Abstract
Rapid antibiotic susceptibility tests (AST) are vital for the effective treatment of disease, necessitating the development of analytical tools to address unmet needs in healthcare. Leveraging the sensitivity of plasmonic nanosensors and isotopic labelling has the potential to accelerate AST. Here, we developed surface-enhanced Raman scattering (SERS)-based nanofibre sensors and heavy water [deuterium oxide (D2O)] labelling (SERS-DIP) for detecting the minimum inhibitory concentration (MIC) and AST for trimethoprim (TMP) against E. coli. SERS-DIP rapidly detected the MIC of TMP for the susceptible strain in 2 h. TMP-resistant cells retained the metabolic activity regardless of TMP levels, confirming the resistance phenotype. Kinetic analysis of D uptake by resistant cells treated with TMP (2 × MIC) revealed increasing D levels proportional to peak redshifts over time, confirmed by machine learning-driven data exploration. Our results demonstrate the utility of nanofibre-enabled SERS-DIP for robust AST, uncovering new spectral biomarkers that may impact clinical medicine.
Similar content being viewed by others
Introduction
The ever-increasing prevalence of antimicrobial resistance (AMR) is a major global health concern, and a crucial step in the effective treatment of microbial infections involves the rapid detection of antibiotic susceptibility profiles of the causative pathogens to guide the timely prescription of appropriate drugs. Antibiotic susceptibility testing (AST) is typically performed with genotypic assays that detect resistant genes or plasmids using tools such as polymerase chain reaction (PCR) to rapidly identify resistant and susceptible strains1. However, genotypic ASTs are prone to false negatives, especially where PCR primers complementary to antibiotic-resistant genes are not systematically applied or are unknown2. In addition, the presence of resistant genes does not always indicate the resistance phenotype, considering that an encoded enzyme can still be inactive even when protein transcription and translation are demonstrated3.
AST based on phenotypic tools offers a more direct measure of antibiotic resistance in which bacterial growth in the presence or absence of antibiotics is monitored through bacterial optical density or zone of inhibition measurements4. Whilst such culture-based phenotypic ASTs are accurate and reliable, they are time-consuming, taking 24–72 h to identify resistant strains and suitable antibiotic dosage5. Such long turnaround times often delay the prescription of targeted therapy and encourage the use of broad-spectrum drugs, which usually exacerbates AMR.
Surface-enhanced Raman scattering (SERS) sensors have attracted huge interest as versatile analytical tools for rapid phenotypic ASTs with the potential to offer solutions to the current AST shortfalls. SERS utilises the near-field electromagnetic (EM) field generated by the localised surface plasmon resonance (LSPR) on nanoparticles (NPs) and molecular vibrational signatures to detect microbial molecules with excellent sensitivity and specificity6. In label-free SERS-based AST (SERS-AST), the information-rich SERS fingerprints unravel drug-induced differential molecular profiles in bacterial cells treated with or without antibiotics over time7,8. Such molecular changes detected by SERS are used to rapidly establish the minimum inhibitory concentration (MIC) of antibiotics to identify susceptible and resistant bacteria.
Recently, Liu et al. used SERS deploying silver NPs (AgNPs) embedded in anodic aluminium oxide to determine the MIC for methicillin-susceptible Staphylococcus aureus and other clinical isolates in 2 h, with SERS yielding AST results tenfold faster than the standard broth microdilution assays9. Similarly, Han et al. successfully identified susceptible and resistant Gram-positive and Gram-negative pathogens related to bacteremia in blood culture samples in 5 h using AgNP-activated SERS substrates10, and the results were strongly correlated (96%) with the VITEK tool commonly applied for clinical AST. Several SERS techniques for bacterial AST have been extensively demonstrated recently with similar levels of success11,12,13, illustrating the robustness of the method. However, it is worth noting that all SERS-AST protocols reported so far identify susceptible and resistant pathogens by computing differential spectral intensity changes induced by drug treatment11. Whilst such SERS designs deliver accurate AST results, they are prone to instrumental factors, such as variations in laser fluency and irregularities in plasmonic substrates/architectures that have considerable impacts on the intensity dynamics of key SERS peaks14.
To overcome these limitations and expand the scope of SERS-AST, deuterium isotope probing (DIP) can be integrated into SERS-AST tools. In DIP, heavy water [deuterium oxide (D2O)] labelling of cells measures the general metabolic activity of bacteria15,16 and has been successfully used in combination with Raman and infrared spectroscopy for spectral signal readouts to probe the physiological behaviour or functions of bacteria as a function of stress17,18. Given that the addition of D2O to a culture medium does not skew the natural substrate reservoir or cellular phenotype19, DIP can be conveniently integrated into the standard AST pipeline. When cells incorporate D2O into biomass via bacterial anabolism, D2O-derived deuterium (D) isotope will replace hydrogen (H) atoms in newly synthesised cellular molecules (e.g., lipids and proteins)16. SERS spectra measured from D2O-labelled bacteria exhibit unique spectral signature bands (C–D vibration modes in lipids and proteins) in the silent region of the SERS spectrum (2000–2400 cm-1)20, which is clearly redshifted from C–H lipid vibrations detected around 3000 cm-1.
