Abstract
Invasive fungal diseases (IFDs) present a growing clinical challenge, underscoring the urgent need for improved diagnostics, therapeutics and mechanistic understanding. This review highlights the key role of innovative imaging techniques across all scales - ranging from whole-body-level diagnostics and therapy monitoring to host-pathogen interactions at cellular resolution in both clinical and preclinical settings. These imaging modalities facilitate translation of preclinical innovations into clinical applications, accelerating research and advancing IFD management.

Similar content being viewed by others
Introduction
Invasive fungal diseases (IFDs) pose a significant, and often underestimated public health threat, underscored by the World Health Organization (WHO)’s Fungal Priority Pathogen List in 20221. The incidence of IFDs continues to rise due to a growing immunocompromised population and newly identified at-risk patients which are not traditionally defined as immunocompromised, has caused re-evaluation of our understanding of how fungal pathogens interact with the host’s immune system2,3. Environmental pressures like climate change and agricultural practices further impact the emergence and spread of fungal pathogens, demonstrating the interconnectedness of human, animal and environmental health as emphasized in the One Health concept4. Moreover, diagnostic tools for IFDs are often invasive, based on a small biopsy or sample, and limited in speed – e.g. awaiting positive culture - and/or specificity. Imaging methods deliver fast, non-invasive, whole-body results and are therefore commonly included in the diagnostic work-up for IFDs, but findings often overlap with other diseases. In clinical practice, signs like pulmonary infiltrates are often presumed to be of viral or bacterial origin and treatment is initiated based on that assumption. Fungal infection is then only considered after failure of these initial therapies, resulting in a critical delay in diagnosis and treatment. Innovating imaging to provide specific biomarkers of fungal infection will improve diagnostic accuracy and inform targeted antifungal treatment5. Currently there are limited available treatment options for IFDs and they are complicated by severe adverse effects6. Furthermore, inappropriate use of antifungals across the One Health spectrum can drive the worldwide emergence of antimicrobial resistant pathogens4, increasing the already high mortality attributed to fungal infections and highlighting an urgent need for novel effective therapeutic strategies to combat IFDs.
Advancing our understanding of IFDs is pivotal to identify new biomarkers for differential diagnosis, evaluate novel treatment strategies and unravel host-pathogen interactions in the search for alternative therapeutic targets including host-directed therapy approaches. For these purposes, basic and preclinical research involving animal models are indispensable, with mice typically being the animal of choice to model aspergillosis, cryptococcosis, candidiasis and many other IFDs7,8,9,10.
Preclinical imaging plays a crucial role in addressing these mechanistic, diagnostic and therapeutic gaps for IFDs with animal models, through complementing traditional readouts of infection that rely on inherently invasive techniques requiring culling of animals at specified experimental timepoints11,12. Histology is prone to sampling errors and colony forming units (CFU) counting suffers from high variability, which can be overcome with more sensitive methods like quantitative PCR (qPCR)12,13. As these are endpoint measurements, studies relying on such snapshot data fail to capture the dynamic nature of infection progression. Survival, body weight and health scoring deliver non-invasive, longitudinal readouts of disease severity, yet only capture indirect effects of infection11. In contrast, preclinical imaging offers a non-invasive approach to investigate infection dynamics in live animals across different scales: monitoring pathogen spread, lesion formation and the host immune response in real-time, opening opportunities for the discovery of novel non-invasive diagnostic biomarkers and therapeutic targets, and all in all a more comprehensive view on infection progression.
Imaging comes with the bonus that each animal is followed over time, providing baseline data before infection and before interventions, reducing the number of animals needed14,15. Imaging can often already pick up pathological changes before the onset of clinical signs, allowing experiments to be conducted before the animals experience severe suffering or succumb to infection to provide survival data. Therefore, preclinical imaging aligns with the principles of the 3Rs by enabling longitudinal monitoring in individual animals, thereby reducing animal numbers, refining data quality and minimizing experimental suffering.
Despite the undeniable value of non-invasive imaging approaches in capturing infection dynamics, their integration into IFD research remains disproportionately limited. This underutilization might stem in part from technical, logistical, and infrastructural barriers—including limited access to imaging infrastructure, high operational costs, anesthesia requirements, reliance on radioactive tracers, and the need for standardized acquisition and analysis protocols. However, the (pre)clinical imaging field is undergoing rapid and strategic advancements aimed at democratizing access, simplifying workflows, and expanding utility. These developments are fueled by the recognized potential of preclinical imaging to overcome the limitations of traditional endpoint-based approaches, ultimately enabling more informative, ethical, and translatable fungal infectious disease research.
In this review, we will dive into how recent innovations in imaging modalities across different resolution and complexity scales can (1) improve accurate differential diagnosis of IFD in the clinic by identifying sensitive, pathogen-specific biomarkers, (2) accelerate the search for novel therapeutic strategies and (3) drive advances in our insights into host-pathogen interactions.
Pixel-perfect pathogen hunting: identifying novel biomarkers for differential diagnosis
Fungal infections can affect different organs, disseminate and become systemic. Whether in a patient or an animal model, understanding the extent of disease starts with a detailed look inside to assess fungal burden and the specifics of the pathogen and host response. The choice of imaging modality may depend on the suspected site of infection, as each site presents unique challenges and opportunities for investigation.
Airways under attack
We inhale fungal spores from the environment daily, making the respiratory tract a common primary infection site, especially in those whose natural defense systems are hampered16. The most common cause of pulmonary IFD is Aspergillus, although many other fungi can also infect the lungs16,17. To diagnose lung pathology, computed tomography (CT) is the imaging modality of choice due to the excellent inherent air-tissue contrast of the lungs17. As contrast in CT images is achieved through differential absorption of X-rays by tissues of different densities, lung CT can be performed without exogenous contrast administration - although in some cases a contrast agent may be used to reveal vascular occlusion and fungal angioinvasion18. Certain CT signs are indicative of invasive pulmonary aspergillosis (IPA) or mucormycosis, e.g. nodules with surrounding halo sign in neutropenic patients or cavitary lesions with air-crescent sign in patients recovering from neutropenia (Fig. 1A, B)17,19,20. Magnetic resonance imaging (MRI), which uses strong magnetic fields and radio waves to generate images based on hydrogen atom signals, may be considered as a radiation-free alternative to visualize lung disease, but MRI of the lungs is technically challenging due to the low proton density in air-filled spaces and the many air-tissue interfaces causing imaging artefacts due to magnetic susceptibility effects19,20. Specialized MRI protocols can overcome these limitations and allow visualization of pathological changes in the lungs19,21,22, but as they cannot reach the high resolution of CT, they are not favored for clinical implementation. CT remains unmatched and the cornerstone for diagnostic imaging of pulmonary IFD as it offers a faster, easier and cost-effective approach compared to other modalities. However, no CT or MRI sign is really specific for pulmonary IFD and the diagnosis remains difficult to make, requiring a multifaceted approach including host factors, clinical features including CT, and microbiological evidence to categorize infections as proven, probable, or possible17,23,24. These criteria provide a standardized framework for identifying IFDs, particularly in immunocompromised patients, and are primarily used for clinical trials and research as a proven diagnosis often still requires invasive sampling, which is far from obvious in these severely ill patients. This challenges current clinical diagnosis and further underscores the need for pathogen-specific differential diagnosis through non-invasive imaging.
Computed tomography (CT) images of a patient with pulmonary aspergillosis, illustrating the transition of dense nodular lesions surrounded by a halo of lower attenuation (halo sign) (A) to an air-crescent sign (B) following neutrophil recovery. Magnetic resonance imaging (MRI) of multiple tumor-like masses in the brain region showcasing: non-enhancing cryptococcomas in the axial plane T1-weighted post-contrast (C), parasagittal post-contrast (D), axial T2-FLAIR (E), and axial T2-weighted (F) planes. Whole-body [18F]FDG (fluorodeoxyglucose) PET (positron emission tomography) scan (G) and PET/CT images of a patient with disseminated candidiasis post-chemotherapy for acute lymphocytic leukemia, revealing intense metabolic activity in the lungs (H), liver, and spleen (I) as bright, high-intensity regions in the chest (H) and abdominal (I) areas. These imaging modalities suggest fungal infections but lack specificity, as similar findings can occur in non-fungal infections and malignancies. Definitive diagnosis is corroborated only through fungal identification, such as by culture or histopathology, underscoring the limitations of these imaging tools and the critical need for fungal-specific biomarkers to enhance diagnostic accuracy. This figure includes images that were either reproduced directly or adapted from the following publications: A, B Maertens et al., Current Opinion in Infectious Diseases, 2009208. C–F Chastain et al., Pathogens, 2022209. G–I Lawal et al., Diagnostics, 2021210.
In an experimental research context, unlike in the clinical context, differential diagnosis is not the prior aim, instead we seek accurate evaluation of an a priori known infection. Micro-CT was developed as the preclinical counterpart of CT with improved spatial resolution suitable for small animal lung imaging25. Micro-CT of the lungs visualizes inflammatory infiltrates and structural damage caused by infection and thereby provides longitudinal information on disease onset, progression, resolution and treatment efficacy21,26,27,28. The few minutes scanning time needed allows easy implementation of micro-CT into the experimental design and using a radio-safe dose allows repeated scanning to monitor the same animal over time without adverse effects29,30. Pulmonary pathology presents on micro-CT as hyperintense regions (nodular lesions, consolidation, ground-glass opacities etc.) caused by fungal growth and inflammatory infiltrates, or as hypointense regions as a result of hyperinflation or emphysema (Fig. 2A)12,31. These hallmarks of lung infection can be either visually scored or quantified to produce imaging-derived biomarkers of pathology in terms of volume and density26,30. These biomarkers reveal the extent of disease burden in a longitudinal manner in mouse models of rapidly progressing IFD such as neutropenic IPA, as well as detect compensatory changes such as expansion of the total lung volume in the lungs during slowly progressing infection such as cryptococcosis, long before the onset of symptoms26. During infection, the volume of hyperintensities and lung density typically increase due to lesion formation and inflammatory infiltrates, whereas hyperinflation and emphysema decrease the lung density. With the aerated lung volume, micro-CT also offers a readout of lung function which correlates to the inspiratory capacity12. Ongoing efforts focus on extracting even more detailed biomarkers on lung pathology from micro-CT scans, that otherwise require additional, dedicated measurements of lung function32.
A Visualization of pulmonary pathology with micro-computed tomography (micro-CT) in healthy lungs (left), and Aspergillus fumigatus infected mice: influenza-associated pulmonary aspergillosis (IAPA, middle) and neutropenic aspergillosis (right). Arrows indicate pathology: consolidation, ground glass opacities and airway dilation. Micro-CT of IAPA lung pathology visualizes the combined viral and fungal pneumonia, without distinction of both infectious etiologies. B T2-weighted brain magnetic resonance (MR) image of two mice with cryptococcosis. Lesions appear as hyperintense regions. C Immuno-PET/MR with the A. fumigatus-specific 64Cu-hJF5 radiotracer to detect invasive pulmonary aspergillosis in a neutropenic mouse model at 48 h p.i. Maximum Intensity Projection (MIP) of the positron emission tomography (PET) image, overlaid with the T2-weighted MR image for anatomical context in the Fusion image. D In vivo bioluminescence imaging (BLI) of lung-to-brain dissemination in a mouse model of cryptococcosis at 3.5 weeks post-infection (p.i.) where BLI signal is limited to the lungs and at 4.5 weeks p.i. signal appears in the brain. This figure includes images that were adapted from the following publications: A neutropenic aspergillosis lungs (right) from Reséndiz-Sharpe et al., Disease Models & Mechanisms, 202228. B Vanherp et al., Antimicrobial Agents and Chemotherapy, 202015. C Henneberg et al., Nature Communications, 202176. D Vanherp et al., Disease Models & Mechanisms, 201989. Figure created in Biorender.
