Abstract
Klebsiella pneumoniae is one of the most important nosocomial pathogens worldwide. In Colombia, K. pneumoniae has been identified as the second most frequent microbial etiologic agent of healthcare-associated infections. We conducted a prospective local study of 335 K. pneumoniae isolates in 26 nationwide hospitals from 2020 to 2021. We found that the spread of carbapenem resistance was mediated by successful clones belonging to sequence types (ST) such as ST11, ST1082, and ST307, related to intra-hospital infections. We observed that blaKPC remains the primary resistance mechanism to carbapenems and that, unlike other countries, ST11 strains commonly carry blaKPC-3. We identified the recent introduction and circulation of new sequence types with different resistance mechanisms and hypervirulence. Besides, we detected possible transmission events closely linked to carbapenemase-carrying strains, mainly in intensive care units. This study helps us understand how K. pneumoniae disseminates in Colombian hospitals and where to direct effective intervention measures.
Similar content being viewed by others
Introduction
Klebsiella pneumoniae is frequently found in various environments, including the mucosal surfaces of animals and humans, where it can exist as a colonizer or as an infection-causing pathogen1,2 and is a common cause of opportunistic infections in hospitalized patients3,4. K. pneumoniae is among the world’s most common nosocomial pathogens and clinical isolates are increasingly resistant to multiple antimicrobials5,6. In recent years, there has been a rising trend in the number of reported deaths attributable to carbapenem resistance. Carbapenem-resistant Klebsiella pneumoniae (CRKP) was the microorganism for which the highest number of related deaths in children under five years could be attributed in 20217. As a result, CRKP is currently a critical WHO Priority Pathogen (among the Enterobacterales, carbapenem-resistant)8.
In Colombia, K. pneumoniae is identified as the second most frequent microorganism causing healthcare-associated infections (HAI) nationwide, with a mortality rate of 58%9. The main carbapenemases reported locally are KPC-type, followed by NDM-type, associated with mobile genetic elements (MGEs), and the expansion of the high-risk clonal groups (CG) CG258, CG147, CG152, and CG14/15. Additionally, the first hypervirulent isolate carrying blaKPC-2 belonging to the ST38010 was described and is currently designated as a lineage of interest to follow the dissemination of CRKP in the country.
The spread of CRKP and carbapenem-susceptible K. pneumoniae (CSKP) significantly impacts the quality of health services in Colombia11,12. This highlights the importance of generating a comprehensive background contributing to the epidemiological understanding of high-risk clones and antimicrobial resistance (AMR)13. The present study aims to enhance knowledge about the genetic diversity and dissemination of K. pneumoniae HAI in Colombia. It uses whole genome sequencing to identify the main resistance mechanisms to carbapenems and their relationship with MGEs, changes in the circulation of high-risk clones, identification of virulence mechanisms, and possible transmission networks of K. pneumoniae strains at the intra- and inter-hospital level.
Results
Bacterial isolates
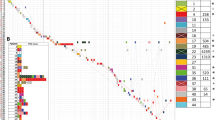
A total of 434 isolates were received for sequencing from the 26 participating institutions distributed in eight cities across the country (Supplementary Figure 1). Of the total isolates 335 met the established criteria: 69 out of the rejected isolates were identified as non-pneumoniae members of the K. pneumoniae species complex. At the same time, seven more corresponded to other bacterial genera, and 23 did not meet the post-sequencing quality control (QC) filtering described in methods. Among the 335 K. pneumoniae isolates, 194 (57.9%) were from males, 140 (41.8%) from female patients, and one isolate lacked this data; most were recovered from older adults ≥60 years old (n = 139; 41.9%), and adults from 18 to 60 years old (n = 132; 39.4%). The most frequent sources of bacterial isolation were urine (n = 98; 29.5%), blood (n = 91; 27.2%), and samples obtained from respiratory origin (n = 79; 24%) (Fig. 1).
a Frequency of CRKP (red) and CSKP (blue) isolates in different sample sources (b) and by hospital unit. c Age and gender distribution of patients from whom CRKP and CSKP were isolated. Dots indicate individual patients. d Relative frequency of phenotypic resistance by drug. (AMK amikacin, CAZ ceftazidime, CIP ciprofloxacin, COL colistin, CRO ceftriaxone, DOR doripenem, ETP ertapenem, FEP cefepime, FOX cefoxitin, GEN gentamicin, IPM imipenem, MEM meropenem, SAM ampicillin-sulbactam, TGC tigecycline, TZP tazobactam-piperacillin).
The phenotypic susceptibility profile showed that 160 (48%) isolates were CRKP and 175 (52%) were CSKP. Within CRKP we found a high rate of resistance to ceftazidime (87.5%, n = 140), piperacillin-tazobactam (98.1%, n = 157), and cefepime (60%, n = 96), compared to tigecycline, which had fewer resistant isolates (15%, n = 24). All resistance data, with their MICs and interpretations, can be found in Supplementary Data 1 and Supplementary Fig. 2. Likewise, Intensive Care Unit (ICU) isolates were more frequently recovered (n = 126; 37.6%), with a distribution of 70 (21%) CRKP and 55 (16%) CSKP (Fig. 1 and Supplementary Table 1).
Sequence types, CG, and serotypes
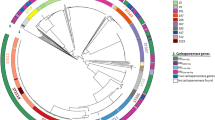
All strains were distributed in 112 known sequence types (ST) and 21 new STs (from ST6690 to ST6710), of which ST11 (n = 47; 14%), ST1082 (n = 15; 4.5%), and ST307 (n = 14; 4.5%) were the most frequent. These STs were classified into 96 CGs and seven singletons, with CG258 being the most prevalent (Fig. 2). This CG primarily included ST11 and ST258. Other CGs at a lower frequency, like CG17/20, CG1082, and CG380, were also found (Table 1).