Cellular D–H isotope exchange and the resultant SERS spectral changes can provide an unambiguous platform to measure MICs and AST of clinically relevant bacteria. Moreover, antibiotic drugs such as trimethoprim (TMP) disrupt DNA biosynthesis by inhibiting the enzymatic production of the essential tetrahydrofolate cofactor, thereby disrupting purine synthesis21. SERS is highly sensitive to the structural and compositional changes in DNA and its related biomolecules, such as purines and pyrimidines22,23. Combining SERS with D2O labelling (SERS-DIP) offers a global picture of intracellular and cell wall (the part of a bacterium in direct contact with a nanoplasmonic surface) molecular dynamics that could yield fast, robust, and accurate bacterial AST24.
However, to the best of our knowledge, SERS-DIP has not been reported for MICs and AST detection to date, perhaps due to the low sensitivity exhibited by plasmonic sensors towards C–D sensing in D2O-enriched cells. Plasmonic nanofibre sensors are poised to revolutionise SERS-AST, allowing the kinetic analysis of metabolic flux in D2O-amended bacteria to establish drug susceptibility profiles. Nanofibres patterned with plasmonic NP arrays deposited on tapered glass fibres exhibit single-molecule detection sensitivity owing to the lens effect25, where intense EM fields are generated at the nanofibre outlet at a point opposite the focal point of the incident light. As such, nanofibres have superior analytical performance to planar and colloidal nanosensors26, which are often deployed for SERS-AST12,14, making nanofibre sensors potential candidates for the sensitive detection of microbial metabolic activity profiles underlying D isotope assimilation by the cells treated with clinically useful antibiotics.
In this study, we developed a novel nanofibre sensor by patterning a block copolymer-templated tapered fibre with a dense layer of AgNPs to explore the applicability of plasmonic nanofibre sensors and D2O labelling as a universal approach for AST via SERS-DIP. AgNPs were firmly attached to a nano-thin poly(styrene)-b-poly(4-vinylpyridine) (PS-b-P4VP) polymer layer coated on the glass fibre, leading to the formation of highly stable arrays of AgNP on the nanofibre sensor. By taking advantage of the high SERS sensitivity of nanofibre sensors, we demonstrate label-free and rapid detection of biomolecular changes underlying bacterial AST in D2O-labelled TMP-susceptible and TMP-resistant Escherichia coli. The application of SERS-DIP allowed us to determine the MIC of TMP for TMP-susceptible and resistant E.coli strains within 2 h. Machine learning-driven exploration of spectral data identified several new spectral biomarkers for SERS-AST: namely, band redshifts at 2180 cm-1 (C–D modes in lipids and proteins) and 724 cm-1 (adenine-related biomolecules), in addition to SERS intensity dynamics at 724 cm-1 peak similar to those implicated in isotope-independent SERS-AST tools reported previously9,10. Thus, our new plasmonic nanofibre sensors and D2O labelling have a huge potential to examine the response of bacteria to antibiotic treatment in order to rapidly establish the MIC and AST and identify susceptible and resistant cells unequivocally via SERS-DIP.