Brain fog
IFDs may not remain restricted to the lungs but disseminate and become systemic. For example, cryptococcosis disseminates to the central nervous system (CNS) causing life-threatening meningoencephalitis33,34. Capturing when and how a fungal pathogen disseminates to the brain is one of the great enigmas of the field. Due to its high contrast between soft tissues, MRI is the imaging modality of choice to visualize the CNS, whereas the intrinsic contrast of CT in the brain will be low35. In patients, as well as in rodent models, the development of meningitis and formation of cryptococcomas is successfully visualized with MRI, with lesions appearing as hyperintense regions on T2-weighted MR images (Figs. 1C–F and 2B)15,34,36,37. Infection progression and response to antifungal treatment can be monitored based on the number, volume and spatial distribution of lesions15.
The differential diagnosis of cryptococcomas or other fungal masses in the brain with anatomical MRI is challenging, as other brain lesions including tumors and bacterial abscesses can present as a hyperintense mass on T2-weighted MR images38,39,40. The required confirmatory tests are often invasive, highlighting the need for new non-invasive diagnostic tools with a higher specificity for fungal pathogen detection. To this end, trehalose, a metabolite produced in high concentrations by cryptococci, was explored as a potential biomarker. In a mouse model, MR spectroscopy (MRS), an MRI-based method to detect and quantify tissue metabolites, non-invasively detected trehalose as a marker for cryptococcomas in the brain41. To improve the poor spatial resolution of MRS and allow the detection of smaller lesions, a chemical exchange saturation transfer (CEST) protocol was optimized to detect Cryptococcus species on MRI42. CryptoCEST uses contrast generated by the high trehalose concentration produced by cryptococcal cells for in vivo detection of cryptococcosis and could differentiate cerebral cryptococcomas from gliomas in the mouse brain. MRI protocols like this have the potential to be translated to the clinic, which would alleviate the need for invasive biopsies and aid the correct diagnosis of IFD in the brain.
MRI is not limited to anatomical imaging and can also provide information on a wide range of tissue properties including hemodynamics and metabolic changes in tissues and brain function8. This is relevant to investigate whether alterations in the brain microvasculature, blood flow, blood volume and blood-brain-barrier (BBB) integrity, contribute to cryptococci entering the CNS, or can serve as biomarkers to estimate permeability of the brain for antifungal treatment37,43,44. Through quantitative MRI (qMRI), biomarkers assessing different tissue properties can be extracted based on their imaging properties45,46,47. For example, diffusion MRI measures the diffusion of water molecules inside tissues. When applied to cryptococcomas, a higher cell density results in a lower apparent diffusion coefficient (ADC) and vice versa, a higher ADC corresponds to lower fungal density due to larger cryptococcal cell size and thicker capsules, which differs between molecular types of C. neoformans and C. gattii46,47.
Lights, tracers, action!
To address the clinical need for specific pathogen detection for differential diagnosis, positron emission tomography (PET) and single-photon emission computed tomography (SPECT) in combination with specific pathogen-targeted radiotracers are attractive alternatives. Depending on the type of radiotracer these molecular imaging techniques rely on, they allow for the whole-body visualization of host cells or pathogens involved in infectious processes through specific targeting mechanisms resulting in local accumulation of the radiotracers.
a) Like sugar tastes, so sweet
The gold standard PET radiotracer, [18F]fluorodeoxyglucose ([18F]FDG), traditionally used in oncology and neurology, has also been investigated for infection imaging (Fig. 1G–I)48,49,50. However, its utility may be limited due to non-specific uptake in areas of high glucose metabolism, which can stem from both pathogens and other highly metabolically active cells, such as immune cells or cancer cells51. While the very high sensitivity of [18F]FDG PET facilitates its application for fungal infection imaging in the clinic, its inability to distinguish between different sources of increased glucose uptake restricts its diagnostic specificity and thus its wide-spread use52. Nonetheless, [18F]FDG has shown promising as a tool for therapy monitoring in patients with previously diagnosed IFD53,54, especially in IPA and invasive candidiasis, allowing clinicians to assess treatment efficacy and adjust therapy accordingly.
To address the specificity limitations of [18F]FDG, radiotracers targeting fungal-specific pathways have been developed and tested preclinically. These fall into two main categories: first, siderophore-based radiotracers, targeting the iron uptake pathways of fungi during growth55. Radiolabeling siderophores with isotopes such as 68Ga or 89Zr enabled specific imaging of IFDs56,57. For instance, 68Ga-triacetylfusarinine C (68Ga-TAFC) and 68Ga-desferrioxamine B (68Ga-DFO-B) have shown high stability and specificity in imaging IPA in rat models, providing high sensitivity and clear differentiation of IFDs58,59 with clinical evaluation underway for 68Ga-TAFC60. Second, radiolabeled sugars exploiting the unique metabolic profile of pathogens, e.g. [18F]fluorodeoxysorbitol ([18F]FDS) has been used for detecting IPA in vivo and Candida infections in vitro61,62,63. Although [18F]FDS primarily targets bacterial infections64,65, its lack of uptake by mammalian cells makes it a promising candidate for general infection imaging. More recently, 2-deoxy-2-[18F]fluorocellobiose has been developed for aspergillosis imaging in mice66. This radiotracer demonstrated high specificity towards Aspergillus fumigatus, with minimal bacterial uptake, highlighting the potential of complex radiolabeled sugars for selective fungal pathogen imaging. Across all metabolic targets for selective fungal imaging, a key consideration is species-specificity of that target. For example, while some siderophore tracers are highly selective for certain fungi, others may also be taken up by bacterial species58,67.
b) Fatal attraction
Radiolabeling clinically validated antifungals has been another avenue of exploration for fungal detection68. The most widespread radiotracers include 18F-fluconazole and 99mTc-fluconazole for PET and SPECT, respectively69,70. These tracers recapitulate the antifungal activity of their parent compounds but have yet to achieve widespread clinical use. Other compounds, such as Amphotericin B and Caspofungin71,72, have also been radiolabeled, as well as antibiotic peptides73,74 with 99mTc-Ubiquicidin representing the most accepted approach, with PET compatible variants appearing in the last decade. However, antimicrobial peptides often are cross-reactive against bacteria, which may be a limiting factor for differential diagnosis during clinical translation. Overall, this approach remains promising, but the chemical modifications required for radiolabeling of small molecules and peptides often lead to changes in target affinity detrimental to their performance as radiotracers.
c) Tag, you’re it!
Antibodies and their fragments offer high specificity for fungal species, making them promising for radiotracer development. 64Cu-radiolabeled JF5, an antibody targeting the Aspergillus-specific β-1,5-galactofuranose, has enabled sensitive in vivo imaging of IPA in mice (Fig. 2C)75. This approach could correlate radiotracer accumulation with fungal load, making it valuable for both diagnosis and therapy monitoring76. Humanized versions of JF5 have recently been used in clinical studies, providing the first clinical PET images of IPA and demonstrating a strong correlation with conventional diagnostic methods77,78. Another promising strategy involves 89Zr radiolabeling anti-β-glucan antibodies and their Fab fragment, which has recently shown potential in PET for aspergillosis79 and would allow for broader IFD detection as β-glucan is present in most clinically relevant fungi. While full-length antibodies showed extended circulation times, leading to lower signal-to-background ratios, the Fab fragments offered improved imaging contrast due to shorter circulation times. Besides filamentous fungi, immuno-PET tracers are also being developed for yeasts, such as 64Cu-labeled MC3 antibody for invasive candidiasis detection80.
The development of pathogen-specific radiotracers continues to evolve, with a focus on improving specificity and sensitivity for IFDs. Advances in antibody engineering and radiolabeling chemistry hold promise to enhance the clinical utility of these tools. Future research should aim to extend towards targeting different fungal pathogens, optimize radiotracer pharmacokinetics including BBB penetration, and validate these agents in clinical settings.
On the bright track: accelerating the preclinical drug development pipeline
While pathogen-specific labeling is crucial in the clinic for improved differential diagnosis, it also holds significant value in preclinical research, albeit with a different objective. In the preclinical context, specific fungal labeling enables direct, non-invasive monitoring of fungal burden over time, which is essential to certain settings, particularly when assessing treatment efficacy in the search for new antifungal strategies.
While MRI and micro-CT offer excellent opportunities to quantify several essential aspects of infection, including treatment response, they cannot distinguish the fungal burden from the host’s inflammatory response and tissue remodeling. A direct readout of fungal burden is therefore indispensable. To achieve this, reporter strains or fungus-specific molecular probes can be employed in animal models, enabling direct and longitudinal quantification of the fungal burden with PET or SPECT, or with optical imaging as a radiation-free and cost-effective alternative to nuclear imaging for high-throughput studies in small animals.
Illuminating the invisible
In vivo bioluminescence imaging (BLI) is a whole-body optical imaging tool based on the detection of photons produced by the enzymatic reaction catalyzed by luciferase, expressed in genetically modified cells or micro-organisms81. In contrast to fluorescence imaging, BLI requires no excitation laser resulting in a low background signal and no phototoxicity or photobleaching issues, enabling longer acquisition times necessary to sensitively image low-metabolically active fungi without compromising tissue integrity82. The most commonly used bioluminescent reporter for eukaryotic cells is the firefly luciferase (FLuc), which uses d-luciferin as its substrate, with new luciferases and substrates continuously being developed for improved BLI sensitivity83,84. The limited tissue imaging depth of the natural Fluc made initial BLI systems suitable for superficial fungal infections, but not for systemic or deep-seated infections83,85,86,87. To improve BLI sensitivity, modified luciferases and substrates were designed towards a red-shifted emission spectrum (600–700 nm) which has less light absorption in mammalian tissues. Thermostable codon-optimized red-shifted FLuc has been successfully integrated in yeasts such as Candida and Cryptococcus species and in filamentous fungi including Aspergillus fumigatus, which improved sensitive detection of fungal burden in vivo (Fig. 2D)28,88,89,90,91. Improving sensitivity is not only important for deep-seated infections but also for systemic infections, with the pathogen being distributed throughout the body in low numbers, or to detect early stages of fungal dissemination89,92.
In the search for novel antifungal strategies, especially with emerging antifungal resistance, BLI offers unprecedented advantages. Since only viable cells express luciferase, the fungal infection kinetics and distribution can be visualized and quantified over time, omitting the need for cross-sectional endpoint measurements of fungal burden. The good sensitivity and real-time readout of fungal viability therefore make BLI the ideal tool to evaluate treatment efficacy in animal models14,27,92,93,94,95,96. Moreover, it allows for the identification of unexpected sites of infection persistence despite otherwise effective antifungal treatment in the primary target organ, which would be missed if only the organ of interest was analyzed at experimental endpoint97. Beyond mice, BLI has also proven essential as proxy for fungal burden in invertebrate models like Galleria mellonella larvae, unlocking this model as an ethical burden-free antifungal screening platform to evaluate promising therapeutic strategies for efficacy under in vivo conditions before stepping-up to mice (Fig. 3)98,99,100,101,102,103. In the context of emerging antifungal resistance, tools such as BLI are indispensable for accelerating the discovery of antifungal treatment while reducing the use of vertebrate models, offering a more ethical and cost-effective approach to combat IFDs.
