Abstract
We present an open-source code for 3D super-resolution ultrasound imaging. Open-3DULM was applied to transcranial imaging in habituated awake mice. Comparative analysis reveals that isoflurane anesthesia induces significant vasodilation and increased cerebral blood flow (CBF) in veins, while only a subset of arteries exhibits these effects compared to the awake state. This method could serve as a reference for developing new types of 3D vascular quantifications, particularly emphasizing the importance of awake and 3D imaging for accurate cerebral blood flow measurement in neuroscience research.
Similar content being viewed by others
Introduction
Ultrasound localization microscopy (ULM) is the leading technique for super-resolution ultrasound imaging1,2,3,4,5 achieving spatial resolutions down to 10 microns. ULM has been widely used to visualize microvascular networks within a variety of organs, including the human brain6, heart7, and kidneys8,9,10. ULM is mostly implemented in 2D which represents various limitations such as user dependency, out of plane motion and imprecise quantification11. Volumetric ULM, or 3D ULM12,13,14, allows appropriate quantification of microbubbles15 and tissue motion, along with organ-wide acquisitions within the time of a single contrast agent’s bolus (minutes).
To prevent pain, stress, and motion artifacts in animal imaging, animals are routinely scanned with various imaging modalities, including ULM, under anesthesia, such as isoflurane. This anesthetic causes vasodilation and altering flows16. Natural physiology, stress response, and neuronal plasticity are altered with anesthesia, hindering the translation of functional imaging17,18. Consequently, the range of relevant biomarkers that could be faithfully extracted from ULM is reduced. The effects of isoflurane-induced anesthesia on cerebral blood flow (CBF) have been extensively studied across various imaging modalities by comparing the anesthetized and awake states. Techniques such as Optical Coherence Tomography have demonstrated significant vasodilation in blood vessels under isoflurane19, albeit with limitations in depth penetration and field of view due to the inherent properties of optical imaging. Similarly, two-photon microscopy has revealed reductions in CBF through measurements of red blood cell flux20, but this method is also invasive, limited by shallow imaging depth, and requires a craniectomy. On the other hand, Magnetic Resonance (MR) techniques, like Arterial Spin Labeling, provide a non-invasive, volumetric approach to assess CBF21, but suffer from poor spatial resolution, particularly at the microvascular level, where precise quantification is critical. Previous work using 2D ULM has demonstrated its potential for awake-state imaging22. This study has relied on initial anesthesia to immobilize the animals for setup, followed by a waiting period for the animal to awaken. However, this approach introduces uncertainty regarding the duration needed for the anesthesia’s effects to fully dissipate, which could significantly impact the accuracy of the awake-state imaging. Additionally, the protocol required invasive procedures such as craniectomies, further complicating their clinical and experimental applicability. Furthermore, the two-dimensional nature of these studies limits their ability to capture the complexity of vascular networks, especially when compared to the whole-brain volumetric coverage needed for comprehensive microvessel characterization. To address the limitations of previous approaches, we introduce an adapted protocol for 3D transcranial ULM on habituated, awake mice under stress-free conditions, eliminating the need for anesthesia prior to imaging22. This method enables high-resolution quantification of microvascular parameters such as vessel diameter and blood flow velocity at the micron scale. The final resolution was 23 ± 2 μm, measured by the Fourier Shell Correlation23. By comparing the results of awake ULM to those obtained from the same mice under isoflurane anesthesia, our approach bridges the gap between optical and MR-based techniques, offering the advantages of non-invasive, deep tissue imaging with high spatial resolution and whole-brain coverage.
Moreover, we also report on Open-3DULM, an openly available algorithm for a full 3D ULM reconstruction from beamformed 3D ultrasound data. Previously, an open-source reconstruction algorithm of 2D ULM has been developed and exploited by several laboratories (PALA24). To extend in the third dimension and adapt to the new challenges linked to data size, 3D localization, and 3D tracking, we propose a Python algorithmic pipeline accessible to a wide range of users. Additionally, we provide beamformed data from brain acquisitions and results to help the users compare to their home-built 3DULM pipeline. Open-3DULM was also optimized for data processing speed through CPU parallelization.