Results and discussion
Fabrication and characterisation of the AgNP-coated nanofibre sensor
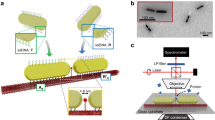
This study designed a novel nanofibre sensor via a low-cost and facile self-assembly process of PS-b-P4VP film and AgNPs on a glass fibre surface. The AgNP-coated nanofibre sensor was developed using a bottom-up approach reported by our group25. First, borosilicate glass rods (diameter 1 mm and length 10 cm) were pulled on one end to create tapered fibres with a distal tip diameter of ~1 µm and washed in piranha to eliminate organic contaminants. Fibres were dip-coated in THF solution containing PS-b-P4VP to create a brush-like polymer film on the fibre, as illustrated in Scheme 1. In previous work, we established that a low polymer concentration (0.05 mg/mL) used for dip-coating produces an optimal brush thickness that allows dense NP monolayers with negligible aggregation27; thus, this concentration was also adopted in this study. The polar pyridine component of P4VP preferentially interacts with the hydroxyl ( − OH) charge density on the borosilicate fibre surface via multiple intermolecular forces (e.g., hydrogen bonding between hydrogen atoms on the fibre and electronegative nitrogen atoms on P4VP)28. The P4VP block adsorbed directly onto the fibre surface, forming a highly stable anchoring or wetting sublayer, while the hydrophobic PS block formed an overlayer covering the P4VP undercoat, with PS chains either stuck out when suspended in THF or collapsed when air-dried at room temperature25. This is consistent with recent findings where PS-b-P4VP formed a brush-like polymer bilayer on polar glass materials and other surfaces when suspended in BCP26. Second, PS-b-P4VP-coated fibre templates were incubated with 3 mL of AgNP colloid (pH 4.52) for 3 h at ambient temperature to allow the deposition of citrate-capped AgNPs on polymer-coated tapered fibres (Scheme 1). The extinction maximum (λmax) of AgNPs detected at 414 nm and TEM images confirmed the mean diameter of AgNPs to be ~80 nm (Supplementary Fig. 1). SEM images in Scheme 1B, C confirmed the formation of a dense AgNP array on nanofibre sensors. One might expect that the energetically unfavourable interactions between negatively charged AgNPs and the nonpolar PS film coupled with steric effects from hydrophobic PS chains would block the electrostatic interactions between AgNPs and P4VP, potentially hindering the adsorption of AgNPs on BCP-coated fibres. To the contrary, SEM images in Scheme 1 illustrate a dense layer of self-assembled AgNPs was formed on nanofibres, with the hemispherical PS-b-P4VP corona straddling AgNPs on the fibre surface when immersed in an AgNP colloid, similar to previous results for BCP-induced AuNP nanopatterning on borosilicate glass fibres25,27. This finding could be driven by both molecular rearrangements and ionisation that, perhaps, occur simultaneously25. The interactions between BCP and AgNPs might induce molecular rearrangements of PS chains in a way that exposes P4VP, resulting in strong P4VP–AgNP electrostatic attractions29. In parallel, an increase in colloidal acidity (pH 4.52 < pKa of P4VP) led to the protonation of P4VP, further increasing the affinity of P4VP for negatively charged aqueous AgNPs. Such results are supported by various studies that successfully fabricated robust sensors on highly curved and 2D plasmonic by depositing citrate-reduced particles on thin films of PS-b-P4VP25,29 or its analogs (e.g., PS-b-P2VP)30, which were electrostatically anchored on glass by P4VP underlayer, with a pendant PS top layer promoting self-assembly of NPs with minimal aggregation.
Bottom-up nano-patterning of a plasmonic nanofibre sensor of 1 µm tip diameter. A tapered glass fibre was initially templated with PS-b-P4VP diblock copolymer before incubation on an AgNP colloid (A), forming a dense AgNP array on the nanofibre sensor (B, C). Scale bars for panels (B, C) are 5 µm and 2 µm, respectively. Panel (A) was created in BioRender with permission.
To confirm the origin of the SERS enhancement leading to sensitive metabolite detection, EM near-fields were simulated using finite-difference time-domain (FDTD) models (Supplementary Fig. 2) in a commercially available FDTD software (Lumerical Solutions, USA). We considered a normally incident Gaussian light beam that induces the LSPR excitation of AgNPs. The reflection spectrum of nanofibre sensors with a tip diameter of ~1 µm exhibited two main peaks at specific wavelengths (λ): the reflection peak (λ1) and dip (λ2) (Fig. 1A, C). The resonance wavelength is at 656 nm [located between 633 nm (excitation wavelength) and 663 nm corresponding to the Raman shift of 733 cm-1 for adenine]. Compared with an electric field distribution at λ1, a stronger electric field enhancement of AgNPs and internal glass fibre cavity was observed at λ2 (Fig. 1B—top panel). Such findings perhaps highlight the contribution of the Fabry-Pérot (FP) resonances in enhancing LSPR on nanofibre sensors, which generated distinct EM interference fringes in the nanofibre cavity, enhancing the LSPR resonance intensity of AgNPs (Fig. 1B—bottom panel). Besides, both 633 nm and 663 nm were located near λ2, so that the LSPR excited by them is also enhanced by FP resonances. Therefore, for the TM-polarised light, the nanofibre sensor can enhance the excitation (incident) light and emitted intensity of the Raman signals. Like TM-polarised light, the EM enhancement for TE-polarised light was stronger at λ2 than λ1 (Fig. 1D). As such, the sensor also exhibited interference patterns related to cavity FP resonances (Fig. 1D—top and bottom panels). The main reason for the shift of the reflection dip position was due to the difference in reflection phase shifts of AgNP arrays for the TM-polarised and TE-polarised light. Despite the blueshift in λ2, the peak remains between 633 and 663 nm, enabling concurrent EM field enhancement for both wavelengths and consequently augmenting the SERS effect. Simultaneous excitation of surface plasmon and FP resonance modes have been reported in photovoltaic sensors, where the co-produced propagating plasmon resonance and electrical current allowed the detection of protein-protein binding within nanocavities31. In another report, coupled resonant cavity structure and LSPR modes were demonstrated to amplify SERS, leading to highly sensitive detection of cortisol in the plasmonic nanogaps at the metal-metal junctions32. In the present study, we leveraged the coupled LSPR and FP effects generated on AgNPs-decorated nanofibre sensors for rapid and label-free AST of D2O-amended bacteria. Prior to AST experiments, the analytical performance of nanofibre sensors was probed through quantitative detection and spectral repeatability assessment of adenine SERS sensing. Regression of the 733 cm-1 band intensity against adenine levels (0–10 µM) yielded a standard curve with a strong linear response shown by a large slope and high coefficient of determination (R2 = 0.982) (inset in Supplementary Fig. 3a), indicating high sensitivity of the sensor. Moreover, the low limit of detection (LOD = 28.7 nM) and high spectral repeatability (relative standard deviation (RSD) = 6.4%)33 (Supplementary Fig. 3b) show that nanofibre sensors had comparable sensitivity and reproducibility to that of well-established analytical tools often used in bacterial metabolite sensing34.