Illustration of BLI-mediated detection of anticryptococcal treatment efficacy from in vitro screening over in vivo screening in Galleria mellonella to mice. A The generation of bioluminescent fungi enables real-time quantification of fungal burden for high-throughput in vitro antifungal efficacy screening. Image shows in vitro BLI of three different bioluminescent Cryptococcus neoformans KN99α transformants compared to the non-bioluminescent wild-type (WT) strain with serial 10-fold dilutions in their corresponding colony-forming units (CFUs). B Quantification of the antifungal effect of amphotericin B (AMB-d), fluconazole (FLC) or sterile water (control) on the in vitro growth of bioluminescent C. neoformans (strain NE1270) liquid cultures from BLI measurements over time. C In vivo BLI of G. mellonella larvae infected with 104 (left) or 106 (right) bioluminescent C. neoformans cells (strain NE1270) on day 2 post infection. D Quantification of the antifungal effect of different amphotericin B (AMB) concentrations in G. mellonella larvae infected with 105 C. neoformans cells (strain NE1270) from in vivo BLI signals over time. E In vivo BLI of mice infected with bioluminescent C. neoformans (strain NE1270) receiving saline, liposomal amphotericin B (L-AMB) or fluconazole (FCZ) treatment on day 5 post infection. F Longitudinal quantification of in vivo BLI photon flux from the brain or abdomen region of individual mice infected with bioluminescent C. neoformans (strain NE1270) receiving saline, liposomal amphotericin B (L-AMB) or fluconazole (FCZ) treatment. This figure includes images that were adapted from the following publications: A Vanherp et al., Disease Models & Mechanisms, 201989. B, E, F Vanherp et al., Antimicrobial Agents and Chemotherapy, 202015. C, D Vanhoffelen et al., Virulence, 2024101. Figure created in Biorender.
The twilight zone
Fluorescence imaging (FLI) could serve as a substrate-free alternative to BLI, and fluorescent reporter strains or fluorescently labeled fungus-specific probes are often commercially available, offering more flexibility than BLI in this regard104. However, whole-body FLI is rarely applied in vivo due to the high signal absorption and background autofluorescence of mammalian tissues at the excitation/emission wavelengths of conventional fluorophores like green fluorescent protein (GFP)105. Although efforts are made to develop brighter, near-infrared fluorophores for deeper tissue imaging, its use for in vivo fungal burden quantification remains widely underexplored106,107,108. Considering FLI’s current limitations on a whole-body imaging scale, its strengths are better suited for microscopy.
While these optical imaging tools are undoubtedly indispensable in studying the dynamics and distribution of fungal burden and screening for antifungal efficacy in preclinical research, the need for genetically modified reporter strains excludes the translation of BLI or FLI for clinical use. However, FLI with fungal-specific fluorescently labeled probes or dyes opens potential clinical applications109. We thereby think about combining intraoperative FLI, now primarily used for guided surgical tumor resection110,111, with fungal labeling towards guided removal of abscesses or biofilm-contaminated implants.
Not only light, but also sound can be imaged. Ultrasound imaging, while not yet specifically used for fungal infections, can detect abnormalities in soft tissue structures using soundwaves. Lung ultrasound can aid in diagnosing pneumonia, pulmonary abscess and pleural effusion, but these findings are not specific for fungal infections112,113. In clinical scenarios, ultrasound has been used to screen for hepatosplenic lesions, including fungal infection, yet it lacks specificity and findings are commonly corroborated by CT or MRI114. Nevertheless, the application of ultrasound could be further explored as a supportive tool in specific clinical contexts, especially because it can be carried out at the patient’s bedside112. Photoacoustic imaging (PAI) builds further on this concept by combining optical excitation of a pathogen-specific probe and ultrasound detection115. PAI combined with a bacterial-specific dye has for example been used to differentiate sites of sterile inflammation and sites of bacterial infection in mice116. Since sound waves are less scattered in tissues than photons117, the development of fungal-specific PAI-compatible probes could emerge as a valuable diagnostic imaging tool, especially for infection sites where optical imaging falls short.
It takes two to tango: unraveling host-pathogen interactions
It is becoming more and more evident that we should not only focus on the pathogen itself as the main driver of disease outcome, and that the role of the host immune response should not be overlooked. Indeed, IFD management is currently centered around antifungal treatment, which is challenged by emerging resistance development, toxicity limitations, drug-drug interactions and limited bioavailability6. It is clear that in certain fungal infections, antifungal therapy alone is insufficient, as high mortality rates persist despite recommended treatments. This underscores the need to develop therapeutic strategies that not only target the pathogen but also support or enhance the host immune response to improve clinical outcomes. Host-directed therapy is now being explored to help combat IFDs. For example, in IFD patients with hematological malignancies, the compromised immune system can be stimulated by supplementing G-CSF or GM-CSF to restore neutrophil counts, and prophylactic IFNγ supplementation can be beneficial in preventing IFDs in chronic granulomatous disease patients118,119. Contrarily, in those cases where an overactive immune response may be a driver of disease, dampening the hyperinflammatory response may be valid strategy, as seen in a mouse model of influenza-associated pulmonary aspergillosis, where dampening IFNγ improved outcomes120. In any case, altering the immune system is a delicate balance: we aim to strengthen immune defenses and restore immune functions while also preventing hyperinflammation that may be driving collateral damage. Therefore, we need precise and dynamic insights on the role of the host response during the course of infection and experimental host-directed therapies, which can be offered by imaging.
50 shades of red
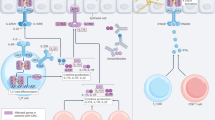
Similar to using genetically modified reporter pathogens, transgenic reporter mouse models expressing luciferases or fluorescent proteins under immune cell-specific promotors enable tracking of immune responses with BLI or FLI. Luciferase can be expressed under promotors of general inflammatory markers, e.g. interleukin (IL)-1β, to quantification the overall inflammatory response, or in specific immune cell populations for in vivo tracking with BLI121,122. For example, in a melanoma mouse model, luciferase-expressing macrophages co-stained with the near-infrared fluorescent dye DiR, were systemically injected to localize and quantify the macrophages at tumor sites with BLI and FLI123. Alternatively, fluorescent dyes can be used to track immune responses, e.g. by staining hypoxia (primarily induced by neutrophils at the infection site in a subdermal C. albicans infection mouse model124.
Different luciferases and substrates are available, with new and brighter alternatives being engineered, each with their own distinct light emission spectrum. This presents the opportunity to perform multiplex BLI by selecting the correct combination of luciferases and substrates, and spectrally unmixing the signals to visualize multiple cell types or pathways simultaneously125,126,127. To illustrate, in the TbiLuc mouse model all T cells constitutively express a green-emitting click beetle luciferase, and activation of the T cells induces the expression of a red-shifted firefly luciferase, allowing dual-color tracking of T cell localization and activation127. Although multiplex BLI for immune cell tracking is primarily pioneered in cancer research, their potential in unraveling the host immune response in (fungal) infections is undeniable. To achieve deep-tissue multiplex BLI, careful spectral planning is needed: the selected luciferases must emit light that is both sufficiently red-shifted or near-infrared for optimal tissue penetration while still being spectrally distinct enough to allow accurate signal unmixing128. Navigating these different ‘shades of red’ is key to unlock the full potential of multiplex BLI in vivo.
Tracer fever
Besides optical imaging methods, radionuclide imaging is another well-established imaging modality to track the immune response. Molecular imaging of infection was initially enabled by 67Ga salts for SPECT ~50 years ago and have more recently been complemented by 68Ga salts for PET129. These radioisotopes follow a similar fate to Iron III and accumulate in regions of high metabolic activity such as inflammation, infection and malignant tumors, and have been used notably for fungal infections in HIV patients130. Nowadays, 67Ga is mostly replaced by [18F]FDG although some niche applications remain131.
To date, [18F]FDG PET remains the gold standard for radionuclide imaging of glucose metabolism, with established applications in oncology and neurology, and an increasingly recognized role in imaging inflammation and infection132. Its clinical approval makes [18F]FDG PET the radiotracer of choice for exploratory clinical research in infection imaging. Moreover, its status as the “gold standard” positions it as a reference in studies of novel radiotracers, facilitating the generation of substantial preclinical data. However, its non-specific uptake in various cell types, including leukocytes, bacteria, malignant cells, and fungi, combined with its high sensitivity, makes [18F]FDG PET a double-edged sword, capable of detecting a broad range of pathological conditions and cellular events at the cost of its specificity52.
To advance beyond these limitations and specifically monitor the host immune response, techniques have been developed for radiolabeling white blood cells (WBCs)133. These approaches enable the tracking of WBCs to localize infection sites and assess their severity and potential therapeutic response. Clinically, SPECT imaging of WBCs, after in vitro radiolabeling of blood samples with tracers such as 111In-oxime or 99mTc-hexamethylpropyleneamine134,135, has demonstrated considerable potential in distinguishing infections from sterile inflammation, with a primary focus on bacterial infections and classically in cases such as diabetic foot infections and osteomyelitis136,137, all being widely accepted in the clinic. The application of these methods to IFDs is however mostly unexplored as certain challenges persist, notably due to variability in leukocyte recruitment at fungal infection sites138.
To address the requirement for in vitro labeling, alternative strategies have been explored, such as the radiolabeling of granulocyte-specific antibodies139. While these approaches have shown success in bone infection and periprosthetic infections, the influence of their pharmacokinetic and biodistribution profiles may lead to results that differ from traditional WBC labeling140,141. These approaches are yet to be applied to IFDs. Ongoing research aims to develop more specific radiotracers, including those targeting neutrophils, with promising preliminary results in candidasis142,143. Macrophage imaging by PET and SPECT has also recently shown potential in a radionuclide imaging approach, although focusing mostly on joint inflammation and cancer144,145. Significant progress in translating these innovations to the clinical imaging of IFD remains to be achieved.
Beyond systemic immune responses, localized changes associated with acute infections offer further opportunities for imaging. For instance, hypoxia, a common feature of localized fungal infections146, presents a viable target for hypoxia-specific radiotracers such as [18F]FMISO and [18F]FAZA, as has been demonstrated in the tumor microenvironment as well as in tuberculosis and odontogenic infections147,148,149. However, caution is warranted when repurposing existing tracers, as their diagnostic specificity may be inherently limited in niche applications.
The best of both worlds
While each imaging modality has its own strengths and applications, no single modality can fully address all gaps in our knowledge of IFDs. To obtain the complete picture, imaging modalities should be combined to acquire complementary information, either across all scales, across models, and from bench to bedside and back. Within a single animal model, combining modalities provides insights on different levels from whole-body BLI as a direct readout of fungal burden, to organ level with anatomical imaging modalities like MRI or micro-CT to visualize structural damage, lesion formation and inflammatory infiltration, even zooming in to cellular resolution with intravital microscopy (IVM)28,93. Evaluating treatment response with multimodal imaging provides spatiotemporal information on fungal viability in response to treatment directly, as well as on the resolution of lesions and inflammation15,27,93,94. Imaging protocols can be adapted across different models, for example from in vitro screening to in vivo testing in G. mellonella larvae and mouse models to facilitate the antifungal discovery pipeline, all with the same modality: BLI100,101. Furthermore, biomarkers identified and validated in animal models can be translated into clinical diagnostic tools like radiotracers77.