The phylogenetic tree includes all the isolates (n = 335) with the ten most abundant CGs, the main STs of epidemiological clinical interest within each CG. Highlighted groups in the tree correspond to the most frequent STs. Terminal nodes are colored as resistant (CRKP) or susceptible (CSKP), correspondingly. Aligned we show the replicons found in the different isolates. The tree was organized by midpoint root in descending order.
Identifying the O antigen (O locus) and different capsule loci (KL) allowed us to establish the most common CRKP profiles and their association with various STs. For instance, the KL105-wzi75:O1/O2v2 profile (n = 45; 13%) was seen in ST11 isolates. The KL107-wzi154:O1/O2v2 profile (n = 10; 3%) was seen in ST258, ST512, and the new ST6694. Other profiles of interest are presented in Table 1. Lastly, 175 (52%) of CSKP isolates were distributed in 104 ST belonging to 79 CGs. ST380 (n = 10; 5.7%) with KL2-wzi203:O1/O2v1 profile held the largest number of CSKP strains (n = 8; 4.5%) (Supplementary Data 1).
Antimicrobial-resistance genes and mobile elements
We found a wide range of carbapenemase genes, extended-spectrum β-lactamase (ESBL) genes, and MGEs among the isolates. Since this study describes two main groups of isolates (CSKP-CRKP), we separated the isolates with similar genetic elements into three groups. Group 1 included resistant isolates containing carbapenemase genes and coproduction of them. Group 2 corresponded to phenotypically CRKP isolates that do not contain carbapenemase genes. Group 3 described all CSKP and CRKP isolates carrying beta-lactamase genes, focusing the analysis mainly on those harboring ESBL-type genes. It should be noted that the groups are not mutually exclusive (see Supplementary Fig. 3).
Group 1
This group included 151 isolates. The blaKPC was identified in 141 (88%) of them, with most isolates carrying one of two variants: blaKPC-3 (n = 80) or blaKPC-2 (n = 60). The blaKPC-19 allele was detected in one ST14 strain and is, until now, the first blaKPC-19-bearing strain reported in Colombia (Table 2).
The blaKPC-3 variant was present in isolates from 23 hospitals across seven cities, mainly in CRKP isolates from ST11, ST1082, ST258, and others (Table 2). Notably, the new ST6694 also carries blaKPC-3. The blaKPC-2 allele was found in strains belonging to 31 STs including ST12, ST258, ST307, and the novel ST6693 and ST6696 (Supplementary Table 2).
Several MGEs were found in this study including Tn4401 with the two isoforms Tn4401a and Tn4401b (Supplementary Fig. 4). We evidenced the presence of the Tn4401a element mainly in STs harboring blaKPC-3, while CSKP isolates belonging to the same ST lacked this element. This result is similar for ST11, ST1082, ST307, ST380, ST14, ST39, along with other STs. Some isolates harboring blaKPC-2 were found in association with Tn4401a/b in members of ST258, ST776, and others. Nevertheless, blaKPC-2 was not exclusively associated with Tn4401 for most of the ST307, ST147, ST37, and ST12.
Among other carbapenemases, blaNDM was found in 18 (11%) CRKP isolates with the blaNDM-1 variant. Of all the blaNDM-1 strains, two showed complete coverage for Tn125, while six isolates had percentages >80% for this mobile element. Tn3000 was not detected in any isolate. Likewise, none of the susceptible strains of the STs associated with blaNDM-1 showed evidence of any transposon (Supplementary Data 1).
Seven isolates were co-producers of several carbapenemases, including blaKPC-2+NDM-1 and blaKPC-2+NDM-1+VIM-24. Similarly, blaVIM and blaGES were only found in co-production as shown in Table 1. These isolates were recovered mainly from in-patient wards and ICU units.
The most prevalent plasmid replicon types identified were IncFIB(K) (n = 116; 34.6%), IncFII_1_pKP91 (n = 79; 23.6%), ColRNAI_1 (n = 74; 22%), IncR_1 (n = 47; 14%), and Col440I_1 (n = 20; 6%) in blaKPC-positive isolates. However, all replicons mentioned were also found in some CSKP (Table 3).
Group2
The group included nine phenotypically carbapenem-resistant isolates without known carbapenemases, eight of which carried the ESBL gene blaCTX-M-15, and three of them in combination with the ESBL gene blaSHV-106. The carbapenem resistance in these isolates may be related to the presence of these ESBL genes10 that could act in conjunction with porin defects in ompk36 and Ompk37 (see Supplementary Data 2). The main STs in this group were ST1082 (n = 2) and ST11 (n = 2). In addition, 13 different replicons were found among the isolates; the most frequent were IncFIB(K)_1_Kpn3 (n = 9; 100%), ColRNAI_1 (n = 5; 56%), and IncFII_1_pKP91 (n = 4; 44%) (Table 3).
Group3
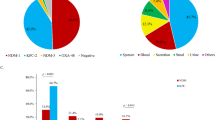
This group, which included genes encoding for class A and D serine β-lactamases (n = 329; 98%), was the most frequent in our dataset. We grouped the variants into three categories based on their molecular characteristics and functional properties according to Bush´s classification14: (i) subgroup 2b and 2 d correspond to beta-lactamases able to hydrolyze penicillin and early cephalosporins; (ii) subgroup 2br, inhibitor-resistant broad-spectrum β-lactamases, and (iii) subgroup 2be with ESBL type genes.