A, C Investigating the effect of transverse magnetic (TM) and transverse electric (TE) light reflectance on electric field enhancement. Simulated spatial electric field (| E |) distributions in x-z (B) and y-z (D) planes for four wavelengths. Bottom panels in (B) and (D) provide localised magnifications for a more detailed analysis of LSPR and FP resonance modes. For (D), the calculated length in the y-direction is \(\sqrt{3}\)P, corresponding to the triangular lattice.
Bacterial AST phenotyping using nanofibre sensors
SERS has been established as a promising tool for drug susceptibility tests due to its ability to determine MICs and AST faster than standard phenotypic assays11. However, the differential intensity ratios (mainly for the SERS peaks between 720 and 745 cm-1) that the current SERS tools employ to determine MICs and AST are vulnerable to instrumental and nanosensor fluctuations. The main goal of this study was to illustrate the applicability of SERS combined with D2O labelling for rapid, non-destructive, and robust detection of the MIC and AST in order to expand the SERS-AST capabilities. We selected two E. coli strains of known susceptibility and resistance to TMP drug as a proof-of-concept study. Wild-type E. coli (BL21) was applied as a TMP-susceptible strain, whilst its knock-in mutant E. coli (DfrB1) strain contained the dfrB1 gene encoding an enzyme that confers resistance to TMP35. The synthetic antibiotic TMP was selected in this work as it is listed among essential medicines for diseases such as urinary tract infections due to its bacteriostatic effect36. The efficacy of TMP is challenged by the steady rise in the prevalence of TMP-resistant bacterial cells carrying dihydrofolate reductase genes providing resistance, including dfrB1, posing a serious health threat36. This necessitates the development of SERS sensors that allow rapid phenotyping of antibiotic susceptibility in susceptible and resistant E. coli cells.
Investigation of bacterial growth and MIC determination using broth dilution
Growth profiles of the TMP-susceptible E. coli strain in nutrient broth supplemented with varying TMP levels with and without 50% D2O were tracked through OD600 measurements. Fig. 2 shows the average growth curves at various TMP concentrations over 16 h of cell culture at 37 oC. It is clear that TMP-susceptible cells exhibited similar growth trends in normal (H2O) (Fig. 2A) and heavy (D2O) (Fig. 2B) water, keeping all other culture conditions constant. This finding suggests that the cells were not significantly affected by 50% D2O present in the growth medium, and so the cells were phenotypically identical. Previous studies show that exposure of bacteria to low concentrations of D2O spiked in culture media, typically ≤80%, has no detectable impact on bacterial growth16,18, which is consistent with our findings in this study (Fig. 2). Next, the MIC of TMP for the susceptible strain was investigated using the growth profiles. Bacteria exposed to increasing TMP concentrations (up to 16 µg/mL) and cultured overnight displayed varying growth phenotypes induced by TMP treatment. Although the susceptible strain grew normally at low TMP amounts (0–0.5 µg/mL) in both H2O- and D2O-enriched nutrient broth, no growth was detected at higher concentrations of antibiotics (≥2 µg/mL). When treated with 1 µg/mL of TMP, susceptible bacteria displayed some growth in the first 3 h, but the OD600 decreased abruptly to the baseline and remained constant throughout 16 h of cell growth (Fig. 2), which is indicative of TMP-induced growth inhibition. Thus, the MIC of TMP for E. coli (BL21) was determined to be 2 µg/mL, which agrees with previous studies37,38.