To unravel host-pathogen interactions, we need to image multiple cell types simultaneously and thus label them differentially. This could be achieved by using fluorophores with different excitation and emission wavelengths, or by using different luciferases and substrates and spectrally unmixing the signals to visualize and quantify different pathways125, e.g. to monitor both the pathogen spread and key host players in fungal dissemination. While multiplexed optical whole-body imaging still faces several challenges and is not yet applied for IFDs, some encouraging examples of multiplex BLI of host-pathogen interactions can be found for mouse models of bacterial meningitis. In combination with tracking bacterial spread, astrogliosis (as a marker for neuronal injury) and general inflammation were quantified in different luciferase-expressing reporter mouse models150,151.
In vivo BLI, FLI, MRI, micro-CT, PET and SPECT provide us with a large field-of-view to visualize aspects of infection at the whole-body level. They do not offer the resolution to visualize host-pathogen interactions at a cellular level152. To add cellular resolution microscopy data to information on infection dynamics at a whole-body scale, dual-labeled fungi are valuable for such multimodal imaging. For example, the Aspergillus-specific monoclonal antibody JF5 was dual-labeled with a radionuclide for in vivo PET and a fluorophore for ex vivo microscopy76. Radiolabeled, fluorescent probes or dyes with fungal uptake present a similar dual-imaging approach153,154. The engineering of fungi to co-express luciferase and a fluorophore is therefore high on the wish list, as that would enable in vivo fungal burden quantification with BLI, and intravital or ex vivo fluorescence microscopy to image host-pathogen interactions.
Zooming in onto the cellular battlefield
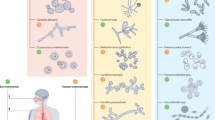
To capture the host-pathogen relationship at cellular level, at key time points during infection identified by whole-body imaging, microscopy plays a central role. While traditional microscopic evaluation of tissue samples is performed post mortem, IVM has been introduced to monitor immune cell recruitment and pathogen behavior in vivo155. IVM can be done at endpoint by exposing the organ of interest in a terminally sedated animal156, but ideally, to obtain longitudinal data, the organ should be accessed non-invasively. This can be achieved by using optical windows, which have been successfully implanted over different organs including the brain and lungs157,158. In the context of IFDs, IVM has been used to study the interaction between immune cells and cryptococci at the BBB and help elucidate the mechanism through which the pathogen enters the brain (Fig. 4A)159,160,161. A challenge presented by IFDs for IVM is that the site of infection is usually a deeper situated organ, which is difficult to access with a microscope objective. Moreover, surgical implantation of the window is technically challenging and this approach is only suited for superficial tissue imaging, with two-photon microscopes reaching up to 1 mm deep105. An alternative IVM approach is fibered confocal fluorescence microscopy (FCFM), in which the microscope objective is replaced by a thin, small probe consisting of a bundle of optical fibers162,163. This flexible probe can endoscopy-wise be inserted into the target organ to gain access to deeper tissues in a minimally invasive manner (Fig. 4B). For example, FCFM of the brain has been done to image fungal cell density in cryptococcomas46. For bronchoscopic FCFM, the probe was inserted in the airways of free-breathing mice, which allowed for repeated imaging to visualize and quantify fungal growth of GFP-expressing A. fumigatus, C. neoformans or C. gattii over time164. A limitation of FCFM is that currently available systems only have one- or two-color channels, meaning that the amount of cell types visualized simultaneously is limited164,165.
To perform intravital microscopy (IVM) of difficult-to-reach organs in mice, two options are presented: A The surgical placement of an optical window over the organ of interest to image cellular trafficking in real-time. Image shows IVM of cryptococci transported to the brain. Blood vessels (red) and FITC-labeled C. neoformans (green). Alternatively, B with fibered confocal microscopy (FCFM) a flexible optical probe can be inserted in the tissue, for example in the airways of mice infected with GFP-expressing Aspergillus fumigatus, showing densely packed hyphae, or GFP-expressing Cryptococcus gattii, showing dispersed round cells. Zebrafish larvae offer a transparent alternative model to mice to study host-pathogen interactions: C optically transparent zebrafish larva 3 days post-fertilization (top) and non-invasive live imaging of cryptococcosis at 1 day post bloodstream infection (bottom). D Real-time interaction of macrophages (green) with cryptococci (blue) in infected zebrafish larva. Advanced ex vivo models to unravel host-pathogen interactions include E light sheet fluorescence microscopy (LSFM) of fixed, optically cleared organs. For example, cleared mouse lung harvested 48 h post-infection (p.i.) with a fluorescent Aspergillus fumigatus to image spatial distribution of the pathogen inside the lung. LSFM image (top), scale bar = 1 mm. Magnified two-photon microscopy image of the same lung (bottom), scale bar = 50 µm. Lung tissue (gray) and fungal biomass (red). F, G A live tissue alternative are precision-cut tissue slices. F Brightfield image of a precision-cut lung slice (PCLS) showing airway structures, scale bar = 500 µm. G Fluorescence microscope image of a PCLS visualizing neutrophils (Ly6G+, red), macrophages (CD11c+, white) and epithelia (Podoplanin, blue), scale bar = 100 µm. This figure includes images that were adapted from the following publications: A Shi et al., Cellular Microbiology, 2012159. B Vanherp et al., Scientific Reports, 2018164. E Henneberg et al., Nature Communications, 202176. Image created in Biorender.
And then it gets fishy…
IVM in optically transparent zebrafish larvae represents an unparalleled model for the study of IFD progression and pathogenesis in an ethically favorable model (Fig. 4C, D). What makes zebrafish larvae so useful for the study of IFDs? The quality of live fluorescent microscopy is equivalent to that achieved in vitro with isolated cells but can be achieved non-invasively in vivo. Furthermore, repeated imaging over the course of infection is uncomplicated to achieve166, something rarely feasible in IVM of mice. The imaging possibilities are constantly evolving with improvements in technology, most notably with the advent of high-resolution selective plane illumination microscopy (also known as light-sheet fluorescence microscopy (LSFM))167. However, without biological relevance there is no benefit to high quality imaging. Here zebrafish excel as an experimental model of IFD. The combination of innate immune system that is very similar to humans168 with the broad host range of many human fungal pathogens, e.g. Aspergillus fumigatus and Mucorales, being as they are environmental organisms, means that the pathogenesis of infection is highly conserved169,170. The exception to environmentally originating fungal pathogens is the human commensal Candida albicans but there is now a significant body of work demonstrating that zebrafish larvae are a suitable and useful model here as well171,172. The live imaging of IFDs has led to several new insights into pathogenesis. For example, the mechanisms of Cryptococcus dissemination and the importance of the early macrophage phagocytic response have been directly demonstrated for the first time in zebrafish166,173,174. Dynamic imaging of immune pathology in parallel to disease progression is possible in fluorescent zebrafish reporter lines for immune cell mediators (e.g. TNFα and IL-1β), validated for IVM175,176. The availability of such transgenic zebrafish models combined with advances in optical imaging provides a powerful platform to address specific research questions on host-pathogen interactions in IFDs177,178.
Besides zebrafish larvae, other non-mammalian alternative animal models are worth considering to study the interaction between fungal pathogens and the host. These include invertebrates such as G. mellonella179, and other examples like Drosophila melanogaster (fruit fly) and C. elegans180,181. However, these models are mainly used in the context of studying virulence factors of fungi and testing efficacy and biocompatibility of antifungals, whereas they are less suited for imaging of host-pathogen interactions due to limited optical access (except for C. elegans), reduced anatomical complexity, and fewer genetic tools. As a vertebrate alternative model, the chicken embryo has been employed to study Aspergillus virulence factors182, however, live cell microscopy is typically restricted to ex ovo conditions, compromising embryo viability and reducing the experimental timeframe183. Ultimately, depending on the research question, these models, combined with a relevant imaging modality, can offer important insights on fungal infections in an ethically responsible manner. Yet, for live cell imaging of the innate host response, zebrafish are unmatched due to their optical transparency and availability of transgenic reporter lines.
Peeling back the layers
While in vivo imaging methods such as IVM have provided profound insights into host-pathogen biology, these methodologies often entail surgical trauma and exhibit lower throughput capabilities184. To mitigate these limitations, ex vivo models have been developed, facilitating the investigation of tissues and organs outside of their native environments while preserving essential biological components. These models can utilize either fixed or live tissues. Fixed tissues retain cellular architecture and are amenable to immunohistochemistry and light microscopy, but also to more advanced microscopy tools. Optical projection tomography and LSFM of optically cleared organs enables the three-dimensional imaging of entire tissue structures to visualize host-pathogen interactions within their preserved spatial context (Fig. 4E)76,152. However, fixed tissues lack the ability to study dynamic, live processes such as cell-to-cell interactions and cellular activity185.
To overcome this and investigate dynamic host-pathogen interactions, live tissue models like precision-cut lung slices (PCLS) offer a versatile platform as they can be derived from human or murine samples that are naïve, treated, or infected. The PCLS technique involves instilling the lung with low-melting-point agarose followed by sectioning the tissue into ~300–400 µm slices. These slices can remain viable for up to 14 days, maintaining vital biological properties, such as smooth muscle contraction and ciliary activity186,187,188,189. Moreover, they exhibit biological activity and responsiveness to external stimuli, including irritants and pathogens, thereby mounting appropriate immune responses189,190,191,192,193,194. PCLS not only retains the structural architecture of the lungs but also retains the spatial organization of various cell types, including ciliated, non-ciliated, secretory, structural, and immune cells. This characteristic allows for the examination of cell-to-cell interactions within a tissue model that closely resembles the respiratory system in a three-dimensional context, thereby facilitating the study of host-pathogen interactions (Fig. 4F, G)192,195,196. Additionally, its multimodal capabilities extend beyond microscopic evaluations, including light and scanning electron microscopy, and enables RNA and protein quantification and analysis, cell marker analysis, and genetic manipulation. Such capabilities facilitate the collection of data that is representative of a single animal, significantly reducing the number of animals required for studies197,198,199,200,201,202,203,204. Collectively, these aspects underscore the importance of the PCLS model in studying lung infections and inflammation, with promising implications for translational research, including drug discovery189,205,206,207.
Discussion
IFDs represent a complex clinical, societal and economical challenge, imposing a substantial burden on healthcare systems and impacting patient morbidity, mortality and quality of life. To help solve this we rely on research in preclinical models to provide us with a better understanding of what drives infection outcome. By implementing imaging modalities in these animal models, infection can be captured as it unfolds, enabling earlier detection, more sensitive treatment monitoring and a deeper understanding of host-pathogen interactions, which eventually also drives better management of IFDs in the clinic. Clinically, IFD management is hindered by diagnostic difficulties and limited therapeutic options. To address this, preclinical imaging plays a crucial role in identifying fungal-specific biomarkers that can be translated into non-invasive, highly sensitive and specific diagnostic tools in the clinic; this is particularly true for PET radiotracers. Additionally, preclinical imaging provides an essential platform to accelerate antifungal testing in animal models through real-time quantification of fungal viability, eventually bringing new therapies to the clinic. Using imaging-derived biomarkers of disease or specific tracers in patients then aids in treatment monitoring in patients, thereby further accelerating clinical trials. Challenges encountered in the clinic, such as newly identified host risk factors or emerging fungal pathogens, can in turn be modeled preclinically, where already established imaging pipelines help us better understand and adapt to these emerging IFDs.
