatilomes were removed, unless otherwise indicated. Integrated peaks were log10 transformed in R, Version 4.0.3 (The R Foundation for Statistical Computing, Vienna, Austria) and missing values were imputed to 0 abundance. Pairwise, two-sided students’ t-tests were applied comparing biological and control VOC abundances across the sampling duration. Abundance observations were assumed to be unpaired, with unequal variance. The resulting p-values were then adjusted according to the Benjamini-Hochberg procedure51 using the p.adjust function in the base R stats library. All experiments were performed in biologically independent triplicate.
Sterilization and re-use Procedure
Following the conclusion of sampling and analysis in the Biodome, the TDT adapter was removed and submerged in a 100% ethanol bath for at least 1 h and subsequently sonicated for 2 min. Removal of the live culture is dependent on the organism considered. For microbiological analysis, a freshly prepared 10% bleach solution was added directly through the outlet of the Biodome and left at room temperature for 15 min. For adherent cell culture, spent media was aspirated and 5 mL of 0.25% Trypsin / 0.53 mM EDTA was added to the Biodome and incubated at 37 °C for 5-10 min to dissociate cells from the surface. Trypsinization was deemed a highly beneficial step to prevent accumulation of leftover cellular debris on the culture surface following autoclaving (later step). In both cases, the liquid culture was then emptied, and the Biodome was subject to two additional wash steps using bleach with vigorous lateral motion. Afterwards, the Biodome was flushed thoroughly with DI water and then autoclaved for 1 h. Following sterilization in the autoclave, the biodome was then placed in an acid bath (3 M HCl) overnight (≥10 h), although 3 h was found to be sufficient. Following the acid bath, the Biodome was flushed with DI water and transferred to a base bath (approximately 150 mM NaOH) for at least 3 h. Finally, to reduce noise from the culture vessel, the Biodome was flushed again with DI water, placed in a glass beaker, and heat treated for >12 h at 100 °C. The authors recommend treating the Biodome in the base bath second, to limit the detection of acid-related contaminants.
Reporting Summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Supplementary datasets 1–3 include all tentative VOCs associated with the flow system and culture media following alignment of experimental replicates using ChromaTOF. Supplementary dataset 4 reports the abundance of common contaminants in a direct comparison of in situ SPME and Biodome sampling methodologies. Supplementary dataset 5 includes all tentative VOCs identified after the inhibition of CPT2 expression in SK-OV-3 mammalian cells. Supplementary dataset 6 presents all tentative VOCs identified in the E. coli and LB Broth volatilomes. Supplementary videos show laminar flow in the Biodome culture vessel and multiplexed component under experimental conditions. All raw chromatographic and mass spectral data have been made publicly available in the MetaboLights99 repository at: https://www.ebi.ac.uk/metabolights/MTBLS8252. All relevant supplementary files not presented in the supplementary datasets, including raw chromatographic data, can be found at: https://figshare.com/authors/Jarrett_Eshima/13813720.
Code availability
Custom R code for the post-processing and statistical analysis of SK-OV-3 and E. coli volatilomes has been made publicly available in the Smith Lab Github repository (https://github.com/BSmithLab/Biodome).
References
Sulway, M. J. & Malins, J. M. Acetone in diabetic ketoacidosis. Lancet 296, 736–740 (1970).
Wang, Z. & Wang, C. Is breath acetone a biomarker of diabetes? A historical review on breath acetone measurements. J breath Res 7, 037109 (2013).
Williams, H. & Pembroke, A. Sniffer dogs in the melanoma clinic? Lancet 333, 734 (1989).
Sponring, A. et al. Release of volatile organic compounds from the lung cancer cell line NCI-H2087 in vitro. Anticancer Res 29, 419–426 (2009).
Abaffy, T., Möller, M. G., Riemer, D. D., Milikowski, C. & DeFazio, R. A. Comparative analysis of volatile metabolomics signals from melanoma and benign skin: a pilot study. Metabolomics 9, 998–1008 (2013).