Results
We performed in-vivo 3D transcranial ULM of the mice brain in awake and anesthetized states (N = 5 per group). Figure 1a shows the schematic of the experimental setup (the detailed awake protocol can be found in the “Methods” section). Habituation was performed during four consecutive days before the 3D ULM experiment to ensure a stress-free environment for awake imaging. The mice were first placed in a body harness, and a 3D-printed mounting plate was attached to the skull with adhesive, allowing it to be securely fastened into the stereotaxic frame, which ensures no head motion for subsequent imaging. As highlighted in a previous study25, a 4-day habituation period was enough to ensure that the mice return to a stress-free state and were then prepared for imaging experiments. imaging experiments. To avoid the need for isoflurane anesthesia on the day of the imaging experiment, which could interfere with awake imaging, the catheter was inserted into the tail vein 1 day prior to the experiment and secured with adhesive tape. On the day of the imaging experiment, the head was mounted on the stereotaxic frame, an incision is made to open the skin, and saline and ultrasound gel are applied to the exposed skull. Mice first underwent 3D ULM acquisition in an entire awake state for 5 continuous minutes at a volume rate of 250 Hz. This acquisition was followed by another 3D ULM acquisition in an isoflurane-anesthetized state for 5 min. The Open-3DULM was applied on 3D ultrasound data that were acquired and beamformed into Brightness mode (B-mode) with a delay and sum algorithm26, i.e., pixel size of [98.5, 150, 150] micrometers in the dorsal-ventral, left-right, and posterior-anterior directions, respectively. Beyond the dataset shared with this article (section “Code and Data availability”), the shared algorithmic pipeline consisted of several steps that can be applied to any 3D ultrasound data acquired in-silico, in-vitro, and in-vivo with microbubble injection. First, each volume sequence underwent filtering to eliminate global tissue motion and skull signals and enhance microbubble signals. Tissue clutter removal was achieved by removing the first 12 among 200 singular vectors using singular value decomposition (SVD) on beamformed 3D data1,27. The localization of the microbubbles was done by extracting the maximum intensity of the image, filtering them by the Full Width Half Maximum (FWHM), the number of local maximum and Signal to Noise Ratio (SNR). Then the sub-wavelength localization of the microbubbles was performed through radial symmetry algorithm24. Finally, the tracking step used for monitoring individual microbubbles is an open-source classical particle tracking algorithm ’simpletracker’28 adapted to Open-3DULM. After generating the tracks of individual microbubbles, 3D density i.e., count of mcirobubbles per pixel and mean velocity maps were created for visualization and quantification purposes. These maps enable the measurement of various 3D ULM metrics, which can be used to assess a range of physiological and pathological biomarkers.
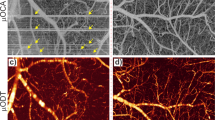
Figure 2a, b show the 3D ULM density rendering of the brain microvasculature in awake (left) and isoflurane (right) states with the same track number and a zoom on the right upper cortex in Fig. 2c, d. Skeletonization of the vessels was also performed to facilitate the measurements of their diameter. As shown in Fig. 2e (awake) and f (isoflurane), most vessels in the whole brain were enlarged under anesthesia, as already highlighted in previous studies16,29. Finally, 3D ULM velocity maps were also projected on the vessel skeletons. Faster blood velocities were observed in the isoflurane state (Fig. 2h) compared to awake (Fig. 2g). Overall, the increase in vessel diameter and blood velocity indicated an overall elevation in CBF consistent with previous findings using 2D ULM22.