At the interface of preclinical testing and clinical application, imaging serves three distinct yet complementary purposes. On the one hand, the development of new imaging biomarkers and modalities addresses a key clinical need, namely the rapid and non-invasive identification of the infectious origin without having to wait on cultures from often difficult to access infection sites, which enables earlier and more targeted treatment decisions. On the other hand, imaging plays a critical role in the antifungal drug development pipeline. It enables sensitive, real-time readouts of fungal burden and treatment efficacy both preclinically and clinically. This not only accelerates the identification of promising drug candidates preclinically, but also refines readouts in clinical trials, eventually speeding up the approval of drugs. Lastly, unraveling host-pathogen interactions through imaging may reveal new therapeutic targets, guiding the discovery of new antifungals or immune-modulatory strategies.
Apart from accelerating clinical translatability of novel diagnostics and antifungal therapeutics, the strength of imaging lies in its translatability across different models within the preclinical phase of research and development. To accelerate the antifungal testing pipeline, BLI offers a rapid, cost-effective approach for in vitro high-throughput drug screening. Promising compounds can then be tested in ethical burden-free in vivo models like Galleria mellonella, after which the results can be validated in murine models, all with the same imaging modality and imaging-compatible strains. Similarly, studies aiming to unravel host-pathogen interaction benefit from the use of imaging across models. Zebrafish models are particularly useful for studying the innate immune response to fungal infection with IVM, while IVM in mice allows imaging of host-pathogen interactions in a more physiologically relevant context. By building on discoveries from simpler animal models and scaling up to mammalian models, imaging can bridge the gap between exploring disease mechanisms and clinical relevance in an ethically responsible manner.
Imaging across multiple scales is central to fitting the different pieces of the puzzle together, enabling real-time tracking of disease progression, treatment response and host-pathogen interactions in vivo. As no single imaging modality captures all these scales, a multimodal approach brings the solution. At whole-body level the fungal burden and pathogen distribution can be monitored over time with optical imaging or PET, which can be combined with organ-level MRI or micro-CT to reveal structural changes caused by the infection, and lastly we can zoom in to a cellular-level resolution through IVM for real-time imaging of the invading pathogen and the host response. Where in vivo methods fall short, either due to limited imaging depth or insufficient cellular resolution, advanced ex vivo tools can further bridge the gap between cellular-level insights and the tissue-level context. By leveraging the full potential of preclinical imaging, the field can accelerate drug discovery from a traditional antifungal point of view, as well as an immune-modulatory approach for which we need to deepen our understanding of the complex host-pathogen relationship driving invasive fungal diseases.
Future perspectives
Imaging plays an essential role in bridging the translational gap between preclinical and clinical research, offering solutions for the three key challenges outlined in this review: (1) improve diagnostic accuracy, (2) accelerate the search for new treatments and (3) advance insights into host-pathogen interactions. An important first step is improving access to preclinical imaging platforms to allow researchers to move away from solely relying on traditional endpoint measurements and access longitudinal information on infection dynamics. To address the need for sensitive, rapid differential diagnosis of IFDs, PET imaging shows the most promise with the development of fungus-specific tracers, yet many of these tracers are still in the early stages of preclinical validation. Tools like BLI, especially when combined with ethical burden-free invertebrate models, could greatly speed up the antifungal drug discovery pipeline by providing real-time readouts on fungal viability. To unravel the role of the host immune response in fungal infections, different imaging modalities can be combined in a multimodal approach. Although advanced optical imaging tools like multiplex BLI hold the promise to investigate multiple processes simultaneously, and would therefore be ideal to sensitively image host-pathogen interactions, this technique is still in its infancy. In order to capture the full complexity of IFDs, the continuous development and validation of innovative imaging techniques in experimental research is essential. Ultimately, besides the development and validation of innovative imaging technologies, the integration of imaging approaches into preclinical studies offers more translatable, comprehensive data to guide clinical trials and researchers, enabling earlier and more sensitive diagnosis, more precise treatment monitoring, and faster development of effective treatment strategies.
Data availability
No datasets were generated or analyzed during the current study.
References
WHO fungal priority pathogens list to guide research, development and public health action. Geneva: World Health Organization; 2022.
Feys, S. et al. Influenza-associated and COVID-19-associated pulmonary aspergillosis in critically ill patients. Lancet Respir. Med. 12, 728–742 (2024).
Denning, D. W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 24, e428–e438 (2024).
Pitt, S. J. & Gunn, A. The one health concept. Br. J. Biomed. Sci. 81, 12366 (2024).
Sanguinetti, M. et al. Diagnosis and treatment of invasive fungal infections: looking ahead. J. Antimicrob. Chemother. 74, ii27–ii37 (2019).
Niño-Vega, G. A., Padró-Villegas, L. & López-Romero, E. New ground in antifungal discovery and therapy for invasive fungal infections: innovations, challenges, and future directions. J. Fungi 10, 871 (2024).
Desoubeaux, G. & Cray, C. Animal models of aspergillosis. Comp. Med. 68, 109–123 (2018).
Roosen, L., Maes, D., Musetta, L. & Himmelreich, U. Preclinical models for cryptococcosis of the CNS and their characterization using in vivo imaging techniques. J. Fungi 10, 146 (2024).
Ben-Ami, R. Experimental models to study the pathogenesis and treatment of mucormycosis. J. Fungi 10, 85 (2024).
Segal, E. & Frenkel, M. Experimental in vivo models of candidiasis. J. Fungi 4, 21 (2018).
Paulussen, C., Boulet, G. A. V., Cos, P., Delputte, P. & Maes, L. J. R. M. Animal models of invasive aspergillosis for drug discovery. Drug Discov. Today 19, 1380–1386 (2014).
Tielemans, B. et al. From mouse to man and back: closing the correlation gap between imaging and histopathology for lung diseases. Diagnostics 10, 636 (2020).
Resendiz-Sharpe, A. et al. Quantitative PCR effectively quantifies triazole-susceptible and triazole-resistant aspergillus fumigatus in mixed infections. J. Fungi 8, 1120 (2022).
Vande Velde, G., Kucharíková, S., Van Dijck, P. & Himmelreich, U. Bioluminescence imaging increases in vivo screening efficiency for antifungal activity against device-associated Candida albicans biofilms. Int. J. Antimicrob. Agents 52, 42–51 (2018).
Vanherp, L. et al. The added value of longitudinal imaging for preclinical in vivo efficacy testing of therapeutic compounds against cerebral cryptococcosis. Antimicrob. Agents Chemother. 64, e00070–20 (2020).
Smith, J. A. & Kauffman, C. A. Pulmonary fungal infections. Respirology 17, 913–926 (2012).
Palacios, C. F. & Moffarah, A. S. Diagnosis of pneumonia due to invasive molds. Diagnostics 11, 1226 (2021).
Lewis, R. E., Stanzani, M., Morana, G. & Sassi, C. Radiology-based diagnosis of fungal pulmonary infections in high-risk hematology patients: are we making progress?. Curr. Opin. Infect. Dis. 36, 250–256 (2023).
Vande Velde, G. et al. Magnetic resonance imaging for noninvasive assessment of lung fibrosis onset and progression: cross-validation and comparison of different magnetic resonance imaging protocols with micro-computed tomography and histology in the bleomycin-induced mouse model. Invest. Radiol. 49, 691–698 (2014).
Ekinci, A., Uçarkuş, T. Y., Okur, A., Öztürk, M. & Doğan, S. MRI of pneumonia in immunocompromised patients: Comparison with CT. Diagn. Interv. Radiol. 23, 22–28 (2017).
Poelmans, J. et al. Longitudinal, in vivo assessment of invasive pulmonary aspergillosis in mice by computed tomography and magnetic resonance imaging. Lab. Invest. 96, 692–704 (2016).
Yan, C. et al. Lung MRI of invasive fungal infection at 3 Tesla: evaluation of five different pulse sequences and comparison with multidetector computed tomography (MDCT). Eur. Radiol. 25, 550–557 (2015).
Donnelly, J. P. et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 71, 1367 (2019).
Hammer, M. M., Madan, R. & Hatabu, H. Pulmonary mucormycosis: radiologic features at presentation and over time. Am. J. Roentgenol. 210, 742–747 (2018).
Clark, D. P. & Badea, C. T. Advances in micro-CT imaging of small animals. Phys. Med. 88, 175–192 (2021).
Vande Velde, G. et al. Longitudinal micro-CT provides biomarkers of lung disease that can be used to assess the effect of therapy in preclinical mouse models, and reveal compensatory changes in lung volume. Dis. Models Mech. 9, 91–98 (2016).
Seldeslachts, L. et al. Overcome double trouble: baloxavir marboxil suppresses influenza thereby mitigating secondary invasive pulmonary aspergillosis. J. Fungi 8, 1 (2022).
Resendiz-Sharpe, A. et al. Longitudinal multimodal imaging-compatible mouse model of triazole-sensitive and -resistant invasive pulmonary aspergillosis. Dis. Models Mech. 15, dmm049165 (2022).
Berghen, N. et al. Radiosafe micro-computed tomography for longitudinal evaluation of murine disease models. Sci. Rep. 9, 17598 (2019).
Vanhoffelen, E., Resendiz-Sharpe, A. & Vande Velde, G. Microcomputed tomography to visualize and quantify fungal infection burden and inflammation in the mouse lung over time. Methods Mol. Biol. 2667, 211–224 (2023).
Christe, A. et al. CT patterns of fungal pulmonary infections of the lung: comparison of standard-dose and simulated low-dose CT. Eur. J. Radiol. 81, 2860–2866 (2012).
Ahookhosh, K., Vanoirbeek, J. & Vande Velde, G. Lung function measurements in preclinical research: What has been done and where is it headed?. Front. Physiol. 14, 1130096 (2023).
Gonzalez-Lara, M. F. & Ostrosky-Zeichner, L. Fungal infections of the brain in neurological complications of infectious diseases. In Current Clinical Neurology (eds. Hasbun, R., Bloch, K. C., Bhimraj, A.) 201–224 (Humana, Cham, 2021).
Lakhani, D. A., Deng, F. & Lin, D. D. M. Infectious diseases of the brain and spine: fungal diseases. Magn. Reson. Imaging Clin. N. Am. 32, 335–346 (2024).
Essig, M., Dinkel, J. & Gutierrez, J. E. Use of contrast media in neuroimaging. Magn. Reson. Imaging Clin. N. Am. 20, 633–648 (2012).
Lee, W. J. et al. Enlarged periventricular space and periventricular lesion extension on baseline brain MRI predicts poor neurological outcomes in cryptococcus meningoencephalitis. Sci. Rep. 11, 6446 (2021).
Pai, M. P., Sakoglu, U., Peterson, S. L., Lyons, C. R. & Sood, R. Characterization of BBB permeability in a preclinical model of cryptococcal meningoencephalitis using magnetic resonance imaging. J. Cereb. Blood Flow. Metab. 29, 545–553 (2009).
Fernando Nicolas-Cruz, C. et al. Cryptococcoma mimicking a brain tumor in an immunocompetent patient: a case and illustrative report. Interdiscip. Neurosurg. 31, 101688 (2023).
Ulett, K. B., Cockburn, J. W. J., Jeffree, R. & Woods, M. L. Cerebral cryptococcoma mimicking glioblastoma. BMJ Case Rep. https://doi.org/10.1136/bcr-2016 (2017).
Dusak, A., Hakyemez, B., Kocaeli, H. & Bekar, A. Magnetic resonance spectroscopy findings of pyogenic, tuberculous, and cryptococcus intracranial abscesses. Neurochem. Res. 37, 233–237 (2012).