Within category (i), we detected the blaTEM genes belonging to subgroup 2b, mostly represented by blaTEM-1, as well as blaOXA genes mainly distributed along in the blaOXA-1, blaOXA-9 variants. Both blaTEM and blaOXA being widely distributed (65% and 58%) along the CRKP isolates. For the Category (ii) we found the genes blaSHV represented by the variants blaSHV10, blaSHV107 and blaSHV28 distributed in susceptible and resistant isolates (Fig. 3).
The figure includes the complete list of beta-lactamase genes detected in the study. Subgroups formed by functional properties according to the Bush classification: 2b, 2 d. Capacity to hydrolyze penicillinases, 2be: extended-spectrum beta-lactamases, 2br: inhibitor-resistant beta-lactamases. Bar-length describes the percentage of a total of 160 CRKP (red) or 175 CSKP (green). Classification based on https://doi.org/10.1093/jac/dkaa513; https://doi.org/10.3389/fmicb.2016.01374.
Concerning the (iii) subgroup 2be we identified the presence of blaSHV variants and blaCTX-M genes. blaSHV was mainly represented by the blaSHV-106 and blaSHV-27 variants, while blaCTX-M was mostly represented by blaCTX-M-15 (Fig. 3). Notably, 63/78 (81%) of the blaCTX-M-15 genes were harbored in isolates recovered from ICU and hospitalized patients. The main replicon types associated with these isolates were IncFIB(K)_1_Kpn3 (n = 100), ColRI_1 (n = 66), IncFII_1_pKP91 (n = 61), IncR_1 (n = 40) and IncFIA(HI1)_1_HI1 (n = 37) (Supplementary Data 1).
Other resistances and mechanisms
Among all the isolates analyzed, we observed chromosomal mutations in the key porins OmpK35 and Ompk36 associated with restricted antibiotic uptake. We identified two CRKP isolates, ST920 and novel ST6693, having a mutation in the OmpK35 gene that generates a premature termination codon associated with carbapenem resistance15. Likewise, we did not detect the OmpK36 gene in one CRKP ST11 isolate. Other mutations in the OmpK36 and OmpK37 genes are detailed in Supplementary Data 2.
Furthermore, we identified other AMR determinants for aminoglycosides (CRKP = 84%; CSKP = 30%), phenicols (CRKP = 26%; CSKP = 9%), fluoroquinolones (CRKP = 49%; CSKP = 17%), tetracyclines (CRKP = 31%; CSKP = 6%), sulfonamides (CRKP = 77%; CSKP = 26%), macrolides (CRKP = 5%; CSKP = 3%), and trimethoprim (CRKP = 76%; CSKP = 22%). Resistance determinants for the different classes of antibiotics were statistically significant (more frequent in CRKP than in CSKP; chi-square p value < 1.0e–9). The genes related to these resistances and their association with CRKP or CSKP phenotypes can be consulted in supplementary data 1.
Detection of virulence mechanisms and hypervirulence
We found acquired virulence mechanisms in 69 (21%) CRKP and 44 (13%) CSKP isolates. Yersiniabactin (ybt) siderophore was identified in 108 (32%) isolates. In particular, the ybt 1 allele was detected in 62 (76%) isolates of ST11 and ST258; ybt 16 was found in 13 genomes corresponding to all ST380 and ST6697. Colibactin (clb) was found in 16 (5%) isolates distributed along clb 1 (n = 2) grouped in ST2039, clb 2 (n = 7) in ST23, and clb 3 (n = 7) in ST258 and ST6694.
In our study, we identified ten hypervirulent K. pneumoniae (hvKp) isolates distributed as follows: (I) six hypervirulent CSKP isolates of ST23 serotype KL1 that contained ybt 1:ICEKp1, aerobactin (iuc), Salmochelin (iro), clb 1-2 and rmpA/rmpA2 corresponding to cities Bogota, Medellín and Cali; (II) two CSKP isolates ST2039 serotype KL2 carrying ybt 12:ICEKp1, iuc, iro and clb 1-2 without rmpA2 that corresponded to Bogota and Medellin; (III) one CSKP isolate ST23 KL1 with ybt 1:ICEKp1, iro and rmpA/rmpA2 from Medellin; (iv) and finally, one CSKP isolate ST6697 KL2 carrying ybt 16:ICEKp12, iro and iuc from Medellin. No hvKp strains were identified among CRKP (Supplementary Data 1).
Possible transmission events
To identify possible transmissions of isolates at intra- and inter-hospital levels, possible transmission groups (PTGs) were defined by grouping isolates with pairwise single-nucleotide polymorphism (SNPs) differences below 25 SNPs per five Mbp of the whole genome (Garcia-Neris doctoral thesis, https://hdl.handle.net/10550/88748). A total of 35 PTGs, which comprised 91 isolates from 21 hospitals, were obtained. Of these, 32 PTGs harbored isolates producing carbapenemases, ESBL, or both, and the remaining three grouped fully susceptible isolates.
The PTGs consisted of 20 STs, mainly represented by ST11, ST1082, and ST307 (Fig. 4). We observed that blaKPC-3 gene was the most frequent, present in 50 of the isolates forming PTGs and associated with Tn4401 in 26 of them. In contrast, blaKPC-2 was present in 22 isolates, but was contained in Tn4401 in only three. Likewise, blaNDM-1 was present in eight isolates, and four included Tn125. Comparably IncFIB(K), IncFII_1_pKP91, and ColRNAI_1 were the main replicons associated with CRKP strains that formed PTGs (Supplementary Data 1).