For quantification, the vessel area was defined as the number of voxels occupied by a detectable vessel, determined by the presence of microbubbles passing through those voxels30. This metric showed significant differences between the two groups (p ≪ 0.01), i.e., a lower vessel area for awake 3DULM (Fig. 3a–c). Vessel segmentation was achieved thanks to the identification of key vascular structures by two radiologists. Thanks to the Open-3DULM capabilities (Fig. 4), selecting a major vessel automatically included all connected branching vessels irrespective of their spatial conformation. Figure 3 highlights various regions of interest (ROIs), including arteries and veins identification. For each ROI, we presented the Open-3DULM density map alongside the skeletonized data, detailing the diameter and velocity of each branch. Additionally, two boxplots were generated per vessels region to compare the average branch diameter and velocity between the awake and isoflurane-anesthetized states. The results revealed that all selected veins exhibit significantly reduction in vessel size and velocity in the awake state (purple) compared to under isoflurane (blue) (Region 1, 3, and 5 upward). Interestingly, unlike the 2D ULM study22, which found no significant changes in arterial measurements, our 3D analysis showed that some arteries also experienced reductions (region 3). Finally, Fig. 5 demonstrated the robustness of our findings, showing consistent results for vessel size (Fig. 5a) and average velocity (Fig. 5b) across five different mice.
a, b vessel diameter maps with definition of the different regions. c Boxplot of vessel area metric showing greater vessel occupancy in isoflurane state. Five regions of interest include the cortex with arteries (red, downward flow) and veins (blue, upward flow). For each ROI, 3D ULM density maps, vessel diameters, and velocities are displayed. Boxplots for each ROI depict the average radius and velocity of branches, comparing awake (purple) and isoflurane (cyan) states. Lhiv (longitudinal hippocampal vein); Dthv (dorsal thalamus vein); Thpv (thalamoperforating vein); Ach (anterior choroidal artery); Dlth (dorsolateral hypothalamus); MCA (middle cerebral artery).
The watershed-like 3D vascular structure of Open-3DULM enables automatic segmentation of vessels, identifying the connections between branches and the main vessel. a Region 3 (Ach: anterior choroidal artery); Dlth (dorsolateral hypothalamus) and (b) Region 4 (MCA: middle cerebral artery). The identification of the MCA and ACH was performed by two radiologists.
Overall, experiments on awake habituated animals showed drastic differences in blood flow patterns (size and speed) in the animal brain concerning their anesthetized counterpart. These differences were effectively captured using Open-3DULM, which optimizes quantification in the preclinical setting. The Open-3DULM has the potential to serve as a universal platform, enabling the conversion of any 3D ultrasound data acquired with contrast agents into super-resolution ultrasound volumes.
Methods
Animals
All experiments were performed in compliance with the European Community Council Directive of September 22, 2010 (010/63/UE) with ARRIVE guidelines and approved by the protocol APAFIS #35158 validated by the French ethics committee “Comité d’éthique Normandie en matière d’expérimentation animale”. Accordingly, the number of animals in our study was kept to the minimum following the 3Rs (reduce, refine, and replace) guidelines. Experiments were performed on N = 5 male (aged 5–7 weeks) Swiss mice (Janvier Labs), weighing 30–40 g at the beginning of the experiments. Animals (housed two per cage) arrived in the laboratory 1 week before the start of the experiment. They were maintained under controlled conditions (21 °C, 50–60% relative humidity, 12/12h light/dark cycle, food and water ad libitum). All animals included in this study were untreated and were used randomly in the various experiments.
Awake protocol condition
Skull cap fixation
Mice were first anesthetized with isoflurane (induction at 5%, maintenance between 1.5% and 2%) in a gas mixture of N2O/O2 (70%/30%) and given buprenorphine (0.1 mg/kg, subcutaneous) for pain relief. The animals were then placed in a stereotaxic frame, keeping their temperature constant at 37 °C using a temperature-controlled heating pad. After cleaning and locally anesthetizing the area with Xylocaine (subcutaneous), an incision was made to expose the skull for the attachment of the skull cap (Fig. 1a). The skull cap, a 3D-printed PLA structure with an imaging window of 2 cm × 1.5 cm, was then affixed. The skull surface was treated with hydrogen peroxide and 70% ethanol, followed by a primer which was rinsed with saline. The skull cap was secured in place using dental cement25.