Vanherp, L. et al. Trehalose as quantitative biomarker for in vivo diagnosis and treatment follow-up in cryptococcomas. Transl. Res. 230, 111–122 (2021).
Vanherp, L. et al. CryptoCEST: a promising tool for spatially resolved identification of fungal brain lesions and their differentiation from brain tumors with MRI. Neuroimage Clin. 31, 102737 (2021).
Callewaert, B., Jones, E. A. V., Himmelreich, U. & Gsell, W. Non-invasive evaluation of cerebral microvasculature using pre-clinical MRI: principles, advantages and limitations. Diagnostics 11, 926 (2021).
Callewaert, B., Gsell, W., Lox, M., Himmelreich, U. & Jones, E. A. V. A timeline study on vascular co-morbidity induced cerebral endothelial dysfunction assessed by perfusion MRI. Neurobiol. Dis. 202, 106709 (2024).
Abulikemu, N. et al. Mechanism of extracellular space changes in cryptococcal brain granuloma revealed by MRI tracer. Front. Neurosci. 16, 1034091 (2022).
Vanherp, L. et al. In vivo assessment of differences in fungal cell density in cerebral cryptococcomas of mice infected with Cryptococcus neoformans or Cryptococcus gattii. Microbes Infect. 25, 105127 (2023).
Musetta, L. et al. Quantitative MRI of a cerebral cryptococcoma mouse model for in vivo distinction between different cryptococcal molecular types. J. Fungi 10, 593 (2024).
Harsini, S., Werner, T., Revheim, M.-E., Alavi, A. & Saboury, B. 18F-FDG PET/CT imaging of infection and inflammation. J. Nucl. Med. 63, 2669 (2022).
Glaudemans, A. W. J. M. & Signore, A. FDG-PET/CT in infections: the imaging method of choice?. Eur. J. Nucl. Med. Mol. Imaging 37, 1986 (2010).
Kung, B. T. et al. An update on the role of 18F-FDG-PET/CT in major infectious and inflammatory diseases. Am. J. Nucl. Med. Mol. Imaging 9, 255 (2019).
Sambuceti, G. et al. 18F-fluoro-2-deoxy-d-glucose (FDG) uptake. What are we looking at?. Eur. J. Nucl. Med. Mol. Imaging 48, 1278–1286 (2021).
Pijl, J. P. et al. Limitations and pitfalls of FDG-PET/CT in infection and inflammation. Semin. Nucl. Med. 51, 633–645 (2021).
Ankrah, A. O. et al. Role of FDG PET/CT in monitoring treatment response in patients with invasive fungal infections. Eur. J. Nucl. Med. Mol. Imaging 46, 174–183 (2019).
Gutiérrez, A. et al. Determining the usefulness of systematic 18F-FDG PET/CT for the management of invasive fungal infection (PETIFI project): a prospective national multicentre cohort study protocol. BMJ Open 13, e074240 (2023).
Choi, S., Kronstad, J. W. & Jung, W. H. Siderophore biosynthesis and transport systems in model and pathogenic fungi. J. Microbiol. Biotechnol. 34, 1551–1562 (2024).
Petrik, M. et al. In vitro and in vivo evaluation of selected 68Ga-siderophores for infection imaging. Nucl. Med. Biol. 39, 361–369 (2012).
Petrik, M. et al. In vitro and in vivo comparison of selected Ga-68 and Zr-89 labelled siderophores. Mol. Imaging Biol. 18, 344–352 (2016).
Petrik, M. et al. 68Ga-triacetylfusarinine C and 68Ga-ferrioxamine E for Aspergillus infection imaging: uptake specificity in various microorganisms. Mol. Imaging Biol. 16, 102–108 (2014).
Misslinger, M. et al. Desferrioxamine B-mediated pre-clinical in vivo imaging of infection by the mold fungus Aspergillus fumigatus. J. Fungi 7, 734 (2021).
Sim, B. Z. et al. P-2126. ‘SPECIFIC’ First-In-Human Clinical Trial of 68Ga-triacetylfusarinine (TAFC) Siderophore PET/CT for Detection of Invasive Aspergillosis. Open Forum Infect. Dis. 12, ofae631.2282 (2025).
Kim, D. Y. et al. In vivo imaging of invasive aspergillosis with 18F-fluorodeoxysorbitol positron emission tomography. Nat. Commun. 13, 1926 (2022).
Lai, J. et al. Evaluation of 2-[18 F]-fluorodeoxysorbitol PET imaging in preclinical models of Aspergillus infection. J. Fungi 8, 25 (2022).
Rua, M. et al. Infection-specific PET imaging with 18F-fluorodeoxysorbitol and 2-[18 F]F-ρ-aminobenzoic acid: an extended diagnostic tool for bacterial and fungal diseases. Front. Microbiol. 14, 1094929 (2023).
Weinstein, E. A. et al. Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci. Transl. Med. 6, 259ra146 (2014).
Ordonez, A. A. et al. A systematic approach for developing bacteria-specific imaging tracers. J. Nucl. Med. 58, 144–150 (2017).
Shah, S. et al. Development and preclinical validation of 2-deoxy 2-[18 F]fluorocellobiose as an Aspergillus-specific PET tracer. Sci. Transl. Med. 16, eadl5934 (2024).
Lai, J., Wang, B., Petrik, M., Beziere, N. & Hammoud, D. A. Radiotracer development for fungal-specific imaging: past, present, and future. J. Infect. Dis. 228, S259–S269 (2023).
Lupetti, A., Welling, M., Pauwels, E. & Nibbering, P. Detection of fungal infections using radiolabeled antifungal agents. Curr. Drug Targets 6, 945–954 (2005).
Fischman, A. J. et al. Pharmacokinetics of 18F-labeled fluconazole in rabbits with Candida infections studied with positron emission tomography. J. Pharmacol. Exp. Ther. 259, 1351–1359 (1991).
de Assis, D. N. et al. Biodistribution of free and encapsulated 99 mTc-fluconazole in an infection model induced by Candida albicans. Biomed. Pharmacother. 99, 438–444 (2018).
Page, L., Ullmann, A. J., Schadt, F., Wurster, S. & Samnick, S. In vitro evaluation of radiolabeled amphotericin B for molecular imaging of mold infections. Antimicrob. Agents Chemother. 64, e02377–19 (2020).
Reyes, A. L., Fernandez, L., Rey, A. & Teran, M. Development and evaluation of 99 mTc-tricarbonyl-caspofungin as potential diagnostic agent of fungal infections. Curr. Radiopharm. 7, 144–150 (2015).
Welling, M. M. et al. Tc-99m-labeled antimicrobial peptides for detection of bacterial and Candida albicans infections. J. Nucl. Med. 42, 788–794 (2001).
Das, S. S., Wareham, D. W. & Britton, K. E. Tc-99m-labeled antimicrobial peptides for detection of bacterial and Candida albicans infections. J. Nucl. Med. 43, 1376–1376 (2002).
Rolle, A. M. et al. ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proc. Natl. Acad. Sci. USA. 113, E1026–E1033 (2016).
Henneberg, S. et al. Antibody-guided in vivo imaging of Aspergillus fumigatus lung infections during antifungal azole treatment. Nat. Commun. 12, 1707 (2021).
Davies, G. et al. Towards translational immunoPET/MR imaging of invasive pulmonary aspergillosis: the humanised monoclonal antibody JF5 detects Aspergillus lung infections in vivo. Theranostics 7, 3398 (2017).
Schwenck, J. et al. Antibody-guided molecular imaging of Aspergillus lung infections in leukemia patients. J. Nucl. Med. 63, 1450–1451 (2022).
Lai, J. et al. PET imaging of Aspergillus infection using Zirconium-89 labeled anti-β-glucan antibody fragments. Eur. J. Nucl. Med. Mol. Imaging 51, 3223–3234 (2024).
Morad, H. O. J. et al. Pre-clinical imaging of invasive candidiasis using immunoPET/MR. Front. Microbiol. 9, 1996 (2018).
Hutchens, M. & Luker, G. D. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 9, 2315–2322 (2007).
Love, A. C. & Prescher, J. A. Seeing (and using) the light: recent developments in bioluminescence technology. Cell Chem. Biol. 27, 904–920 (2020).
Avci, P. et al. In-vivo monitoring of infectious diseases in living animals using bioluminescence imaging. Virulence 9, 28 (2018).
Zambito, G., Chawda, C. & Mezzanotte, L. Emerging tools for bioluminescence imaging. Curr. Opin. Chem. Biol. 63, 86–94 (2021).
Brock, M. Application of bioluminescence imaging for in vivo monitoring of fungal infections. Int. J. Microbiol. 2012, 956794 (2012).
Doyle, T. C., Nawotka, K. A., Kawahara, C. B., Francis, K. P. & Contag, P. R. Visualizing fungal infections in living mice using bioluminescent pathogenic Candida albicans strains transformed with the firefly luciferase gene. Microb. Pathog. 40, 82–90 (2006).
Enjalbert, B. et al. A multifunctional, synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect. Immun. 77, 4847–4858 (2009).
Dorsaz, S., Coste, A. T. & Sanglard, D. Red-shifted firefly luciferase optimized for Candida albicans in vivo bioluminescence imaging. Front. Microbiol. 8, 1478 (2017).
Vanherp, L. et al. Sensitive bioluminescence imaging of fungal dissemination to the brain in mouse models of cryptococcosis. Dis. Model. Mech. 12, dmm039123 (2019).
Schrevens, S., Torelli, R., Sanguinetti, M. & Sanglard, D. Using bioluminescence to image Candida glabrata urinary tract infections in mice. Methods Mol. Biol. 2658, 239–247 (2023).
Resendiz-Sharpe, A., Vanhoffelen, E. & Vande Velde, G. Bioluminescence imaging, a powerful tool to assess fungal burden in live mouse models of infection. Methods Mol. Biol. 2667, 197–210 (2023).
Van Dyck, K., Van Dijck, P. & Vande Velde, G. Bioluminescence imaging to study mature biofilm formation by Candida spp. and antifungal activity in vitro and in vivo. Methods Mol. Biol. 2081, 127–143 (2020).
Poelmans, J. et al. A multimodal imaging approach enables in vivo assessment of antifungal treatment in a mouse model of invasive pulmonary aspergillosis. Antimicrob. Agents Chemother. 62, e00240–18 (2018).
Seldeslachts, L. et al. Early oseltamivir reduces risk for influenza-associated aspergillosis in a double-hit murine model. Virulence 12, 2493–2508 (2021).
Galiger, C. et al. Assessment of efficacy of antifungals against Aspergillus fumigatus: value of real-time bioluminescence imaging. Antimicrob. Agents Chemother. 57, 3046 (2013).
Zeng, Q. et al. In vitro and in vivo efficacy of a synergistic combination of itraconazole and verapamil against Aspergillus fumigatus. Front. Microbiol. 10, 1266 (2019).
Jacobsen, I. D., Lüttich, A., Kurzai, O., Hube, B. & Brock, M. In vivo imaging of disseminated murine Candida albicans infection reveals unexpected host sites of fungal persistence during antifungal therapy. J. Antimicrob. Chemother. 69, 2785–2796 (2014).
Delarze, E., Ischer, F., Sanglard, D. & Coste, A. T. Adaptation of a Gaussia princeps Luciferase reporter system in Candida albicans for in vivo detection in the Galleria mellonella infection model. Virulence 6, 684–693 (2015).