Each dot represents an isolate (n = 335), and the color corresponds to the hospital (H) it was isolated. Dots were assembled in space using a force-directed network that linked dots with less than 25 SNPs closer to PTGs and rearranged the layout by hospitals and city. PTGs are represented by rhomboids colored by resistance. Balloons represent each city and are colored accordingly. Most PTGs are of two or three isolates that share identical or nearly identical AMR genes (data not shown). PTGs shared between cities could not be traced to unveil the underlying transmission mechanism, which could be geographical, patient-transfer, or others. H1: Hospital 1, H2: Hospital 2, H3: Hospital 3, H4: Hospital 4, H5: Hospital 5, H6: Hospital 6, H7: Hospital 7, H8: Hospital 8, H9: H9, H10: Hospital 10, H11: Hospital 11, H12: Hospital 13, H14: Hospital 14, H15: Hospital 16, H17: Hospital 17, H18: Hospital 18, H19: Hospital 19, H20: Hospital 20, H21: Hospital 21, H22: Hospital 22, H23: Hospital 23, H24: Hospital 24, H25: Hospital 25, H26: Hospital 26.
The largest PTG grouped six ST12-KL35:O3/O3a isolates carrying blaKPC-2. This PTG was found primarily in the ICU of one of the hospitals in the city of Pasto (HUDN). We identified two other smaller PTGs (each with two strains) in this hospital. Other hospitals also included multiple PTGs of different STs: the HUN (n = 5) and HKE (n = 4) in Bogota, the HUEM (n = 6) in Cucuta, and the LHE (n = 4) in Bucaramanga. These results suggest frequent patient-to-patient transmission in these hospitals. We did not identify PTGs in four out of the 26 hospitals. However, since this study did not include all K. pneumoniae isolates found in the hospital, we cannot rule out undetected transmission events (Supplementary Data 3).
In total, we identified 26 PTGs (70 isolates) associated with intra-hospital transmissions, and nine PTGs (21 isolates) were detected in more than one hospital. This could be linked to inter-hospital transmissions in the same city (Supplementary Data 3), except for three PTGs (two isolates in each PTG), which grouped strains from hospitals of different cities (CSO/Medellin-HUN/Bogota, LHE/Bucaramanga-HIRHT/Manizales, and HUEM/Cucuta-FVL/Cali). This suggests infrequent inter-hospital transmission.
Discussion
The evolution of carbapenem resistance challenges the effective treatment of Gram-negative bacterial infections. It is mainly driven by the acquisition of carbapenemase genes on MGEs such as plasmids and transposons16. Globally, high rates of AMR in K. pneumoniae and other Enterobacterales, and consequent high mortality, arise largely from HAIs17,18. In Colombia, similar trends in HAI are found, with a country-wide distribution of CRKP19. Even so, there are knowledge gaps around the national distribution of carbapenemase producers, the nature of the genes they carry, and the hospital transmission of AMR. In this article, we describe the diversity and dissemination of the main K. pneumoniae STs carrying carbapenemase and ESBL genes: ST11, ST1082, ST258, and ST307 (Table 2), and their likely patterns of intra-hospital and inter-hospital transmission. Previous work determined a high circulation of multidrug-resistant strains in hospitals associated with diverse factors, including inappropriate use of antimicrobials, deficiencies in infection control, high density of vulnerable patients, and cross-transmission between patients and staff16,20,21,22,23.
Our study evaluated 335 K. pneumoniae isolates from eight cities and 26 hospitals from 2020 to 2021. Most of the CRKP held carbapenemases or ESBL genes. Our dataset’s carbapenemase genes of epidemiological importance included blaKPC-2, blaKPC-3, and blaNDM-1. The most abundant gene was blaKPC-3, mainly present in ST11 and ST1082 isolates. The ST1082 lineage was restricted to the geographical area of Bogota, concordant with the first reports in 201924. The high-risk clone ST11, the most predominant in some regions of the globe25,26, gradually displaced ST258 considered the most prevalent in the country between 2013 and 201910,23,25. To highlight, ST11 was initially reported to have the blaKPC-2 gene, with a gradual replacement by the blaKPC-3 gene, inferred from temporal data of previous reports (temporal data from10), possibly mediated by the introduction of an IncFIB-plasmid carrying the blaKPC-3 gene27. The observed high diversity of STs carrying the blaKPC-2 gene is consistent with reports from around the world28, and in the country10. The less abundant blaNDM-1 linked to ST147 and ST307 mimics other global reports where blaNDM-positive strains are increasingly connected to high-risk lineages of importance in clinical settings29,30. The abovementioned highlights the importance of genomic surveillance in monitoring the spread of CRKP in Colombian hospitals.
Co-location of carbapenemases has become an increasingly recognized phenomenon globally31. Strains possessing multiple carbapenemase genes pose a therapeutic challenge and are typically resistant to most available antimicrobials. From 2010 to 2021, the proportion of CRKP strains co-carrying two carbapenemases rose significantly from 0.40 to 9.67%, with the frequency of strains carrying both blaKPC and blaNDM increasing from 0 to 4.4%31. In Colombia, the presence of blaKPC-2,NDM-1 has been reported previously10, but the blaKPC-3,VIM-24, blaKPC-2,GES-2, and the blaKPC-2,NDM-1,VIM-24 co-carrying isolates found in this study are, to our knowledge, the first reports in the Americas.