Habituation
After a 3-day recovery period, a 4-day habituation protocol was implemented to ensure the mice remained still during the ultrasound imaging sessions. Each day, the mice were placed in the body harness for progressively longer duration to acclimate them to the restraint conditions. We selected a 4-day habituation period based on prior research involving functional magnetic resonance imaging in mice25. In this study, they demonstrated that after 4 days, mice exhibited normalized heart rates, respiratory rates, and corticosterone levels, indicating a return to a stress-free state.
Catheter placement
To avoid the effects of isoflurane anesthesia on ultrasound imaging, the catheter was placed a day in advance. The day before ultrasound imaging, the mice were anesthetized with isoflurane (5% induction, 1.5–2% maintenance) in a gas mixture of N2O/O2 (70%/30%). They were given buprenorphine (0.05 mg/kg, subcutaneous) for pain relief and then positioned in a stereotaxic frame within their body harness. A regulated heating pad kept their body temperature stable at 37 °C. A catheter was then inserted into the tail vein and secured with adhesive tape to enable intravenous (IV) injections of microbubbles.
Preparation for imaging
The animals were secured in a small animal stereotaxic frame (David Kopf stereotaxic instrument, Tujunga, CA, USA). Imaging was first performed on awake animals, followed by applying an anesthesia mask delivering a mixture of isoflurane (2%) and nitrous oxide. Heart and respiratory rates were continuously monitored to maintain stable anesthesia (Labchart, AD Instruments). Two milliliters of saline were gently applied to the brain, preserving the dura mater’s integrity, and followed by ultrasound gel (Neo Jelly US, France). The ultrasonic probe was then precisely positioned using the stereotaxic electrode manipulator (0.1 mm resolution, David Kopf stereotaxic instrument) fitted with a custom 3D-printed probe holder.
Microbubble contrast agent injections
During the 5-min imaging period, 25 μL of SonoVue microbubbles, prepared using the standardized clinical protocol, were injected into the tail vein catheter every minute, resulting in a total volume of 150 μL. To prevent the microbubbles from floating to the top of the syringe, gentle agitation was achieved using two magnets, one placed inside the syringe and one outside. If the microbubbles solution became clear, the syringe was replaced at least once during the imaging session.
3D ultrasound imaging
Ultrasound data was acquired using a single Verasonics 256-channel research scanner (Kirkland, USA) coupled with a 1024-elements matrix probe from Vermon (Tours, France) with a central frequency of 7.8 MHz and 56% bandwidth at −6 dB. The probe connected to the Vantage 256 system through a Verasonics UTA 1024-MUX adapter. The imaging protocol employed an ultrafast sequence of five plane waves, oriented at ±5∘ in both elevation and lateral directions, with a maximum pulse repetition frequency (PRF) of 16 kHz. Each pulse was a 2-cycle emission at 67% duty cycle, driven at 10 volts, producing a Peak Negative Pressure (PNP) of 298 kPa and a mechanical index (MI) of 0.1. The probe was divided into four 256-element panels, and during transmission, each panel received signals from its neighboring panels, requiring ten emissions per angle13. Five hundred sequences, each containing two hundred continuous volumes at 250 Hz, were recorded over ~4 min, resulting in about 250 GB of data. Each block required ~0.5 s to be saved continuously. The acquired volumes were sampled with a 50% bandwidth.
3D beamforming
The data were then reconstructed with a classical delay and sum beamforming26 on a [98.5, 150, 150] μm grid, before reaching the final 3D ULM resolution of [9.85, 9.85,9.85] μm. The final ULM resolution was 23.2 +- 2 µm (see Fig. 6).