Milhomem Cruz-Leite, V. R. et al. Bioluminescence imaging in Paracoccidioides spp.: a tool to monitor the infectious processes. Microbes Infect. 24, 104975 (2022).
Vanhoffelen, E. et al. Powerful and real-time quantification of antifungal efficacy against triazole-resistant and -susceptible Aspergillus fumigatus Infections in Galleria mellonella by longitudinal bioluminescence imaging. Microbiol. Spectr. 11, https://doi.org/10.1128/spectrum.00825-23 (2023).
Vanhoffelen, E. et al. Sensitive bioluminescence imaging of cryptococcosis in Galleria mellonella improves antifungal screening under in vivo conditions. Virulence 15, 2327883 (2024).
Kavanagh, K. & Sheehan, G. The use of Galleria mellonella larvae to identify novel antimicrobial agents against fungal species of medical interest. J. Fungi 4, 113 (2018).
Vanhoffelen, E. et al. Combinations of posaconazole and tacrolimus are effective against infections with azole-resistant Aspergillus fumigatus. Front. Cell. Infect. Microbiol. 15, 1550457 (2025).
Van Dyck, K., Rogiers, O., Vande Velde, G. & Van Dijck, P. Let’s shine a light on fungal infections: a noninvasive imaging toolbox. PLoS Pathog. 16, e1008257 (2020).
Chapuis, A. F., Ballou, E. R. & Maccallum, D. M. A bright future for fluorescence imaging of fungi in living hosts. J. Fungi 5, 29 (2019).
Ryan, L. K. et al. A novel immunocompetent mouse model for testing antifungal drugs against invasive Candida albicans infection. J. Fungi 6, 1–12 (2020).
Bergeron, A. C. et al. Candida albicans and Pseudomonas aeruginosa interact to enhance virulence of mucosal infection in transparent zebrafish. Infect. Immun. 85, e00475–17 (2017).
Pócsi, I. et al. Use of red, far-red, and near-infrared light in imaging of yeasts and filamentous fungi. Appl. Microbiol. Biotechnol. 106, 3895 (2022).
Mendive-Tapia, L. et al. Spacer-free BODIPY fluorogens in antimicrobial peptides for direct imaging of fungal infection in human tissue. Nat. Commun. 7, 10940 (2016).
Ge, Y. & O’Shea, D. F. Review of clinically assessed molecular fluorophores for intraoperative image guided surgery. Molecules 29, 5964 (2024).
Ullah, Z. et al. Fluorescence imaging-guided surgery: current status and future directions. Biomater. Sci. 12, 3765–3804 (2024).
Díez-Vidal, A., Martínez-Martín, P., González-Muñoz, B. & Tung-Chen, Y. Point-of-care ultrasound in infectious diseases: current insights and future perspectives. Clin. Infect. Dis. 79, 420–429 (2024).
Alamdaran, S. A., Bagheri, R., Darvari, S. F., Bakhtiari, E. & Ghasemi, A. Pulmonary invasive fungal disease: ultrasound and computed tomography scan findings. Thorac. Res. Pract. 24, 292 (2023).
Zhang, W., Nie, W., Li, B. & Wang, H. Ultrasonography is an effective method for evaluating hepatosplenic fungal infections in pediatric onco-hematological patients. J. Clin. Ultrasound 52, 866–874 (2024).
Pérez-Liva, M. et al. Dual photoacoustic/ultrasound technologies for preclinical research: current status and future trends. Phys. Med. Biol. 70, 07TR01 (2025).
Swann, R. et al. Photoacoustic imaging of a cyanine dye targeting bacterial infection. Sci. Rep. 14, 18322 (2024).
Weber, J., Beard, P. C. & Bohndiek, S. E. Contrast agents for molecular photoacoustic imaging. Nat. Methods 13, 639–650 (2016).
Sam, Q. H., Yew, W. S., Seneviratne, C. J., Chang, M. W. & Chai, L. Y. A. Immunomodulation as therapy for fungal infection: are we closer?. Front. Microbiol. 9, 357398 (2018).
Armstrong-James, D. Antifungal chemotherapies and immunotherapies for the future. Parasite Immunol. 45, e12960 (2022).
Seldeslachts, L. et al. Damping excessive viral-induced IFN-γ rescues the impaired anti-Aspergillus host immune response in influenza-associated pulmonary aspergillosis. EBioMedicine 108, 105347 (2024).
Li, L. et al. Functional imaging of interleukin 1 beta expression in inflammatory process using bioluminescence imaging in transgenic mice. BMC Immunol. 9, 49 (2008).
Suff, N. & Waddington, S. N. The power of bioluminescence imaging in understanding host-pathogen interactions. Methods 127, 69–78 (2017).
Zambito, G., Mishra, G., Schliehe, C. & Mezzanotte, L. Near-infrared bioluminescence imaging of macrophage sensors for cancer detection in vivo. Front. Bioeng. Biotechnol. 10, 867164 (2022).
Lopes, J. P. & Urban, C. F. Visualizing hypoxia in a murine model of Candida albicans infection using in vivo biofluorencence. Bio Protoc. 9, e3326 (2019).
Mezzanotte, L., van ‘t Root, M., Karatas, H., Goun, E. A. & Löwik, C. W. G. M. In vivo molecular bioluminescence imaging: new tools and applications. Trends Biotechnol. 35, 640–652 (2017).
Gibbons, A. E., Luker, K. E. & Luker, G. D. Dual reporter bioluminescence imaging with NanoLuc and firefly luciferase. Methods Mol. Biol. 1790, 41–50 (2018).
McMorrow, R. et al. Whole-body bioluminescence imaging of T-cell response in PDAC models. Front. Immunol. 14, 1207533 (2023).
Stowe, C. L. et al. Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin. Elife 8, e45801 (2019).
Vorster, M., Buscombe, J., Saad, Z. & Sathekge, M. Past and future of Ga-citrate for infection and inflammation imaging. Curr. Pharm. Des. 24, 787–794 (2018).
Parker, K. M., Nicholson, J. K., Cezayirli, R. C. & Biggs, P. J. Aspergillosis of the sphenoid sinus: presentation as a pituitary mass and postoperative gallium-67 imaging. Surg. Neurol. 45, 354–358 (1996).
Palestro, C. J. The current role of gallium imaging in infection. Semin. Nucl. Med. 24, 128–141 (1994).
Abikhzer, G. et al. EANM/SNMMI guideline/procedure standard for [18 F]FDG hybrid PET use in infection and inflammation in adults v2.0. Eur. J. Nucl. Med. Mol. Imaging 52, 510–538 (2025).
Signore, A., Soroa, V. & De Vries, E. Radiolabeled white blood cells or FDG for imaging on inflammation and infection?. Q. J. Nucl. Med. Mol. Imaging 53, 23–25 (2009).
Roca, M., De Vries, E. F. J., Jamar, F., Israel, O. & Signore, A. Guidelines for the labelling of leucocytes with (111)In-oxine. Inflammation/Infection Taskgroup of the European Association of Nuclear Medicine. Eur. J. Nucl. Med. Mol. Imaging 37, 835–841 (2010).
De Vries, E. F. J., Roca, M., Jamar, F., Israel, O. & Signore, A. Guidelines for the labelling of leucocytes with (99 m)Tc-HMPAO. Inflammation/Infection Taskgroup of the European Association of Nuclear Medicine. Eur. J. Nucl. Med. Mol. Imaging 37, 842–848 (2010).
Erdman, W. A. et al. Indexing severity of diabetic foot infection with 99 mTc-WBC SPECT/CT hybrid imaging. Diab. Care 35, 1826–1831 (2012).
Love, C. & Palestro, C. J. Nuclear medicine imaging of bone infections. Clin. Radiol. 71, 632–646 (2016).
Haseman, M. K., Blake, K. & McDougall, I. R. Indium 111 WBC scan in local and systemic fungal infections. Arch. Intern. Med. 144, 1462–1463 (1984).
Signore, A. et al. Clinical indications, image acquisition and data interpretation for white blood cells and anti-granulocyte monoclonal antibody scintigraphy: an EANM procedural guideline. Eur. J. Nucl. Med. Mol. Imaging 45, 1816–1831 (2018).
Hotze, A. L. et al. Technetium-99m-labeled anti-granulocyte antibodies in suspected bone infections. J. Nucl. Med. 33, 526–531 (1992).
Xing, D. et al. Use of anti-granulocyte scintigraphy with 99mTc-labeled monoclonal antibodies for the diagnosis of periprosthetic infection in patients after total joint arthroplasty: a diagnostic meta-analysis. PLoS One 8, e69857 (2013).
Meng, Y., Sun, J., Zhang, G., Yu, T. & Piao, H. Approaches for neutrophil imaging: an important step in personalized medicine. Bioengineered 13, 14844–14855 (2022).
Wilks, M., Yuan, H., Viens, A., Mansour, M. & Normandin, M. PET imaging of neutrophil trafficking fungal infection. J. Nucl. Med. 63, 2511–2511 (2022).
Toner, Y. C. et al. Macrophage PET imaging in mouse models of cardiovascular disease and cancer with an apolipoprotein-inspired radiotracer. npj Imaging 2, 12 (2024).
Verweij, N. J. F. et al. First in man study of [18 F]fluoro-PEG-folate PET: a novel macrophage imaging technique to visualize rheumatoid arthritis. Sci. Rep. 10, 1047 (2020).
Puerner, C., Vellanki, S., Strauch, J. L. & Cramer, R. A. Recent advances in understanding the human fungal pathogen hypoxia response in disease progression. Annu. Rev. Microbiol. 77, 403–425 (2023).
Choen, S. et al. Kinetic evaluation of the hypoxia radiotracers [18 F]FMISO and [18 F]FAZA in dogs with spontaneous tumors using dynamic PET/CT imaging. Nucl. Med. Mol. Imaging 57, 16–25 (2023).
Belton, M. et al. Hypoxia and tissue destruction in pulmonary TB. Thorax 71, 1145–1153 (2016).
Liu, R. S. et al. Detection of anaerobic odontogenic infections by fluorine-18 fluoromisonidazole. Eur. J. Nucl. Med. 23, 1384–1387 (1996).
Kadurugamuwa, J. L. et al. Reduction of astrogliosis by early treatment of pneumococcal meningitis measured by simultaneous imaging, in vivo, of the pathogen and host response. Infect. Immun. 73, 7836–7843 (2005).
Liu, Y. H. et al. Group A Streptococcus subcutaneous infection-induced central nervous system inflammation is attenuated by blocking peripheral TNF. Front. Microbiol. 10, 265 (2019).
Munck, S., Swoger, J., Coll-Lladó, M., Gritti, N. & Vande Velde, G. Maximizing content across scales: Moving multimodal microscopy and mesoscopy toward molecular imaging. Curr. Opin. Chem. Biol. 63, 188–199 (2021).
Pfister, J. et al. Live-cell imaging with Aspergillus fumigatus-specific fluorescent siderophore conjugates. Sci. Rep. 10, 15519 (2020).
Pfister, J. et al. Hybrid imaging of Aspergillus fumigatus pulmonary infection with fluorescent, 68Ga-labelled siderophores. Biomolecules 10, 168 (2020).
Giampetraglia, M. & Weigelin, B. Recent advances in intravital microscopy for preclinical research. Curr. Opin. Chem. Biol. 63, 200–208 (2021).
Sun, D. et al. Fungal dissemination is limited by liver macrophage filtration of the blood. Nat. Commun. 10, 4566 (2019).