SERS sensors unravel TMP susceptibility tests via D uptake from D2O in active cells
After establishing the growth phenotype in TMP-spiked cell cultures with and without D2O (Fig. 2), SERS was used to investigate the MIC and AST for susceptible and resistant E. coli strains. SERS generates information-rich whole-organism spectral fingerprints measured from multiple cellular molecules, such as lipids, proteins, and amino acids, which are used to identify bacteria39. The SERS spectral features measured from bacteria were different from those acquired from the blank nanofibre (Supplementary Fig. 4), confirming that background signals from the sensor did not have an impact on the SERS data. According to SERS spectra in Fig. 3A, TMP-susceptible cells were metabolically active when cultivated in a nutrient broth spiked with low concentrations of TMP (0–0.5 µg/mL) and 50% D2O for 2 h as indicated by spectral redshifts and/or intensity decrease of many SERS bands, such as 2180 and 724 cm-1 peaks, which are annotated in Fig. 3A and quantified in the regression plots presented in Supplementary Fig. 5A, B. The 724 cm-1 SERS vibrational mode was zoomed in further in Supplementary Fig. 6 to highlight spectral intensity dynamics and redshifts as functions of isotope assimilation into TMP-susceptible cells grown in LB media supplemented with D2O and normal water.
Determination of the MIC and AST for (A, B) TMP-susceptible and (C, D) TMP-resistant E. coli strains cultivated in LB nutrient broth containing varying TMP levels with (A, C) or without (B, D) 50% D2O. Metabolically active susceptible cells grown in low TMP levels (0–0.5 µg/mL) and resistant bacteria exposed to TMP in D2O exhibited an apparent spectral peak at 2180 cm-1 assigned to the C–D vibrations in lipids and proteins (A, C). For the susceptible bacterial strain, the C–D band disappeared completely after bacterial exposure to 2 µg/mL TMP within 2 h of cell cultivation (A), which was determined as the MIC using our nanofibre sensor-based SERS-DIP approach. The impact of TMP on active susceptible bacteria was further characterised by the reduction in the intensity of several spectral bands above the MIC ( ≥ 2 µg/mL) with increasing TMP (B). In contrast, SERS intensities for resistant cells did not significantly change across TMP concentrations (D). The corresponding intensity regression plots are computed in Supplementary Figs. 5 and 7.
However, the unique peak at 2180 cm-1 is the most apparent differential spectral feature, which is redshifted from the C–H band detected at 2927 cm-1 for bacteria assimilating D2O-derived D isotope into cell biomass (Fig. 3A). This spectral band is assigned to C–D vibrations in lipids or proteins40, and such a clear peak redshift was due to an increase in the reduced mass, resulting in the decrease in the vibrational frequency of the chemical bond in cellular biomolecules upon assimilation of heavier D isotopes41. During cellular lipid and protein biosynthesis, hydrogen (H) from normal water (H2O) is incorporated into fatty acids and amino acids anabolic biosynthesis pathways16. As such, when D2O is used in place of H2O and is incorporated into the physiologically active bacteria, D2O-derived D heavy isotopes effectively replace the lighter H atoms, forming deuterated lipids and proteins that our sensitive nanofibre-based SERS rapidly detected. As can be seen in Fig. 3A and Supplementary Fig. 5A, the C–D peak and D-associated redshifts at 724 cm-1 decreased significantly nearly down to the baseline (i.e., spectra of bacteria that are not incorporating D) when the susceptible strain was treated with 1 µg/mL of TMP. This perhaps explains the intermittent increase in optical densities (OD600) in the initial 3 h of cell growth (Fig. 2)—above 1 µg/mL TMP, C–D bands disappeared completely, and D-related band shifts were undetectable after susceptible bacterial cells were treated with ≥2 µg/mL TMP (Fig. 3A and Supplementary Fig. 5A). This, together with the apparent decrease in SERS characteristic peaks (Supplementary Fig. 5B) indicates that the TMP-susceptible E. coli strain was physiologically inactive at higher TMP levels, which is also reflected by the low cell density (OD600) detected in Fig. 2. Hence, SERS data in Fig. 3A and Supplementary Fig. 5A confirmed that the MIC of TMP for the susceptible strain was 2 µg/mL, which was in complete agreement with the MIC calculated using broth dilution.
