Abstract
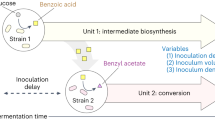
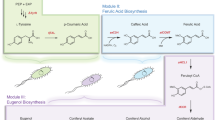
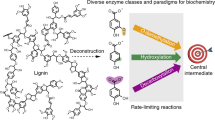
Aromatic esters possess flavor and fragrance qualities that are widely used in the food, pharmaceutical and cosmetic industries. However, microbial production of these compounds is hampered by a limited understanding of the natural biosynthetic pathway and the relatively low titer and yield. This study establishes a microbial platform for the efficient production of various aromatic esters. A systematic engineering strategy was developed, involving reshaping the substrate access tunnel to enhance enzyme substrate specificity, shifting acetyl coenzyme A metabolic pathways to improve cofactor supply and engineering a dynamic regulation system to redistribute the carbon flux from cell growth toward product synthesis. The implementation of these approaches led to the production of 10.4 g l–1 benzyl benzoate, representing a 4,700-fold increase in titer compared with the initial strain. This work showcases a bacterial platform for the efficient production of aromatic esters and offers insights into overcoming challenges in microbial cell factory construction.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data supporting the results are provided in the article and its Supplementary Information. Related materials, including the protein structures, tunnel analyses, docking simulations, ASMD simulations, MD simulations, CAVER Dock analyses and a video showing the dynamic changes of all four tunnels throughout the MD simulations are available via Zenodo at https://doi.org/10.5281/zenodo.12784348 (ref. 78). Source data are provided with this paper.
References
Nielsen, J. & Keasling, J. D. Engineering cellular metabolism. Cell 164, 1185–1197 (2016).
Lee, S. Y. et al. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2, 18–33 (2019).
Huang, J. et al. Complete integration of carbene-transfer chemistry into biosynthesis. Nature 617, 403–408 (2023).
Zhang, J. et al. A microbial supply chain for production of the anti-cancer drug vinblastine. Nature 609, 341–347 (2022).
Qin, J. et al. Engineering yeast metabolism for the discovery and production of polyamines and polyamine analogues. Nat. Catal. 4, 498–509 (2021).
Xu, Y. et al. A light-driven enzymatic enantioselective radical acylation. Nature 625, 74–78 (2024).
Wu, L., Qin, L., Nie, Y., Xu, Y. & Zhao, Y.-L. Computer-aided understanding and engineering of enzymatic selectivity. Biotechnol. Adv. 54, 107793 (2022).
Qu, G., Li, A., Acevedo-Rocha, C. G., Sun, Z. & Reetz, M. T. The crucial role of methodology development in directed evolution of selective enzymes. Angew. Chem. Int. Ed. 59, 13204–13231 (2020).
Amin, S. R., Erdin, S., Ward, R. M., Lua, R. C. & Lichtarge, O. Prediction and experimental validation of enzyme substrate specificity in protein structures. Proc. Natl Acad. Sci. USA 110, E4195–E4202 (2013).
Chong, W. et al. Computer-aided tunnel engineering: a promising strategy for improving lipase applications in esterification reactions. ACS Catal. 14, 67–83 (2024).
Chen, R. et al. Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast. Nat. Chem. Biol. 18, 520–529 (2022).
Vuoristo, K. S., Mars, A. E., Sanders, J. P. M., Eggink, G. & Weusthuis, R. A. Metabolic engineering of TCA cycle for production of chemicals. Trends Biotechnol. 34, 191–197 (2016).
Meng, W. et al. 2,3-Butanediol synthesis from glucose supplies NADH for elimination of toxic acetate produced during overflow metabolism. Cell Discov. 7, 43 (2021).
Krivoruchko, A., Zhang, Y., Siewers, V., Chen, Y. & Nielsen, J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab. Eng. 28, 28–42 (2015).
Weitzman, P. D. J. Regulation of citrate synthase activity in Escherichia coli. Biochim. Biophys. Acta 128, 213–215 (1966).