Davis, T. J. et al. Pseudomonas aeruginosa volatilome characteristics and adaptations in chronic cystic fibrosis lung infections. Msphere 5, e00843–e00820 (2020).
Weisskopf, L., Schulz, S. & Garbeva, P. Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat Rev Microbiol 19, 391–404 (2021).
Bitas, V., Kim, H. S., Bennett, J. W. & Kang, S. Sniffing on microbes: diverse roles of microbial volatile organic compounds in plant health. Mol Plant-Microbe Interact 26, 835–843 (2013).
Song, G. C. et al. Plant growth-promoting archaea trigger induced systemic resistance in Arabidopsis thaliana against Pectobacterium carotovorum and Pseudomonas syringae. Environ Microbiol 21, 940–948 (2019).
Ditengou, F. A. et al. Volatile signalling by sesquiterpenes from ectomycorrhizal fungi reprogrammes root architecture. Nat Commun 6, 6279 (2015).
Becher, P. G. et al. Developmentally regulated volatiles geosmin and 2-methylisoborneol attract a soil arthropod to Streptomyces bacteria promoting spore dispersal. Nat Microbiol 5, 821–829 (2020).
Ponnusamy, L. et al. Identification of bacteria and bacteria-associated chemical cues that mediate oviposition site preferences by Aedes aegypti. Proc Natl Acad Sci 105, 9262–9267 (2008).
Schulz-Bohm, K. et al. The prey’s scent–volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J 11, 817–820 (2017).
Baldwin, I. T., Halitschke, R., Paschold, A., Von Dahl, C. C. & Preston, C. A. Volatile signaling in plant-plant interactions:” talking trees” in the genomics era. Science 311, 812–815 (2006).
Dudareva, N., Klempien, A., Muhlemann, J. K. & Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. N. Phytologist 198, 16–32 (2013).
Traxler, S. et al. VOC breath profile in spontaneously breathing awake swine during Influenza A infection. Sci Rep. 8, 14857 (2018).
Ebadzadsahrai, G., Higgins Keppler, E. A., Soby, S. D. & Bean, H. D. Inhibition of fungal growth and induction of a novel volatilome in response to Chromobacterium vaccinii volatile organic compounds. Front Microbiol 11, 1035 (2020).
Farag, M. A. et al. Biological and chemical strategies for exploring inter-and intra-kingdom communication mediated via bacterial volatile signals. Nat Protoc 12, 1359–1377 (2017).
Kesarwani, M. et al. A quorum sensing regulated small volatile molecule reduces acute virulence and promotes chronic infection phenotypes. PLoS Pathog 7, e1002192 (2011).
Kim, K. S., Lee, S. & Ryu, C. M. Interspecific bacterial sensing through airborne signals modulates locomotion and drug resistance. Nat Commun 4, 1809 (2013).
Schallschmidt, K. et al. Investigation of cell culture volatilomes using solid phase micro extraction: Options and pitfalls exemplified with adenocarcinoma cell lines. J Chromatogr B 1006, 158–166 (2015).
Jia, Z., Patra, A., Kutty, V. K. & Venkatesan, T. Critical review of volatile organic compound analysis in breath and in vitro cell culture for detection of lung cancer. Metabolites 9, 52 (2019).
Groenhagen, U., Maczka, M., Dickschat, J. S. & Schulz, S. Streptopyridines, volatile pyridine alkaloids produced by Streptomyces sp. FORM5. Beilstein J Org Chem 10, 1421–1432 (2014).
Thriumani, R. et al. A study on volatile organic compounds emitted by in-vitro lung cancer cultured cells using gas sensor array and SPME-GCMS. BMC cancer 18, 1–7 (2018).
Reese, K. L., Rasley, A., Avila, J. R., Jones, A. D. & Frank, M. Metabolic profiling of volatile organic compounds (VOCs) emitted by the pathogens Francisella tularensis and Bacillus anthracis in liquid culture. Sci Rep. 10, 1–7 (2020).