Alizadeh-Tabrizi, N., Hall, S. & Lehmann, C. Intravital Imaging of Pulmonary Immune Response in Inflammation and Infection. Front. Cell Dev. Biol. 8, 620471 (2021).
Shi, M. & Mody, C. H. Fungal infection in the brain: what we learned from intravital imaging. Front. Immunol. 7, 292 (2016).
Shi, M., Calaruso, P. & Mody, C. H. Real-time in vivo imaging of fungal migration to the central nervous system. Cell. Microbiol. 14, 1819–1827 (2012).
Zhang, M. et al. Real-time in vivo imaging reveals the ability of neutrophils to remove Cryptococcus neoformans directly from the brain vasculature. J. Leukoc. Biol. 99, 467–473 (2016).
Sun, D. et al. VCAM1/VLA4 interaction mediates Ly6C^low^ monocyte recruitment to the brain in a TNFR signaling dependent manner during fungal infection. PLoS Pathog. 16, e1008361 (2020).
Al-Gubory, K. H. Shedding light on fibered confocal fluorescence microscopy: applications in biomedical imaging and therapies. J. Biophotonics 12, e201900146 (2019).
Morisse, H. et al. In vivo and in situ imaging of experimental invasive pulmonary aspergillosis using fibered confocal fluorescence microscopy. Med. Mycol. 50, 386–395 (2012).
Vanherp, L. et al. Bronchoscopic fibered confocal fluorescence microscopy for longitudinal in vivo assessment of pulmonary fungal infections in free-breathing mice. Sci. Rep. 8, 3009 (2018).
Van Dyck, K. et al. Probe-based intravital microscopy: filling the gap between in vivo imaging and tissue sample microscopy in basic research and clinical applications. J. Phys. Photonics 3, 032003 (2021).
Bojarczuk, A. et al. Cryptococcus neoformans intracellular proliferation and capsule size determines early macrophage control of infection. Sci. Rep. 6, 21489 (2016).
Huisken, J. & Stainier, D. Y. R. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608 (2007).
Renshaw, S. A. & Trede, N. S. A model 450 million years in the making: zebrafish and vertebrate immunity. Dis. Model. Mech. 5, 38–47 (2012).
Voelz, K., Gratacap, R. L. & Wheeler, R. T. A zebrafish larval model reveals early tissue-specific innate immune responses to Mucor circinelloides. Dis. Model. Mech. 8, 1375–1388 (2015).
Knox, B. P. et al. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot. Cell 13, 1266–1277 (2014).
Ho, J. et al. Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nat. Commun. 10, 2297 (2019).
Brothers, K. M., Newman, Z. R. & Wheeler, R. T. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot. Cell 10, 932–944 (2011).
Gibson, J. F. et al. Blood vessel occlusion by Cryptococcus neoformans is a mechanism for haemorrhagic dissemination of infection. PLoS Pathog. 18, e1010389 (2022).
Gilbert, A. S. et al. Vomocytosis of live pathogens from macrophages is regulated by the atypical MAP kinase ERK5. Sci. Adv. 3, e1700898 (2017).
Ogryzko, N. V. et al. Hif-1α-induced expression of Il-1β protects against mycobacterial infection in zebrafish. J. Immunol. 202, 494–502 (2019).
Lewis, A. & Elks, P. M. Hypoxia induces macrophage tnfa expression via cyclooxygenase and prostaglandin E2 in vivo. Front. Immunol. 10, 2321 (2019).
Chalakova, Z. P. & Johnston, S. A. Zebrafish Larvae as an experimental model of cryptococcal meningitis. Methods Mol. Biol. 2667, 47–69 (2023).
Van Leeuwen, L. M. et al. A transgenic zebrafish model for the in vivo study of the blood and choroid plexus brain barriers using claudin 5. Biol. Open 7, bio030494 (2018).
Pereira, T. C. et al. Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J. Fungi 4, 128 (2018).
Arvanitis, M., Glavis-Bloom, J. & Mylonakis, E. Invertebrate models of fungal infection. Biochim. Biophys. Acta 1832, 1378–1383 (2013).
Fusco-Almeida, A. M. et al. Alternative non-mammalian animal and cellular methods for the study of host–fungal interactions. J. Fungi 9, 943 (2023).
Jacobsen, I. D. et al. Embryonated eggs as an alternative infection model to investigate Aspergillus fumigatus virulence. Infect. Immun. 78, 2995–3006 (2010).
Sarnella, A. et al. The chicken embryo: an old but promising model for in vivo preclinical research. Biomedicines 12, 2835 (2024).
Germain, R. N., Miller, M. J., Dustin, M. L. & Nussenzweig, M. C. Dynamic imaging of the immune system: progress, pitfalls and promise. Nat. Rev. Immunol. 6, 497–507 (2006).
Xia, J. Y., Zeng, Y. F., Wu, X. J. & Xu, F. Short-term ex vivo tissue culture models help study human lung infections: a review. Medicine 102, e32589 (2023).
Wohlsen, A., Hirrle, A., Tenor, H., Marx, D. & Beume, R. Effect of cyclic AMP-elevating agents on airway ciliary beat frequency in central and lateral airways in rat precision-cut lung slices. Eur. J. Pharmacol. 635, 177–183 (2010).
Pouwels, S. D. et al. LL-37 and HMGB1 induce alveolar damage and reduce lung tissue regeneration via RAGE. Am. J. Physiol. Lung Cell Mol. Physiol. 321, L641–L652 (2021).
Delmotte, P. & Sanderson, M. J. Ciliary beat frequency is maintained at a maximal rate in the small airways of mouse lung slices. Am. J. Respir. Cell Mol. Biol. 35, 110–117 (2006).
Koziol-White, C., Gebski, E., Cao, G. & Panettieri, R. A. Precision cut lung slices: an integrated ex vivo model for studying lung physiology, pharmacology, disease pathogenesis and drug discovery. Respir. Res. 25, 231 (2024).
Booth, J. L., Coggeshall, K. M., Gordon, B. E. & Metcalf, J. P. Adenovirus type 7 induces interleukin-8 in a lung slice model and requires activation of Erk. J. Virol. 78, 4156–4164 (2004).
Ebsen, M. et al. Infection of murine precision cut lung slices (PCLS) with respiratory syncytial virus (RSV) and Chlamydophila pneumoniae using the Krumdieck technique. Pathol. Res. Pract. 198, 747–753 (2002).
Viana, F., O’Kane, C. M. & Schroeder, G. N. Precision-cut lung slices: a powerful ex vivo model to investigate respiratory infectious diseases. Mol. Microbiol. 117, 578–588 (2022).
Neuhaus, V. et al. Assessment of long-term cultivated human precision-cut lung slices as an ex vivo system for evaluation of chronic cytotoxicity and functionality. J. Occup. Med. Toxicol. 12, 13 (2017).
Wronski, S. et al. Rhinovirus-induced human lung tissue responses mimic chronic obstructive pulmonary disease and asthma gene signatures. Am. J. Respir. Cell Mol. Biol. 65, 544–554 (2021).
Alberro-Brage, A. et al. Influenza virus decreases albumin uptake and megalin expression in alveolar epithelial cells. Front. Immunol. 14, 1260973 (2023).
Liu, G. et al. Use of precision cut lung slices as a translational model for the study of lung biology. Respir. Res. 20, 162 (2019).
Guarino, V. & Ambrosio, L. Temperature-driven processing techniques for manufacturing fully interconnected porous scaffolds in bone tissue engineering. Proc. Inst. Mech. Eng. H. 224, 1389–1400 (2010).
Alsafadi, H. N. et al. An ex vivo model to induce early fibrosis-like changes in human precision-cut lung slices. Am. J. Physiol. Lung Cell Mol. Physiol. 312, L896–L902 (2017).
Michalaki, C., Dean, C. & Johansson, C. The use of precision-cut lung slices for studying innate immunity to viral infections. Curr. Protoc. 2, e505 (2022).
Temann, A. et al. Evaluation of inflammatory and immune responses in long-term cultured human precision-cut lung slices. Hum. Vaccin. Immunother. 13, 351–358 (2017).
Lyons-Cohen, M. R., Thomas, S. Y., Cook, D. N. & Nakano, H. Precision-cut mouse lung slices to visualize live pulmonary dendritic cells. J. Vis. Exp. 2017, 55465 (2017).
Ruigrok, M. J. R. et al. siRNA-mediated RNA interference in precision-cut tissue slices prepared from mouse lung and kidney. AAPS J. 19, 1855–1863 (2017).
Ruigrok, M. J. R. et al. siRNA-mediated protein knockdown in precision-cut lung slices. Eur. J. Pharm. Biopharm. 133, 339–348 (2018).
Klouda, T. et al. From 2D to 3D: promising advances in imaging lung structure. Front. Med. 7, 343 (2020).
Lam, M., Lamanna, E., Organ, L., Donovan, C. & Bourke, J. E. Perspectives on precision cut lung slices-powerful tools for investigation of mechanisms and therapeutic targets in lung diseases. Front. Pharmacol. 14, 1162889 (2023).
Danov, O. et al. Rupintrivir reduces RV-induced TH-2 cytokine IL-4 in precision-cut lung slices (PCLS) of HDM-sensitized mice ex vivo. Respir. Res. 20, 228 (2019).
Liu, R. et al. Mouse lung slices: An ex vivo model for the evaluation of antiviral and anti-inflammatory agents against influenza viruses. Antivir. Res. 120, 101–111 (2015).
Maertens, J., Meersseman, W. & Van Bleyenbergh, P. New therapies for fungal pneumonia. Curr. Opin. Infect. Dis. 22, 183–190 (2009).
Chastain, D. B. et al. Cerebral cryptococcomas: a systematic scoping review of available evidence to facilitate diagnosis and treatment. Pathogens 11, 205 (2022).
Lawal, I. O. et al. Radionuclide imaging of invasive fungal disease in immunocompromised hosts. Diagnostics 11, 2057 (2021).
Acknowledgements
The authors acknowledge support by the Flemish Research Foundation (FWO, grant number G057721N), KU Leuven Internal Funds (C2E/23/036, C3/23/005) and the FAILSAFE PROJECT at the University of Exeter, co-funded by the UK Department of Health and Social Care's (DHSC) Global AMR Innovation Fund (GAMRIF). The views expressed in this publication are those of the author(s) and not necessarily those of the UK DHSC. L.M. and E.V. received an FWO aspirant mandate (1199825N and 1SF2224N). S.A.J. was supported by Medical Research Council Fellowship MR/J009156/1 and Grant APP20535. M.S. was supported by a studentship from the Medical Research Council Discovery Medicine North (DiMeN) Doctoral Training Partnership (MR/N013840/1). N.B. acknowledges funding by the Werner Siemens Stiftung and the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project Number 512332979.
Author information
Authors and Affiliations
Contributions
Conceptualization: G.V.V.; Writing – original draft and figure preparation: L.M., M.S., E.V., A.R.S., S.A.J., N.B. and G.V.V.; Writing – review and editing: L.M., M.S., E.V., A.R.S., S.A.J., N.B. and G.V.V. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Michiels, L., Shameem, M., Vanhoffelen, E. et al. Fungal infections in focus: accelerating non-invasive imaging from preclinical insights to clinical breakthroughs. npj Imaging 3, 42 (2025). https://doi.org/10.1038/s44303-025-00105-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44303-025-00105-y
This article is cited by
-
Effect of antibiotic combination on secondary invasive fungal infection in critically ill sepsis patients
BMC Infectious Diseases (2026)