He, L. et al. Deciphering flux adjustments of engineered E. coli cells during fermentation with changing growth conditions. Metab. Eng. 39, 247–256 (2017).
Sá, A. G. A., de Meneses, A. C., de Araújo, P. H. H. & de Oliveira, D. A review on enzymatic synthesis of aromatic esters used as flavor ingredients for food, cosmetics and pharmaceuticals industries. Trends Food Sci. Technol. 69, 95–105 (2017).
Lee, J.-W. & Trinh, C. T. Towards renewable flavors, fragrances, and beyond. Curr. Opin. Biotechnol. 61, 168–180 (2020).
Kalpaklioglu, A. F., Ferizli, A. G., Misirligil, Z., Demirel, Y. S. & Gürbüz, L. The effectiveness of benzyl benzoate and different chemicals as acaricides. Allergy 51, 164–170 (1996).
Forton, F. M. N. & De Maertelaer, V. Treatment of rosacea and demodicosis with benzyl benzoate: effects of different doses on Demodex density and clinical symptoms. J. Eur. Acad. Dermatol. Venereol. 34, 365–369 (2020).
Meyersburg, D., Kaiser, A. & Bauer, J. W. Loss of efficacy of topical 5% permethrin for treating scabies: an Austrian single-center study. J. Dermatol. Treat. 33, 774–777 (2022).
Xue, F. et al. Codon-optimized Rhodotorula glutinis PAL expressed in Escherichia coli with enhanced activities. Front. Bioeng. Biotechnol. 8, 610506 (2021).
Dippe, M. et al. Coenzyme A-conjugated cinnamic acids—enzymatic synthesis of a CoA-ester library and application in biocatalytic cascades to vanillin derivatives. Adv. Synth. Catal. 361, 5346–5350 (2019).
Luo, Z. W. & Lee, S. Y. Metabolic engineering of Escherichia coli for the production of benzoic acid from glucose. Metab. Eng. 62, 298–311 (2020).
Finnigan, W. et al. Characterization of carboxylic acid reductases as enzymes in the toolbox for synthetic chemistry. ChemCatChem 9, 1005–1017 (2017).
Kunjapur, A. M., Tarasova, Y. & Prather, K. L. J. Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J. Am. Chem. Soc. 136, 11644–11654 (2014).
Altenschmidt, U., Oswald, B. & Fuchs, G. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J. Bacteriol. 173, 5494–5501 (1991).
Boatright, J. et al. Understanding in vivo benzenoid metabolism in petunia petal tissue. Plant Physiol. 135, 1993–2011 (2004).
Chedgy, R. J., Köllner, T. G. & Constabel, C. P. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113, 149–159 (2015).
Shen, X. et al. Establishment of novel biosynthetic pathways for the production of salicyl alcohol and gentisyl alcohol in engineered Escherichia coli. ACS Synth. Biol. 7, 1012–1017 (2018).
Shen, X. et al. Elevating 4-hydroxycoumarin production through alleviating thioesterase-mediated salicoyl-CoA degradation. Metab. Eng. 42, 59–65 (2017).
Lu, L. et al. Establishing biosynthetic pathway for the production of p-hydroxyacetophenone and its glucoside in Escherichia coli. Metab. Eng. 76, 110–119 (2023).
Gosset, G., Yong-Xiao, J. & Berry, A. A direct comparison of approaches for increasing carbon flow to aromatic biosynthesis in Escherichia coli. J. Ind. Microbiol. 17, 47–52 (1996).
Liu, Y. et al. Genetic engineering of Escherichia coli to improve l-phenylalanine production. BMC Biotechnol. 18, 5 (2018).
Bang, H. B., Son, J., Kim, S. C. & Jeong, K. J. Systematic metabolic engineering of Escherichia coli for the enhanced production of cinnamaldehyde. Metab. Eng. 76, 63–74 (2023).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2023).