Krall, L., Huege, J., Catchpole, G., Steinhauser, D. & Willmitzer, L. Assessment of sampling strategies for gas chromatography–mass spectrometry (GC–MS) based metabolomics of cyanobacteria. J Chromatogr B 877, 2952–2960 (2009).
Bolling, C. & Fiehn, O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol 139, 1995–2005 (2005).
Franchina, F. A., Purcaro, G., Burklund, A., Beccaria, M. & Hill, J. E. Evaluation of different adsorbent materials for the untargeted and targeted bacterial VOC analysis using GC× GC-MS. Analytica Chim acta 1066, 146–153 (2019).
Schulz, S., Fuhlendorff, J. & Reichenbach, H. Identification and synthesis of volatiles released by the myxobacterium Chondromyces crocatus. Tetrahedron 60, 3863–3872 (2004).
Blom, D. et al. Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ Microbiol 13, 3047–3058 (2011).
Ahmed, W. M. et al. Development of an adaptable headspace sampling method for metabolic profiling of the fungal volatome. Analyst 143, 4155–4162 (2018).
Franchina, F. A., Zanella, D., Lazzari, E., Stefanuto, P. H. & Focant, J. F. Investigating aroma diversity combining purge-and-trap, comprehensive two-dimensional gas chromatography, and mass spectrometry. J Sep Sci 43, 1790–1799 (2020).
Ochiai, N., Tsunokawa, J., Sasamoto, K. & Hoffmann, A. Multi-volatile method for aroma analysis using sequential dynamic headspace sampling with an application to brewed coffee. J Chromatogr A 1371, 65–73 (2014).
Oliver-Pozo, C., Trypidis, D., Aparicio, R., García-González, D. L. & Aparicio-Ruiz, R. Implementing dynamic headspace with SPME sampling of virgin olive oil volatiles: optimization, quality analytical study, and performance testing. J Agric food Chem 67, 2086–2097 (2019).
Ha, J., Wang, Y., Jang, H., Seog, H. & Chen, X. Determination of E, E-farnesol in Makgeolli (rice wine) using dynamic headspace sampling and stir bar sorptive extraction coupled with gas chromatography–mass spectrometry. Food Chem 142, 79–86 (2014).
Maurer, D. L. et al. Screening of microbial volatile organic compounds for detection of disease in cattle: development of lab-scale method. Sci Rep. 9, 12103 (2019).
Traxler, S., Bischoff, A. C., Trefz, P., Schubert, J. K. & Miekisch, W. Versatile set-up for non-invasive in vitro analysis of headspace VOCs. J Breath Res 12, 041001 (2018).
Rosenthal, K. et al. Volatile atmospheric pressure chemical ionisation mass spectrometry headspace analysis of E. coli and S. aureus. Anal Methods 13, 5441–5449 (2021).
Bunge, M. et al. On-line monitoring of microbial volatile metabolites by proton transfer reaction-mass spectrometry. Appl Environ Microbiol 74, 2179–2186 (2008).
Lavra, L. et al. Investigation of VOCs associated with different characteristics of breast cancer cells. Sci Rep. 5, 1–2 (2015).
Silva, R. C., Aguiar, P. M. & Augusto, F. Coupling of dynamic headspace sampling and solid phase microextraction. Chromatographia 60, 687–691 (2004).
Dias, R. P. et al. Improved sample storage, preparation and extraction of blueberry aroma volatile organic compounds for gas chromatography. J Chromatogr Open 3, 100075 (2023).
Stefanuto, P. H. et al. Advanced method optimization for volatile aroma profiling of beer using two-dimensional gas chromatography time-of-flight mass spectrometry. J Chromatogr A 1507, 45–52 (2017).
Leemans, M., Bauër, P., Cuzuel, V., Audureau, E. & Fromantin, I. Volatile organic compounds analysis as a potential novel screening tool for breast cancer: a systematic review. Biomarker Insights, (2022).
Keppler, E. A., Jenkins, C. L., Davis, T. J. & Bean, H. D. Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC Trends Anal Chem 109, 275–286 (2018).