Several researchers have previously illustrated that SERS can detect bacterial antibiotic susceptibility in 5 h10,12. In this study, where we integrated SERS with D2O isotopic labelling (so-called SERS-DIP), D incorporation into susceptible E. coli cells was detected in 2 h, highlighting that our SERS-DIP tool accomplished AST results eight times faster than the standard dilution bioassays. Notice that the C–D chemical bond vibration was not detected in bacterial cells cultured in the growth medium supplemented with normal H2O only (Fig. 3B and Supplementary Fig. 5C) since D2O was absent. However, the impact of TMP on the growth of susceptible cells was manifested in the apparent decrease in the intensity of several peaks, including SERS spectral biomarkers at 724 and 1331 cm-1 assigned to ring vibrations in adenine-related molecules (e.g., purines, adenosine triphosphate) and 956 cm-1 (C–N modes in proteins, nucleotides, etc.)39,42 (Fig. 3B and Supplementary Fig. 5D). This is consistent with a study by Liu et al., who detected a similar decrease in intensity spectral ratios in key SERS-AST bands for susceptible Staphylococcus aureus treated with oxacillin drug concentrations that were greater than the MIC9. It is well established that metabolically active bacterial cells in contact with a plasmonic surface secrete purines, pyrimidines, nucleotides, and other adenine-related compounds that, in addition to the cell wall molecular components, give rise to the strong SERS bands43,44. When cells are inactive (either by drug treatment or other means), the secretion of SERS-active molecules decreases, leading to weaker spectral bands43. This explains the decrease in SERS intensity observed in Fig. 3A, B, given that TMP inhibits cell growth by inhibiting DNA synthesis, significantly halting the secretion of SERS-active cellular biomolecules37. On the other hand, DfrB1 cells exhibited strong C–D vibrations at 2180 cm-1 and spectral redshifts 724 cm-1 of similar intensities when treated with varying TMP levels probed in this study (Fig. 3C and Supplementary Fig. 7A, B), confirming the high resistance of DfrB1 cells against TMP (MIC of 600 µg/mL38), even at high TMP levels (i.e., 3 × MIC)45. Similarly, TMP-resistant cells grown in the medium with normal water had no C–D peaks (Fig. 3D), and the peak ratios and intensities, including SERS spectral markers at 724 and 1331 cm-1, did not significantly change throughout the cell growth period (Supplementary Figs. 7C, D, and 8). Indeed, this result provides further evidence that the DfrB1 strain is highly resistant to TMP treatment.
SERS sensors reveal the kinetics of deuterium (D) uptake in TMP-resistant E. coli strain
Understanding the kinetics of heavy isotope incorporation by metabolically active bacteria is essential to assess antibiotic susceptibility or resistance evolution over time. To determine the kinetics of AST for TMP, the susceptible and resistant strains were treated with 4 µg/mL TMP (i.e., 2 × MIC) with and without 50% heavy water. This experiment was performed to ensure adequate suppression of metabolic activity in the susceptible strain whilst also warranting high enough TMP against resistant bacteria to obtain detailed information about time-dependent biomolecular dynamics underpinning the TMP resistance phenotype. At a TMP concentration of 4 µg/mL, the susceptible cells did not exhibit any C–D spectral features nor any decline in band intensity related to D2O uptake or TMP treatment over time (Fig. 4A). To probe this further, the rate of D assimilation by cells grown in the medium enriched with D2O was quantified by regression analysis of intensity ratios for the characteristic peaks at 724 and 2180 cm-1 (C–D ratios were computed by the formula [(C–D/(C–D + C–H)])20 over time compared to cell replication rates (OD600) across 4 h). For the susceptible strain, no spectral band shifts or changes in OD600 were recorded in 4 h (Fig. 4B), confirming the lack of metabolic activity and cell division due to the bacteriostatic effects of TMP. In contrast, SERS spectra of TMP-resistant cells exhibited significant and increasing incorporation rates of D as shown by the emergence of the C–D peak at 2180 cm-1 as well as the spectral redshift and increase in intensity of the adenine-related band at 724 cm-1 over time (Fig. 4C). Interestingly, D assimilation and the resulting spectral shifts were clearly observed in just 1 h for the resistant bacterial cells, which increased proportionally between 0 to 4 h owing to the additive effect of D uptake by cells, which was also evidenced by the steady increase in cell density (OD600) over time (Fig. 4D). This illustrates that treatment of the resistant strain with high TMP levels (2 × MIC) had no significant impact on bacterial cell metabolism and proliferation. To confirm this finding, control experiments were performed where DfrB1 was cultivated in LB media enriched with D2O in the absence of TMP to examine the impact of TMP on cell growth. Supplementary Fig. 9 shows an increase in the C–D vibrations (2180 cm-1) and concomitant redshifts in the adenine band (724 cm-1) characteristic of time-dependent D2O uptake by bacteria, providing further evidence that DfrB1 maintained its metabolic activity in the presence or absence of TMP.
SERS spectra for TMP-susceptible strain did not exhibit any C–D modes and intensity changes or redshifts attributed to TMP treatment (A, B), which was also reflected in static OD600 values throughout 4 h of cell cultures (B). The C–D and various peak shifts were clearly detected in TMP-resistant cells incorporating D isotopes within 1 h and progressively increased over time (C, D), which increased proportionally with cell replication (OD600) up to 4 h (D). Insets in (A, C) highlight non-shifting (A) and increasing band shifts (C) in the SERS spectral biomarker at 724 cm-1 for the susceptible (A) and resistant (C) cells. Error bars represent the standard deviation of the mean computed from ≥6 repeated measurements for each experiment.