To overview the data regarding possible hospital transmission of carbapenem resistance in our dataset, we analyzed the formation of PTGs. Based on their genomics, we found they consisted mainly of resistant isolates, such as those belonging to the ST11, ST1082, and ST307 clones. Strains belonging to these STs hold at least one carbapenemase and one ESBL gene (Supplementary Table 2). According to clinical information, most of the PTGs suggested an intra-hospital transmission of the AMR in the ICU. In contrast, inter-hospital transmission was often restricted to the same city or geographic area.
Considering both genomics and clinical information, the existence of PTGs allows inferences about how the resistance is transmitted intra-hospital in the ICU and general medical units as described previously16,17,32. These mechanisms included infection prevention and control shortfalls22, environmental contamination of beds and instruments, and patient transfer within the hospital or between hospitals22,32. Without clinical and epidemiological data, assessing the underlying mechanisms of K. pneumoniae transmission is impossible. Nevertheless, intra-hospital transmission is still an important route for CRKP and K. pneumoniae dissemination given the number of intra-hospital PTGs formed.
A limitation of using PTGs to infer transmission is establishing the SNP threshold; this can lead to false-positive or false-negative interpretations of AMR transmission. False-positive in two cases; the first, clones from patients of closely related healthcare networks32, leading to sampling the same patient in two geographical locations, for example, in inter-hospital PTGs. The second case is when the ST is acquired from a common source, such as food that is supplied to different hospitals17. In our results, the inter-hospital ST661 PTG, alongside the intra-hospital ST177 PTG, are described as STs frequently found in animals for human consumption17. Thus, collecting additional metadata to evaluate CRKP transmission through food consumption and other routes would be important. False-negative transmission could result from hypermutator clones found in colonized patients with many SNPs above the established threshold so some PTGs may not be detectable32. A further limitation is the sampling method used, which provides us with only isolates from infected patients and the first CRKP or CSKP found on each patient by the hospital.
Despite these limitations, the number of PTGs detected was higher than expected and supports the implementation of effective infection control measures in the studied hospitals and for active surveillance in specific wards, such as the ICU, from where most of the PTGs were detected. Active surveillance, including sequencing of CRKP strains, could limit the transmission of such strains, possibly decreasing unnecessary costs for hospitals and patient complications including mortality. Several studies have shown that the presence of infections caused by CRKP is related to an increase in mortality33,34,35.
Even though AMR research focuses on CRKPs, the addition of CSKP provides relevant information about the overall population structure and ecological diversity of K. pneumoniae. This diversity of CSKP was observed in 104 STs; these susceptible isolates were distributed throughout the study population without presenting any particular clustering or grouping, except for the hypervirulent STs, showcasing that CRKPs are indeed under strong selection. Highly diverse CSKP populations, considering the hypothesis that colonization often precedes infection, can potentially acquire and transmit AMR16,17,36,37.
Currently, circulation of hvKp, encoding essential factors, such as the rmp genes, siderophores, salmochelins, capsular serotypes, and ICEKp elements, is commonly reported38,39. We identified significant associations between K-locus types and clb variants; for example, the KL1/clb2 and KL2/clb1 profiles were associated with hypervirulent clones ST23 and ST2039, reported previously in Ecuador by Valdiviezo et al. (http://www.dspace.uce.edu.ec/bitstream/25000/18020/1/T-UCE-0008-CQU-093.pdf) and globally39,40. Contrarily, clb3 was associated with non-hypervirulent CRKP strains related to ST258 and ST6694, aligning with previous reports in Colombia10 and worldwide41. Our data also confirm the presence of a recently circulating hypervirulent clone (CG380) in the country10 and the new hvKP strain ST6697, a single-locus variant of the ST380 clone. We observed that most hvKP strains were mainly associated with respiratory samples and blood cultures. Currently, the coexistence of virulence and AMR is becoming a widespread problem, which should guide surveillance programs42.
The present study provided recent epidemiological information on the behavior of CRKP and CSKP in Colombia. We found that carbapenem resistance was mediated by disseminating successful clones, e.g., ST11 and ST1082, related to intra-hospital transmission. We found no relationship between hypervirulent clones and CRKP clones. The K. pneumoniae population found in Colombia is a carrier of different STs, mainly from susceptible isolates, capturing a genetic diversity with the potential for possible events of infection, acquisition, and transmission of AMR. In summary, we gained a better knowledge of the population structure, spread, and expansion of antibiotic resistance in K. pneumoniae in the country. These results highlight the importance of implementing a genomic surveillance program for AMR in America’s region.
Methods
Ethical considerations
This study complies with all relevant ethical regulations. The research ethics committees of each of the participating hospitals approved the collection of clinical isolates and associated data.
Bacterial isolates
Between February 2020 and December 2021, K. pneumoniae isolates were collected in Cary-Blair transport medium from 26 participating institutions. Samples were sent to the Global Health Research Unit (GHRU) on the Genomic Surveillance of AMR group at the Colombian Corporation for Agricultural Research (AGROSAVIA). Isolates sent by each health institution complied with the following criteria: (1) be associated with hospital infections; (2) with positive confirmation at the genus and species level by the automated Vitek2 Compact® equipment (bioMérieux); and (3) that half of the isolates were carbapenem-resistant, and the other half were carbapenem-susceptible.