Filipiak, W. et al. A compendium of volatile organic compounds (VOCs) released by human cell lines. Curr Med. Chem 23, 2112–2131 (2016).
de Lacy Costello, B. et al. A review of the volatiles from the healthy human body. J breath Res 8, 014001 (2014).
Yamaguchi, M. S. et al. Headspace sorptive extraction-gas chromatography–mass spectrometry method to measure volatile emissions from human airway cell cultures. J Chromatogr B 1090, 36–42 (2018).
Sumner, L. W. et al. Proposed minimum reporting standards for chemical analysis: chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3, 211–221 (2007).
Harshman, S. W. et al. Storage stability of exhaled breath on Tenax TA. J breath Res 10, 046008 (2016).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Ser B (Methodol) 57, 289–300 (1995).
Kolb, H. et al. Ketone bodies: from enemy to friend and guardian angel. BMC Med 19, 1–15 (2021).
Rizzo, W. B. Fatty aldehyde and fatty alcohol metabolism: review and importance for epidermal structure and function. Biochim. et Biophys. Acta (BBA)-Mol Cell Biol Lipids 1841, 377–389 (2014).
Zakhari, S. Overview: how is alcohol metabolized by the body? Alcohol Res health 29, 245 (2006).
Walsh, C. T., Garneau-Tsodikova, S. & Howard-Jones, A. R. Biological formation of pyrroles: nature’s logic and enzymatic machinery. Nat Prod Rep. 23, 517–531 (2006).
Eshima, J. et al. Monitoring changes in the healthy female metabolome across the menstrual cycle using GC× GC-TOFMS. J Chromatogr B 1121, 48–57 (2019).
Drabińska, N., Starowicz, M. & Krupa-Kozak, U. Headspace solid-phase microextraction coupled with gas chromatography–mass spectrometry for the determination of volatile organic compounds in urine. J Anal Chem 75, 792–801 (2020).
Wang, Y., Li, Y., Yang, J., Ruan, J. & Sun, C. Microbial volatile organic compounds and their application in microorganism identification in foodstuff. TrAC Trends Anal Chem 78, 1–16 (2016).
Wishart, D. S. et al. HMDB 5. 0: the human metabolome database for 2022. Nucleic acids Res 50, D622–D631 (2022).
IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some chemicals that cause tumours of the urinary tract in rodents. Lyon (FR): International Agency for Research on Cancer; 2019. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 119.) 4, Mechanistic and Other Relevant Data. Available from: https://www.ncbi.nlm.nih.gov/books/NBK546961/.
Amal, H. et al. Detection of precancerous gastric lesions and gastric cancer through exhaled breath. Gut 65, 400–407 (2016).
Xu, Z. Q. et al. A nanomaterial-based breath test for distinguishing gastric cancer from benign gastric conditions. Br J cancer 108, 941–950 (2013).
Gomez-Arroyo, S. & Souza, S. V. In vitro and occupational induction of sister-chromatid exchanges in human lymphocytes with furfuryl alcohol and furfural. Mutat Res/Genet Toxicol 156, 233–238 (1985).
Dighe, S. U., Juliá, F., Luridiana, A., Douglas, J. J. & Leonori, D. A photochemical dehydrogenative strategy for aniline synthesis. Nature 584, 75–81 (2020).
Preti, G., Labows, J. N., Kostelc, J. G., Aldinger, S. & Daniele, R. Analysis of lung air from patients with bronchogenic carcinoma and controls using gas chromatography-mass spectrometry. J Chromatogr B: Biomed Sci Appl 432, 1–11 (1988).
Cho, T. M., Rose, R. L. & Hodgson, E. In vitro metabolism of naphthalene by human liver microsomal cytochrome P450 enzymes. Drug Metab Disposit. 34, 176–183 (2006).
O’neill, H. J., Gordon, S. M., O’neill, M. H., Gibbons, R. D. & Szidon, J. P. A computerized classification technique for screening for the presence of breath biomarkers in lung cancer. Clin Chem 34, 1613–1618 (1988).
Ibrahim, S. R. & Mohamed, G. A. Naturally occurring naphthalenes: chemistry, biosynthesis, structural elucidation, and biological activities. Phytochem. Rev 15, 279–295 (2016).