Statistical analysis
Statistical analyses were conducted to compare brain regions between awake and isoflurane-anesthetized states, utilizing a two-way ANOVA in GraphPad Prism 9 software. The model included an interaction term to assess the effects of condition, ROI, and their interaction. Intra-animal data dependencies were accounted for by by treating animal ID as a repeated measure. The significance levels of the findings are reported as follows: ns = P ≥ 0.05, ⋆ = P ≤ 0.05, ⋆⋆ = P ≤ 0.01, ⋆⋆⋆ = P ≤ 0.001, ⋆⋆⋆⋆ = P ≤ 0.0001.
Open 3DULM: algorithm
Clutter filter
After obtaining a 3D ultrasound data and beamformed into Brightness mode (B-mode) volumes, the first step of the Open 3DULM algorithm is to remove the tissue signal, tissue motion as well as the skull signal to enhance microbubbles signal. Clutter filtering was performed using the SVD technique1,27. The user could specify the number of singular vectors to remove. In this study, we removed 12 out of 200. This threshold was empirically determined and applied uniformly across all animals.
Localization
The second step in the Open-3DULM algorithm was the localization of the Point Spread Function (PSF) of the microbubbles. We specified the number of microbubbles (\({}^{{\prime} }number\,\_of\_\,particle{s}^{{\prime} }\)) present in one volume at a specific frame. In this work, we set this parameter to 900. The 3D full-width-half-maximum (“fwhm”) of the PSF for the microbubbles was defined as fwhm = [5 × 5 × 5] as data was beamformed at half of a wavelength in all directions. We used fwhm = [5 × 5 × 5]. After selecting the number of microbubbles per frame and the PSF size, the algorithm localized the regional maxima of the filtered data (3D+time) by applying the “maximumFilter” function from “scipy.ndimage”, arranging the pixels from the highest intensity value to the lowest. To ensure that the high-intensity pixels were likely to be microbubbles, the algorithm computed the local Signal-to-Noise Ratio (SNR) within a user-defined patch size (“patch_size”). In this work, the patch size was set to [7 × 7 × 7] to consider an area larger than the microbubble spot (fwhm). We also specified the minimum SNR (“min_snr”) for an accepted microbubble PSF and the number of local maxima within the chosen patch (“nb_local_max”). The algorithm selects the (\({}^{{\prime} }number\,\_of\_\,particle{s}^{{\prime} }\)) meeting the minimum SNR and (“nb_local_max”) conditions, starting with the localized microbubbles with the highest SNR.
Sub-wavelength localization
For each localized PSF within the patches, 3D radial symmetry12 was employed to pinpoint the microbubble’s position with sub-wavelength precision. A “localization” matrix was then generated, containing five columns: the local SNR of the microbubble, the z, x, and y positions, and the frame number.
3D tracking
Once the subwavelength localization step was completed, the “localization” matrix was sent to the 3D tracking final step in the Open-3DULM algorithm. The 3D tracking algorithm is a library Python-adapted (“PeasyTracker”) version of the open-source “SimpleTracker”28. It uses the Hungarian algorithm to minimize the Euclidean distance between points in consecutive frames. Table 1 summarizes the localization and tracking parameters, their explanations, and the values used in this work. The interpolation step was done using the “interp” function from the Numpy library with an interpolation factor of 1 over the resolution factor ’res’ that should be defined by the user. Here, we used a factor of 10 (res = 10). Finally, the “tracks” structure was then computed with all the tracks, i.e., the raw and the interpolated ones.