The isolates received were cultured on MacConkey Agar plates (OXOID CM-115) and incubated for 18–24 h at 37 °C. Next, single colonies were plated in Luria-Bertani Agar. The genus-species and antimicrobial susceptibility profile were confirmed using the Vitek 2 Compact (bioMérieux, FRA) with GN (Ref. 21341) and AST-N272 cards (Ref. 414164). The minimum inhibitory concentration (MIC) results were interpreted according to the Clinical and Laboratory Standards Institute criteria (CLSI, 2020)43.
Whole genome sequencing
Bacterial DNA was extracted using the PureLink™ Genomic DNA Mini Kit (Invitrogen, USA), following the manufacturer’s instructions. DNA quality and quantity were assessed with a NanoDrop™ 2000 spectrophotometer (Thermo Scientific, USA) and a Qubit 4.0 fluorometer (Invitrogen, USA), respectively. Library preparation was conducted using the NEBNext Ultra II FS DNA Library Kit, and sequencing was performed on the Illumina HiSeq-X10 platform (San Diego, California, USA) at the Wellcome Sanger Institute (Hinxton, United Kingdom).
Data analysis
Sequencing results were processed following GHRU-AMR protocols, utilizing versioned Nextflow workflows and corresponding Docker containers (https://gitlab.com/cgps/ghru/pipelines/dsl2/pipelines)44. The study included assemblies that met the following QC parameters: raw reads with good sequencing quality measured by FastqC, assemblies with less than 150 contigs, an N50 above 50,000 pb, a GC content close to 57%, a genome size between 4.5–7 Mbp as expected for K. pneumoniae, and no contamination from other species or clones of the same species. The first protocol, covering de novo assembly, began with sequence quality assessment using FastQC (v0.11.8), followed by gentle quality trimming and adapter clipping with Trimmomatic (v0.38). Next, species identification was performed with BactInspector (v0.1.3), and potential contamination from other species was checked using ConFinder (v0.7.2). De novo assembly was then generated using SPAdes (v3.12.0). Finally, the assemblies were evaluated for quality with QUAST (v5.2.0), utilizing ContigTools for further analysis44.
The second protocol uses good-quality assemblies to determine the Multilocus sequence typing (MLST) through alleles of housekeeping genes found in the PubMLST database45 using the ARIBA software (v2.14.4)46. The third protocol allowed searching for AMR marker genes by annotating the sequences with the resistance genes from the NCBI database through the ARIBA software. The last workflow performed variant calling mapping to the reference genome K. pneumoniae strain K2044 (NCBI RefSeq NZ_CP026011.1), and a maximum likelihood phylogeny was inferred using IQ_TREE (v1.6.8)47 software with GTR + G model and 1000 bootstrap replicates.
The virulence factors associated with integrative and conjugative elements (ICE), K-locus (capsule), and O antigen (LPS) serotype prediction via wzi alleles, were all defined by Kleborate (v2.3.2) and Kaptive41. The CG were determined by BIGSdb-Pasteur25,48 and using core genome SNP analysis using Prokka v1.14.5 gene annotation and a 70–90% ANI between isolates10. The search for plasmid replicons was performed using the PlasmidFinder database49. The variants of Tn4401a, Tn4401b, Tn125 and Tn3000 were identified through BLAST local alignment using the reference sequences KT378596.1, KT378598.1, KT965092, and KR822246, respectively. PTGs were obtained by using the alignment-based variant calling tool named snippy v4.5.1 (https://github.com/tseemann/snippy)50, followed by calculating the paired SNP distances between samples using SNP-dists v0.7.0 (https://github.com/tseemann/snp-dists)51 and clustering, with a threshold of 25 SNPs over 5 Mbp (approximately the entire genome). Cluster calculation and visualization were done using the hclust function in the stats package and igraph v2.0.2 in R v3.4.052, respectively. We also evaluated the presence of potential mutations in the genes encoding the porins ompK35, ompK36, and ompK37 using ResFinder v4.7.2 with the Pointfinder database53. In this analysis, we implemented a minimum coverage threshold of 0.6 and an identity threshold of 0.9 to identify genes and detect recognized mutations.
Statistical analysis
The resultant genomic data was analyzed using R v4.3.1 (Vienna, Austria) through RStudio. Quantitative variables were checked for normality, and the Shapiro-Wilk test showed a non-normal distribution. Categorical variables were evaluated with the Fisher exact test by groups of CSKP and CRKP, followed by the Mantel-Haenszel chi-square test to assess significant categories within each variable and co-occurring category. Significant categories for different combinations of variables were also evaluated with the Fisher exact test and added to the results tables. Variables included resistance, location, unit, source, age, gender, STs, CGs, carbapenemase genes, ESBL genes, K-locus, O-locus, and virulence genes.
Data availability
All data is merged and available in the Microreact project https://microreact.org/project/mmEih4RswDx27LkymNVwSi-plhkpn. Raw sequence data were deposited at the European Nucleotide Archive (ENA) under the bio-project accession PRJEB80567.
Code availability
GHRU-AMR protocols are available at https://gitlab.com/cgps/ghru/pipelines/dsl2/pipelines.
References
Ludden, C. et al. A one health study of the genetic relatedness of Klebsiella pneumoniae and their mobile elements in the East of England. Clin. Infect. Dis. 70, 219–226 (2020).
Ramatla, T. et al. One health” perspective on prevalence of co-existing extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae: a comprehensive systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 22, 88 (2023).
Roach, D. J., Sridhar, S. & Oliver, E. Clinical and genomic characterization of a cohort of patients with Klebsiella pneumoniae bloodstream infection. Clin. Infect. Dis. 78, 31–39 (2024).