Gahloth, D. et al. Structures of carboxylic acid reductase reveal domain dynamics underlying catalysis. Nat. Chem. Biol. 13, 975–981 (2017).
Stourac, J. et al. Caver Web 1.0: identification of tunnels and channels in proteins and analysis of ligand transport. Nucleic Acids Res. 47, W414–W422 (2019).
Schwendenwein, D. et al. Random mutagenesis-driven improvement of carboxylate reductase activity using an amino benzamidoxime-mediated high-throughput assay. Adv. Synth. Catal. 361, 2544–2549 (2019).
Jurcik, A. et al. CAVER Analyst 2.0: analysis and visualization of channels and tunnels in protein structures and molecular dynamics trajectories. Bioinformatics 34, 3586–3588 (2018).
Kaushik, S. et al. Impact of the access tunnel engineering on catalysis is strictly ligand-specific. FEBS J. 285, 1456–1476 (2018).
Case, D. A. et al. AmberTools. J. Chem. Inf. Model. 63, 6183–6191 (2023).
Case, D. A. et al. Amber 2024 (University of California San Francisco, 2024).
Bernal, V., Castaño-Cerezo, S. & Cánovas, M. Acetate metabolism regulation in Escherichia coli: carbon overflow, pathogenicity, and beyond. Appl. Microbiol. Biotechnol. 100, 8985–9001 (2016).
Parimi, N. S., Durie, I. A., Wu, X., Niyas, A. M. M. & Eiteman, M. A. Eliminating acetate formation improves citramalate production by metabolically engineered Escherichia coli. Microb. Cell Fact. 16, 114 (2017).
Trichez, D. et al. Engineering of Escherichia coli for Krebs cycle-dependent production of malic acid. Microb. Cell Fact. 17, 113 (2018).
Lee, J.-W. & Trinh, C. T. Controlling selectivity of modular microbial biosynthesis of butyryl-CoA-derived designer esters. Metab. Eng. 69, 262–274 (2022).
Feng, J. et al. Renewable fatty acid ester production in Clostridium. Nat. Commun. 12, 4368 (2021).
Layton, D. S. & Trinh, C. T. Microbial synthesis of a branched-chain ester platform from organic waste carboxylates. Metab. Eng. Commun. 3, 245–251 (2016).
Choi, K. R., Luo, Z. W., Kim, G. B., Xu, H. & Lee, S. Y. A microbial process for the production of benzyl acetate. Nat. Chem. Eng. 1, 216–228 (2024).
Rodriguez, G. M., Tashiro, Y. & Atsumi, S. Expanding ester biosynthesis in Escherichia coli. Nat. Chem. Biol. 10, 259–265 (2014).
Horton, C. E., Huang, K.-X., Bennett, G. N. & Rudolph, F. B. Heterologous expression of the Saccharomyces cerevisiae alcohol acetyltransferase genes in Clostridium acetobutylicum and Escherichia coli for the production of isoamyl acetate. J. Ind. Microbiol. Biotechnol. 30, 427–432 (2003).
Akhtar, M. K., Turner, N. J. & Jones, P. R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl Acad. Sci. USA 110, 87–92 (2013).
Clomburg, J. M., Crumbley, A. M. & Gonzalez, R. Industrial biomanufacturing: the future of chemical production. Science 355, aag0804 (2017).
Scesa, P. D., Lin, Z. & Schmidt, E. W. Ancient defensive terpene biosynthetic gene clusters in the soft corals. Nat. Chem. Biol. 18, 659–663 (2022).
Nett, R. S., Lau, W. & Sattely, E. S. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 584, 148–153 (2020).
Tu, L. et al. Genome of Tripterygium wilfordii and identification of cytochrome P450 involved in triptolide biosynthesis. Nat. Commun. 11, 971 (2020).
Zhou, Y. J. et al. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 7, 11709 (2016).
Chen, Z. et al. Establishing an artificial pathway for de novo biosynthesis of vanillyl alcohol in Escherichia coli. ACS Synth. Biol. 6, 1784–1792 (2017).