Open 3DULM: Image analysis
Once the tracks are generated, they were used to create a 3DULM density rendering (output_density), which was a 3D matrix where each voxel’s value represents the number of times a track passed through that voxel. Additionally, the tracks were used to create 3DULM velocity maps, including the norm of the 3D velocity (output_velocity) and the velocity with directionality (output_directivity), indicating upward and downward velocity direction in the axial z direction. Open-3DULM rendering were computed by loading the output_density into Amira (Amira 2019 software, FEI) and using the Volume Rendering function for 3DULM density rendering. To skeletonize the vasculature31, a Gaussian filter (σ = [1, 1, 1] px) was first applied, followed by the “auto-skeleton” function in Amira (Fig. 3e, f). For 3D velocity rendering, output_velocity was loaded into Amira and used as a colormap on the skeletonized structure (Fig. 3g, h).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
370 blocks of B-mode 3D cineloop for awake state was shared with this publication. The data are available at https://zenodo.org/records/14289690. Due to the large size of the dataset (2.5 TB), the remaining data will be shared upon request.
Code availability
All codes are available under the Open-3DULM pipeline presented on a GitHub repository (https://github.com/Lab-Imag-Bio/Open-3DULM).
References
Errico, C. et al. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature 527, 499–502 (2015).
Christensen-Jeffries, K. et al. Super-resolution ultrasound imaging. Ultrasound Med. Biol. 46, 865–891 (2020).
Couture, O., Hingot, V., Heiles, B., Muleki-Seya, P. & Tanter, M. Ultrasound localization microscopy and super-resolution: A state of the art. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 65, 1304–1320 (2018).
Song, P., Rubin, J. M. & Lowerison, M. R. Super-resolution ultrasound microvascular imaging: Is it ready for clinical use? Z. Med. Phys. 33, 309–323 (2023).
Dencks, S. & Schmitz, G. Ultrasound localization microscopy. Z. Med. Phys. 33, 292–308 (2023).
Demené, C. et al. Transcranial ultrafast ultrasound localization microscopy of brain vasculature in patients. Nat. Biomed. Eng. 5, 219–228 (2021).
Yan, J. et al. Transthoracic ultrasound localization microscopy of myocardial vasculature in patients. Nat. Biomed. Eng. 8, 689–700 (2024).
Bodard, S. et al. Ultrasound localization microscopy of the human kidney allograft on a clinical ultrasound scanner. Kidney Int. 103, 930–935 (2023).
Denis, L. et al. Sensing ultrasound localization microscopy for the visualization of glomeruli in living rats and humans. EBioMedicine 91, 104578 (2023).
Bodard, S. et al. Visualization of renal glomeruli in human native kidneys with sensing ultrasound localization microscopy. Investig. Radiol. 59, 561–568 (2024).
Naji, M. A., Taghavi, I., Thomsen, E. V., Larsen, N. B. & Jensen, J. A. Underestimation of flow velocity in 2-d super-resolution ultrasound imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 71, 1844–1854 (2024).
Heiles, B. et al. Volumetric ultrasound localization microscopy of the whole rat brain microvasculature. IEEE Open J. Ultrason. Ferroelectr. Freq. Control 2, 261–282 (2022).
Chavignon, A. et al. 3d transcranial ultrasound localization microscopy in the rat brain with a multiplexed matrix probe. IEEE Trans. Biomed. Eng. 69, 2132–2142 (2021).
Chabouh, G. et al. Whole organ volumetric sensing ultrasound localization microscopy for characterization of kidney structure. IEEE Trans. Med. Imaging 43, 4055–4063 (2024).
Chavignon, A. et al. Deep and complex vascular anatomy in the rat brain described with ultrasound localization microscopy in 3d. IEEE Open J. Ultrason. Ferroelectr. Freq. Control 3, 203–209 (2023).
Sullender, C. T., Richards, L. M., He, F., Luan, L. & Dunn, A. K. Dynamics of isoflurane-induced vasodilation and blood flow of cerebral vasculature revealed by multi-exposure speckle imaging. J. Neurosci. Methods 366, 109434 (2022).