The next analysis stage was to further visualise the kinetics of D uptake in TMP-resistant cells to draw insights into and confirm the molecular dynamics linked to TMP resistance and reproducibility of our nanofibre sensor-based SERS-DIP scheme. To achieve this, machine learning based on DFA was deployed to minimise the within-class variance while also maximising the between-class variance in parallel46 (class implies the spectral dataset measured at individual time points), leading to the formation of distinct sample groups along DF axes. The visual output of DFA modelling is a scores plot presented in Fig. 5, with each sphere representing a full-length signal (400–3200 cm-1). Spectra measured from the same samples are assigned to the same group, whereas data from different samples are grouped into different classes based on their unique spectral fingerprints. Samples were clustered along the positive or negative coefficients of the DFA scores plots depending on whether peak redshifts or intensities were increasing or decreasing when bacteria assimilate heavy water into biomass at various time intervals. According to Fig. 5, four individual clusters (for cells sampled at 0, 1, 2, and 4 h) clearly separated from one another can be seen along DF1 due to the increasing incorporation of the D isotope by bacteria. The spectral data for individual sample classes clustered closer, indicating high repeatability of signals from the sensors.
Discriminant function analysis (DFA) of the kinetic incorporation of D2O-derived D isotope by metabolically active TMP-resistant cells cultured in the growth medium containing 4 µg/mL TMP (2 × MIC) in 50% D2O, highlighting progressive incorporation of heavy deuterium atoms into cellular biomass from 0, 1, 2, and 4 h.
The largest separations were observed for D-enriched TMP-resistant bacteria collected at 0 and 4 h, as shown along DF1. A sharp change was observed along the DF2 axis for cells harvested at 1 and 2 h time points that were discriminated from the rest of the samples along DF2. This dramatic change could be due to the change in bacterial phenotypes upon the assimilation of D2O into biomass, and this observation is consistent with time-course metabolic patterns detected for 13C-enriched E. coli that were probed by Raman/IR spectroscopy and DFA15.
The next question was to ascertain the origin(s) of the clustering trends observed in Fig. 5. Our DFA model is explainable by nature as it can compute the DF loading plots to reveal the significant spectral bands underlying the clustering patterns detected in the DFA scores biplots. Fig. 6 illustrates DF loading plots computed from DFA scores with the major explanatory SERS spectral bands that were significantly affected by D2O assimilation into cellular biomass annotated along DF1 and DF2.
According to the DF1 loadings, which explains the largest variance in the dataset, the main explanatory spectral bands related to the clustering trends detected in DFA scores plots are the apparent redshifts at 724 cm-1 (adenine-like molecules) and 2180 cm-1 (C–D vibrations), and other SERS spectral peaks, such as 953 cm-1 (C–N in proteins/nucleotides) and 1331 cm-1 (adenine-like compounds)47 were also affected by uptake of D. DF2 loadings mainly highlighted an increase in peak intensity at 724 cm-1 and 2180 cm-1 (C–D) as the most important spectral vibrations underlying the DF2 scores (Fig. 5). Thus, kinetic exploration of the SERS data for the D2O-enriched TMP-resistant E. coli (DfrB1) bacterial cells identified two new spectral biomarkers for SERS-AST: spectral redshifts at 2180 cm-1 (C–D vibrations mainly in lipids and proteins) and 724 cm-1 (ring modes in adenine-related molecules), and the intensity dynamics at 724, 956, and 1331 cm-1 similar to those implicated in isotope-free SERS-AST11,12.
In summary, we have shown the utility of nanofibre-based SERS coupled with heavy water (deuterium oxide (D2O)) labelling (SERS-DIP) for rapid and robust detection of bacterial susceptibility to trimethoprim (TMP). SERS spectral data of TMP-susceptible E. coli cultivated in D2O-enriched growth media revealed a loss of metabolic activity when treated with 2 µg/mL TMP, which was rapidly determined as the minimum inhibitory concentration (MIC) by SERS-DIP in 2 h via the disappearance of C–D biomarker bands and an apparent reduction in the intensity of adenine-related peaks. Such an observation starkly contrasted the spectral profiles for the TMP-resistant DfrB1 strain that retained the C–D band at all TMP levels, confirming the TMP resistance phenotype of bacteria expressing DfrB1 protein. Bacterial susceptibility to TMP drug was further monitored through the incorporation kinetics of D2O-derived D by TMP-resistant cells exposed to 4 µg/mL TMP over time. In such a case, the C–D spectral biomarker was detected in 1 h for the resistant E. coli strain following exposure to TMP and increased proportionally to the quantity of D assimilated into cellular biomass at different time points, with these findings confirmed by the machine learning modelling of the kinetics data. This demonstrates the high sensitivity of our nanofibre-based SERS-DIP for time-course nanosensing of TMP resistance. Our findings may pave the way for fast and robust antibiotic susceptibility tests (ASTs), and given the small size of our nanofibre sensors, such modalities can be employed for in vitro and real-time AST to accelerate the treatment of bacterial infections.