Wang, J., Li, C., Zou, Y. & Yan, Y. Bacterial synthesis of C3-C5 diols via extending amino acid catabolism. Proc. Natl Acad. Sci. USA 117, 19159–19167 (2020).
Chen, X., Li, S. & Liu, L. Engineering redox balance through cofactor systems. Trends Biotechnol. 32, 337–343 (2014).
Cress, B. F. et al. CRISPRi-mediated metabolic engineering of E. coli for O-methylated anthocyanin production. Microb. Cell Fact. 16, 10 (2017).
Luo, Z. W., Cho, J. S. & Lee, S. Y. Microbial production of methyl anthranilate, a grape flavor compound. Proc. Natl Acad. Sci. USA 116, 10749–10756 (2019).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Li, Y. et al. Metabolic engineering of Escherichia coli using CRISPR–Cas9 meditated genome editing. Metab. Eng. 31, 13–21 (2015).
Eberhardt, J., Santos-Martins, D., Tillack, A. F. & Forli, S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 61, 3891–3898 (2021).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
O’Boyle, N. M. et al. Open Babel: an open chemical toolbox. J. Cheminf. 3, 33 (2011).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Schrödinger, L. The PyMOL Molecular Graphics System, Version 1.8 (2015).
Maier, J. A. et al. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015).
Sprenger, K. G., Jaeger, V. W. & Pfaendtner, J. The general AMBER force field (GAFF) can accurately predict thermodynamic and transport properties of many ionic liquids. J. Phys. Chem. B 119, 5882–5895 (2015).
Mark, P. & Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960 (2001).
Götz, A. W. et al. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J. Chem. Theory Comput. 8, 1542–1555 (2012).
Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Lin, H.-Y. et al. Rational redesign of enzyme via the combination of quantum mechanics/molecular mechanics, molecular dynamics, and structural biology study. J. Am. Chem. Soc. 143, 15674–15687 (2021).
Lu, L. & Wang J. Data for “Establishing a bacterial platform for production of aromatic esters from glycerol.” Zenodo https://doi.org/10.5281/zenodo.12784348 (2024).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (grant no. 2022YFC2106100) and the National Natural Science Foundation of China (grant nos. 22378016, 22078011 and 22238001). We thank Z. Sun and C. Li (Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences) for their guidance on the ASMD simulation and Y. Zhou (Dalian Institute of Chemical Physics, Chinese Academy of Sciences) for his guidance on paper preparation.
Author information
Authors and Affiliations
Contributions
L.L. and J.W. conceived the study and wrote the paper. L.L., X.W. and T.W. performed the experiments. X. Shen and X. Sun participated in the research. J.W., P.T., Y.Y., J.N. and Q.Y. revised the paper. J.W. and Q.Y. directed the research. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Engineering thanks Guo-Qiang Chen, Yong Hwan Kim, Cong T. Trinh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Supplementary information
Supplementary Information
Supplementary Figs. 1–21, Tables 1–3 and references.
Supplementary Data 1
Oligonucleotides used in this study.
Supplementary Data 2
Nonsense sites in the E. coli genome and corresponding sgRNA (N20).
Supplementary Data 3
Raw NMR data.
Source Data for Supplementary Figures
Statistical source data for Supplementary Figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig./Table 1
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, L., Wang, X., Wang, T. et al. A bacterial platform for producing aromatic esters from glycerol. Nat Chem Eng 1, 751–764 (2024). https://doi.org/10.1038/s44286-024-00148-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s44286-024-00148-9
This article is cited by
-
Chemoenzymatic platform with coordinated cofactor self-circulation for lignin valorization
Nature Synthesis (2025)
-
Economic and sustainable revolution to facilitate one-carbon biomanufacturing
Nature Communications (2025)
-
Streamlined aromatic ester process via tunnel engineering
Nature Chemical Engineering (2024)