Maekawa, T., Tommasino, C., Shapiro, H. M., Keifer-Goodman, J. & Kohlenberger, R. W. Local cerebral blood flow and glucose utilization during isoflurane anesthesia in the rat. Anesthesiology 65, 144–151 (1986).
Miller, R. D. et al. Miller’s Anesthesia E-Book (Elsevier Health Sciences, 2014).
Rakymzhan, A., Li, Y., Tang, P. & Wang, R. K. Differences in cerebral blood vasculature and flow in awake and anesthetized mouse cortex revealed by quantitative optical coherence tomography angiography. J. Neurosci. Methods 353, 109094 (2021).
Lyons, D. G., Parpaleix, A., Roche, M. & Charpak, S. Mapping oxygen concentration in the awake mouse brain. Elife 5, e12024 (2016).
Li, C.-X., Patel, S., Auerbach, E. J. & Zhang, X. Dose-dependent effect of isoflurane on regional cerebral blood flow in anesthetized macaque monkeys. Neurosci. Lett. 541, 58–62 (2013).
Wang, Y. et al. Longitudinal awake imaging of deep mouse brain microvasculature with super-resolution ultrasound localization microscopy. eLife 13, RP95168 (2024).
Hingot, V., Chavignon, A., Heiles, B. & Couture, O. Measuring image resolution in ultrasound localization microscopy. IEEE Trans. Med. Imaging 40, 3812–3819 (2021).
Heiles, B. et al. Performance benchmarking of microbubble-localization algorithms for ultrasound localization microscopy. Nat. Biomed. Eng. 6, 605–616 (2022).
Tsurugizawa, T. et al. Awake functional mri detects neural circuit dysfunction in a mouse model of autism. Sci. Adv. 6, eaav4520 (2020).
Perrot, V., Polichetti, M., Varray, F. & Garcia, D. So you think you can das? a viewpoint on delay-and-sum beamforming. Ultrasonics 111, 106309 (2021).
Desailly, Y. et al. Contrast enhanced ultrasound by real-time spatiotemporal filtering of ultrafast images. Phys. Med. Biol. 62, 31 (2016).
Tinevez, J.-Y. et al. Trackmate: an open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017).
Slupe, A. M. & Kirsch, J. R. Effects of anesthesia on cerebral blood flow, metabolism, and neuroprotection. J. Cereb. Blood Flow. Metab. 38, 2192–2208 (2018).
Hingot, V. et al. Microvascular flow dictates the compromise between spatial resolution and acquisition time in ultrasound localization microscopy. Sci. Rep. 9, 2456 (2019).
Demeulenaere, O. et al. Coronary flow assessment using 3-dimensional ultrafast ultrasound localization microscopy. Cardiovasc. Imaging 15, 1193–1208 (2022).
Acknowledgements
We thank Oscar Demeulenaere for his help regarding skeletenization on AMIRA software.
Author information
Authors and Affiliations
Contributions
G.C., M.A., D.V., C.O., and O.C. designed the research. G.C., L.D., and M.A. did the experiments. G.C., L.D., A.C., and B.H. developed the acquisition sequence, ULM reconstruction, and analysis framework. G.C., L.D., B.P., S.B., and O.C. analyzed the processed data. J.B.D., A.C, J.B., G.C., and L.D. made the code available on Gitlab. G.C., L.D. made the data available on the ZENODO platform. All the authors discussed the results and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
O.C. is the holder of patents concerning super-resolution ultrasound imaging. He is also a co-founder and share-holder in the startup ResolveStroke.
Peer review
Peer review information
Communications Engineering thanks Jianbo Tang, Ruud van Sloun, and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: [Anastasiia Vasylchenkova]. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chabouh, G., Denis, L., Abioui-Mourgues, M. et al. 3D transcranial ultrasound localization microscopy in awake mice: protocol and open-source pipeline. Commun Eng 4, 102 (2025). https://doi.org/10.1038/s44172-025-00415-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44172-025-00415-4