Snitkin, E. S., Won, S. & Pirani, A. Integrated genomic and interfacility patient-transfer data reveal the transmission pathways of multidrug-resistant Klebsiella pneumoniae in a regional outbreak. Sci. Transl. Med. 9, eaan0093 (2017).
Pendleton, J. N., Gorman, S. P. & Gilmore, B. F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti Infect. Ther. 11, 297–308 (2013).
Asri, N. A. M., Ahmad, S. & Mohamud, R. Global prevalence of nosocomial multidrug-resistant Klebsiella pneumoniae: a systematic review and meta-analysis. Antibiotics 10, 1508 (2021).
Naghavi, M., Vollset, S. E. & Ikuta, K. S. Global burden of bacterial antimicrobial resistance 1990-2021: a systematic analysis with forecasts to 2050. Lancet 404, 1199–1226 (2024).
World Health Organization. Bacterial priority pathogens list, 2024: bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance (WHO, 2024).
Instituto Nacional de Salud de Colombia. IAAS Infecciones Asociadas a la Atencion en Salud, Colombia 2023–2024 (INS, 2024).
Saavedra, S. Y., Bernal, J. F. & Montilla-Escudero, E. Complexity of genomic epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Colombia urges the reinforcement of whole genome sequencing-based surveillance programs. Clin. Infect. Dis. 73, S290–S299 (2021).
Ocampo, A. M., Chen, L. & Cienfuegos, A. V. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob. Agents Chemother. 60, 332–342 (2016).
Ovalle, M. V., Saavedra, S. Y. & González, M. N. Results of the national surveillance of antimicrobial resistance of Enterobacteriaceae and Gram negative bacilli in health care-associated infections in Colombia, 2012-2014. Biomedica 37, 473–483 (2017).
Yang, J., Zhang, K. & Ding, C. Exploring multidrug-resistant Klebsiella pneumoniae antimicrobial resistance mechanisms through whole genome sequencing analysis. BMC Microbiol. 23, 245 (2023).
Bush, K. & Jacoby, G. A. Updated functional classification of β-lactamases. Agents Chemother. 54, 969–976 (2010).
Ruiz, E. et al. Acquisition of carbapenem resistance in multiresistant Klebsiella pneumoniae strains harbouring blaCTX-M-15, qnrS1 and aac(6’)-Ib-cr genes. J. Med. Microbiol. 61, 672–677 (2012).
David, S., Reuter, S. & Harris, S. R. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929 (2019).
Gorrie, C. L. Genomic dissection of Klebsiella pneumoniae infections in hospital patients reveals insights into an opportunistic pathogen. Nat. Commun. 13, 3017 (2022).
Tosi, M. et al. Multidrug resistant bacteria in critically ill patients: a step further antibiotic therapy. J. Emerg. Crit. Care Med. 2, 377–390 (2018).
Diaz, M. H. Protocolo de Vigilancia de Resistencia Bacteriana a Los Antimicrobianos en el áMbito Hospitalario (Instituto Nacional de Salud, 2022).
Shu, L. et al. Sources of Klebsiella pneumoniae isolated in a hospital in the past decade and trends of in vitro drug susceptibility. Sichuan Da Xue Xue Bao Yi Xue Ban. 53, 696–700 (2022).
Guh, A. Y. et al. Epidemiology of carbapenem-resistant Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 314, 1479–1487 (2015).
Wei, L. et al. Spread of carbapenem-resistant Klebsiella pneumoniae in an intensive care unit: a whole-genome sequence-based prospective observational study. Microbiol. Spectr. 9, e0005821 (2021).
Rojas, L. J. et al. An analysis of the epidemic of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: convergence of two evolutionary mechanisms creates the “perfect storm”. J. Infect. Dis. 217, 82–92 (2017).
Gamboa, A. C. Evaluación del comportamiento epidemiológico de aislamientos clínicos de Klebsiella pneumoniae mediante secuenciación de genoma completo (WGS). (Universidad Nacional de Colombia, 2023).
Argimón, S., David, S. & Underwood, A. Rapid genomic characterization and global surveillance of Klebsiella using Pathogenwatch. Clin. Infect. Dis. 73, S325–S335 (2021).
Sampaio, J. L. & Gales, A. C. Antimicrobial resistance in Enterobacteriaceae in Brazil: focus on β-lactams and polymyxins. Braz. J. Microbiol. 47, 31–37 (2016).
Garcia-Fulgueiras, V. et al. First characterization of K. pneumoniae ST11 clinical isolates harboring blaKPC-3 in Latin America. Rev. Argent. Microbiol. 52, 211–216 (2020).
Wang, Q. et al. Expansion and transmission dynamics of high-risk carbapenem-resistant Klebsiella pneumoniae subclones in China: an epidemiological, spatial, genomic analysis. Drug Resist. Updat. 74, 101083 (2024).
Nordmann, P., Naas, T. & Poirel, L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17, 1791–1798 (2011).
Gao, H. et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 51, 102599 (2020).
Guo, H. et al. Global emergence of carbapenem-resistant Klebsiella pneumoniae co-carrying multiple carbapenemases. Comput. Struct. Biotechnol. J. 21, 3557–3563 (2023).
Hawken, S. E. et al. Threshold-free genomic cluster detection to track transmission pathways in healthcare settings: a genomic epidemiology analysis. Lancet Microbe 3, e652–e662 (2022).
Ramos-Castañeda, J. A. et al. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: systematic review and meta-analysis. J. Infect. 76, 438–448 (2018).