Longo, V. et al. In vitro profiling of endothelial volatile organic compounds under resting and pro-inflammatory conditions. Metabolomics 15, 1–16 (2019).
Filipiak, W. et al. TD-GC-MS analysis of volatile metabolites of human lung cancer and normal cells in vitro. Cancer Epidemiol, Biomark Prev 19, 182–195 (2010).
Rodrigues, D. et al. Volatile metabolomic signature of bladder cancer cell lines based on gas chromatography–mass spectrometry. Metabolomics 14, 1–15 (2018).
Schivo, M. et al. Volatile emanations from in vitro airway cells infected with human rhinovirus. J breath Res 8, 037110 (2014).
Kwak, J. et al. Volatile biomarkers from human melanoma cells. J Chromatogr B 931, 90–96 (2013).
Cai, C. M., Zhang, T., Kumar, R. & Wyman, C. E. Integrated furfural production as a renewable fuel and chemical platform from lignocellulosic biomass. J Chem Technol Biotechnol 89, 2–10 (2014).
Adebesin, F. et al. Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science 356, 1386–1388 (2017).
Liao, P. et al. Emission of floral volatiles is facilitated by cell-wall non-specific lipid transfer proteins. Nat Commun 14, 330 (2023).
Newton, W. A. & Snell, E. E. Catalytic properties of tryptophanase, a multifunctional pyridoxal phosphate enzyme. Proc Natl Acad Sci 51, 382–389 (1964).
Newton, W. A., Morino, Y. & Snell, E. E. Properties of crystalline tryptophanase. J Biol Chem 240, 1211–1218 (1965).
Li, G. & Young, K. D. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159, 402–410 (2013).
Dixon, B., Ahmed, W. M., Mohamed, A. A., Felton, T. & Fowler, S. J. Metabolic phenotyping of acquired ampicillin resistance using microbial volatiles from Escherichia coli cultures. J Appl Microbiol 133, 2445–2456 (2022).
Bos, L. D., Sterk, P. J. & Schultz, M. J. Volatile metabolites of pathogens: a systematic review. PLoS Pathog 9, e1003311 (2013).
Wang, D., Ding, X. & Rather, P. N. Indole can act as an extracellular signal in Escherichia coli. J Bacteriol 183, 4210–4216 (2001).
Di Martino, P., Fursy, R., Bret, L., Sundararaju, B. & Phillips, R. S. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol 49, 443–449 (2003).
Chimerel, C., Field, C. M., Piñero-Fernandez, S., Keyser, U. F. & Summers, D. K. Indole prevents Escherichia coli cell division by modulating membrane potential. Biochim. et Biophys. Acta (BBA)-Biomembr. 1818, 1590–1594 (2012).
Hirakawa, H., Inazumi, Y., Masaki, T., Hirata, T. & Yamaguchi, A. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol Microbiol 55, 1113–1126 (2005).
Sousa, M. et al. Volatilomes reveal specific signatures for contamination of leafy vegetables with Escherichia coli O157: H7. Food Control 146, 109513 (2023).
Almeida, O. A. et al. Bacterial volatile organic compounds (VOCs) promote growth and induce metabolic changes in rice. Front Plant Sci 13, 1056082 (2023).
Schulz, S. & Dickschat, J. S. Bacterial volatiles: the smell of small organisms. Nat Prod Rep. 24, 814–842 (2007).
Plyuta, V. A., Popova, A. A., Koksharova, O. A., Kuznetsov, A. E. & Khmel, I. A. The ability of natural ketones to interact with bacterial quorum sensing systems. Mol Genet, Microbiol Virol 29, 167–171 (2014).
Yu, K., Hamilton-Kemp, T. R., Archbold, D. D., Collins, R. W. & Newman, M. C. Volatile compounds from Escherichia coli O157: H7 and their absorption by strawberry fruit. J Agric food Chem 48, 413–417 (2000).
Rees, C. A. et al. Volatile metabolic diversity of Klebsiella pneumoniae in nutrient-replete conditions. Metabolomics 13, 1–11 (2017).
Devaraj, H., Pook, C., Swift, S., Aw, K. C. & McDaid, A. J. Profiling of headspace volatiles from Escherichia coli cultures using silicone-based sorptive media and thermal desorption GC–MS. J Sep Sci 41, 4133–4141 (2018).
Jünger, M. et al. Ion mobility spectrometry for microbial volatile organic compounds: a new identification tool for human pathogenic bacteria. Appl Microbiol Biotechnol 93, 2603–2614 (2012).
Tait, E., Perry, J. D., Stanforth, S. P. & Dean, J. R. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC–MS. J chromatographic Sci 52, 363–373 (2014).
Drabińska, N. et al. Application of a solid-phase microextraction-gas chromatography-mass spectrometry/metal oxide sensor system for detection of antibiotic susceptibility in urinary tract infection-causing Escherichia coli–a proof of principle study. Adv Med Sci 67, 1–9 (2022).
Yan, Q. et al. Metabolic engineering of β-oxidation to leverage thioesterases for production of 2-heptanone, 2-nonanone and 2-undecanone. Metab Eng 61, 335–343 (2020).
Hummadi, E. H. et al. Antimicrobial volatiles of the insect pathogen Metarhizium brunneum. J Fungi 8, 326 (2022).
Bean, H. D., Rees, C. A. & Hill, J. E. Comparative analysis of the volatile metabolomes of Pseudomonas aeruginosa clinical isolates. J breath Res 10, 047102 (2016).
Haug, K. et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic acids Res 48, D440–D444 (2020).
Acknowledgements
The authors would like to acknowledge Drs. Heather Bean and Trenton Davis in the School of Life Sciences at Arizona State University for their project input and initial discussions as to design feasibility. The authors would like to give a special thanks to Christine Roeger for her glassblowing expertise and fabrication of the Biodome culture vessels utilized in this study.
Author information
Authors and Affiliations
Contributions
J.E. helped design the tool, analyzed all samples by GC×GC-TOFMS, performed the knockdown of CPT2 and transformation of E. coli, developed the code to perform statistical analysis of the results, and wrote the manuscript. T.R.P. cultured ampicillin-resistant E. coli, collected all bacterial volatilome samples, performed the knockdown of CPT2, and edited the manuscript. Y.A. performed the transient inhibition assay following CPT2 knockdown, collected RNAi volatilome samples, analyzed gene expression by RT-qPCR, and helped write the manuscript. A.P.O. supported the collection of samples from various experiments, performed the manual assessment of mass spectral data, and helped write the manuscript. J.F.L. played a key role in the design of the tool, established early methodology, and reviewed the manuscript. B.D.A. performed the characterization of fluid flow in the Biodome using ANSYS Fluent®, performed live-dead fluorescence assays, collected morphological images following live actin staining, and reviewed the manuscript. E.M. developed high-quality CAD renders and schematics of the system and tool, fabricated the custom tray device to enable imaging in the tool, and reviewed the manuscript. C.M. contributed to early methodological development used throughout the study, developed code for determination of chemical functional group, and reviewed the manuscript. P.P. and C.A. performed live-dead fluorescence assays, assisted with RNA inhibition experiments, and reviewed the manuscript. B.S.S. devised the idea, designed the tool and coordinated fabrication by glassblowing, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.E., J.F.L., and B.S.S. hold a non-provisional patent, filed by Arizona State University, related to the device described in the manuscript. Non-Provisional US Patent Application No. 17/715,352, “Devices and Systems for Non-Destructive Collection and Monitoring of Biological Volatiles”. All other authors declare no competing interests.
Peer review
Peer review information
Communications Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: [Mengying Su, Saleem Denholme]. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Eshima, J., Pennington, T.R., Abdellatif, Y. et al. An engineered culture vessel and flow system to improve the in vitro analysis of volatile organic compounds. Commun Eng 4, 162 (2025). https://doi.org/10.1038/s44172-025-00364-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44172-025-00364-y