Hauck, C. et al. Spectrum of excess mortality due to carbapenem-resistant Klebsiella pneumoniae infections. Clin. Microbiol. Infect. 22, 513–519 (2016).
Stewardson, A. J. et al. Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect. Dis. 19, 601–610 (2019).
Zhou, H. et al. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Klebsiella pneumoniae. Front. Microbiol. 8, 371 (2017).
Wyres, K. L., Lam, M. M. C. & Holt, K. E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 18, 344–359 (2020).
Lam, M. M. et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9, 2703 (2018).
Chen, J., Zhang, H. & Liao, X. Hypervirulent Klebsiella pneumoniae. Infect. Drug Resist. 16, 5243–5249 (2023).
Liu, Y. et al. Clinical characteristics and molecular epidemiology of ST23 Klebsiella pneumoniae in China. Infect. Drug Resist. 16, 7597–7611 (2023).
Lam, M. M. C., Wick, R. R. & Watts, S. C. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12, 4188 (2021).
Choby, J. E., Howard-Anderson, J. & Weiss, D. S. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287, 283–300 (2020).
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing 30th edn (CLSI, 2020).
Afolayan, A. O., Bernal, J. F. & Gayeta, J. M. Overcoming data bottlenecks in genomic pathogen surveillance. Clin. Infect. Dis. 73, S267–S274 (2021).
Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3, 124 (2018).
Hunt, M., Mather, A. E. & Sánchez-Busó, L. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Micro. Genom. 3, e000131 (2017).
Minh, B. Q., Schmidt, H. A. & Chernomor, O. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Jolley, K. A. & Maiden, M. C. J. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinforma. 11, 595 (2010).
Carattoli, A., Zankari, E. & Garcìa-Fernandez, A. In silico detection and typing of plasmids using PlasmidFinder and pMLST. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Seemann, T. snippy: rapid haploid variant calling and core genome alignment. GitHub https://github.com/tseemann/snippy (2022).
Seemann, T. snp-dists: pairwise SNP distance matrix from a FASTA sequence alignment. GitHub https://github.com/tseemann/snp-dists (2022).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. Inter. Complex Syst. 1695, 1–9 (2006).
Florensa, A. F., Kaas, R. S. & Clausen, P. T. L. C. ResFinder - an open online resource for identification of antimicrobial resistance genes and prediction of phenotypes from genotypes. Micro. Genom. 8, e000748 (2022).
Acknowledgements
This research was commissioned by the National Institute for Health Research using Official Development Assistance (ODA) funding. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the National Institute for Health Research, or the Department of Health. The authors acknowledge the clinicians and epidemiologists of the participating hospitals for providing the K. pneumoniae isolates obtained under the GHRU on Genomic Surveillance of Antimicrobial Resistance. In detail, we acknowledge by name(s) and surname(s), Lorena Jaramillo Ochoa from Hospital Universitario Departamental de Nariño; Claudia Fajardo Uribe, Ayda María Pardo Cortes, and Claudia Rocio Sierra Parada from Clínica Colombia; Adriana Marín Osorio from Clínica general del Norte; Ingrid Gisell Bustos, Lina Mendez, and Andrea Quintero from Clínica Universidad de la Sabana; Dr Carlos Moreno, Dr. Sai Chinome from Hospital Universitario Erasmo Meoz; Ana Yadira Santana, Nella Sánchez and Gleidis Castillo from Suberd Centro Oriente; Aleyda Montaño, Elizabeth Sierra from Clínica Rosario-Medellín; Leidy Villamil-Vega from Clínica Versalles; Magda Costanza Toledo Garzon from Clínica Bolivariana; Gustavo Roncancio and Liliana Franco from Clinica Cardiovascular; Lina Arteaga, and Carolina Velásquez from Clínica CES; Edwin Federico Rios from Clínica Somer; Juan David Villa from Hospital General de Medellín; Yeni Díaz García, Yanneth Andrea Lopez Vasquez, Alejandro Guzman Benitez, Diana Carolina Suarez Castrillon and Marcela Villegas Garcia from Hospital Manuel Uribe Ángel; Natalia Vásquez-Ramirez from Clínica Avidanti; Claudia Pinilla, and Natalia Gonzales from Hospital Infantil Rafael Henao Toro. We would like to thank the researchers from the University of La Sabana (MED-308-2021) for their support. Besides, we sincerely thank Erik Cristopher Osma Castro for his invaluable support in coordinating the logistics of materials, supplies, and budget, contributing to the development and completion of this article. We thank Maria F. Valencia for her support as a research support professional hired by AGROSAVIA for the GHRU project. This work was supported by ODA funding from the National Institute for Health Research.
Author information
Authors and Affiliations
Consortia
Contributions
V.A.M., A.S.G.-V., F.R., and E.S.T.-G. contributed to data processing and analysis, results interpretation, writing draft, and designing tables and figures. J.A.R., A.L.L., J.C.G., L.F.R., E.A.K., G.L.-W., S.A., and A.P.U. provided extensive reviews and substantial changes to the draft. X.F., D.M.A., M.P.D.-G., I.N.O., and N.C. provided project design and manuscript review. S.A.A. contributed to isolating the processing and review of the draft. All GEAR consortium members assisted with reviewing the draft, isolating the processing, and the metadata.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Medina, V.A., García-Vega, A.S., Rodríguez, F. et al. Epidemiological Genomics of Klebsiella pneumoniae isolates from hospitals across Colombia. npj Antimicrob Resist 3, 64 (2025). https://doi.org/10.1038/s44259-025-00127-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44259-025-00127-x