Methods
Chemicals and materials
Silver nitrate (AgNO3, ≥99%), sodium hydroxide (NaOH), Luria-Bertani, adenine ( ≥ 99%), sodium chloride (NaCl), trimethoprim ( ≥ 99%), and deuterium oxide (D2O, ≥99%) were purchased from Sigma-Aldrich. Trisodium citrate dihydrate ( ≥ 99%), hydrogen peroxide (H2O2, 30%), nitric acid (HNO3, 70%), and sulfuric acid (H2SO4) were acquired from Fischer Scientific. THF was obtained from VWR. PS-P4VP diblock copolymer [Mn (PS) = 41 kDa, Mn (P4VP) = 20 kDa, total Mw/Mn = 1.18] was acquired from Polymer Source. Isopropyl β-D-1-thiogalactopyranoside (IPTG) was obtained from ThermoFisher. Borosilicate glass rods (1 mm diameter and 10 cm length) were purchased from Sutter Instruments.
Synthesis and characterisation of colloidal silver nanoparticles
All glassware used for AgNP synthesis was thoroughly cleaned using aqua regia (3:1 hydrochloric acid: nitric acid) and copious amounts of distilled water. AgNPs with a mean diameter of 80 ± 12 nm (Supplementary Fig. 1) were synthesised at room temperature using previously reported protocols with minor changes48. Aqueous AgNO3 solution (1 mM, 150 mL) was prepared in a 2-neck round bottom flask and heated to boiling in an oil bath under moderate stirring using a magnetic stirrer bar and hot plate in a fume hood. This was followed by the dropwise addition of 6 mL of 1% (w/v) aqueous trisodium citrate (Na3C6H5O7) to the boiling AgNO3. The reaction between AgNO3 and Na3C6H5O7 produced colour changes from the clear solution to green, then milky grey, confirming successful AgNP synthesis. The sol was left to boil for ~55 min before the reaction chamber was removed from the hot plate and cooled to room temperature. AgNPs were washed three times through centrifugation at 10,000 rpm for 10 min, resuspended in an equal volume of de-ionised water, and kept in a fridge at 4 oC until further use.
Fabrication of AgNP-decorated plasmonic nanofibre nanosensors
Tapered fibres were created by pulling borosilicate glass rods on one end (tip diameter ~1 µm) using a CO2 laser beam on the P-2000 micropipette puller instrument (Sutter Instruments, California, USA), as reported previously27. Next, fibres were initially cleaned in ethanol, followed by a 1 h-wash in 90 oC hot piranha solution (7:3 H2SO4:H2O2) in a fume hood to remove adsorbed contaminants from the fibres (Scheme 1A). Piranha-soaked fibres were rinsed with ultrapure water and thoroughly dried under a vacuum. To fabricate plasmonic nanofibre sensors, clean fibre glass rods were first dip-coated in 0.05 mg/mL of PS-b-P4VP block copolymer (BCP) (Mw = 41 kDa for PS and 20 kDa for P4VP) in tetrahydrofuran (THF). Dip-coating was performed at the rate of 50 mm/min with a 2 min pause in BCP, forming a nano-thin brush polymer layer on the fibre, as we showed recently using ellipsometry and atomic force microscopy, using the same dip-coating method as reported in this study25,28. Polymer-coated fibres were dried under room temperature conditions before being immersed in 3 mL of colloidal AgNPs (pH 4.52) on a pristine Petri dish for 3 h to allow for the adsorption of AgNPs on BCP-anchored nanofibres (Scheme 1A). AgNP-decorated nanofibres (so-called nanofibre sensors) were subsequently washed in ultrapure water to remove excess nanoparticles, dried under N2 current, and stored in the dark until use. A Regulus 8220 high-resolution scanning electron microscope (SEM) image confirmed the morphology and AgNP array on the nanofibre sensor (Scheme 1B). Details of the characterisation of LSPR and analytical performance of nanofibre sensors are shown in Supplementary Figs. 1, 2 and 3.
Optimisation and performance analysis of the nanofibre sensor
The sensitivity and repeatability of our nanofibre sensors were optimised using adenine as a model compound. Adenine was selected to examine the analytical performance of nanofibre sensors because it is an important building block for biologically essential molecules, including DNA, flavins, and RNA, which are typically detected in bacterial cells22,47. Serial dilutions of adenine were prepared within the micromolar to nanomolar range in water to mimic the physiological levels of adenine in bacterial growth cultures49. SERS spectral profiles of adenine measured with nanofibre sensors show intense signals with major spectral peaks detected at 733 and 1329 cm-1 (Supplementary Fig. 3a), assigned to adenine in-plane ring vibrational modes.
Finite-difference time-domain (FDTD) simulations of EM fields
In order to explain the enhancement mechanism of SERS on nanofibre sensors, the optical performance of the proposed tapered sensor was characterised by FDTD simulation. In this simulation, the refractive indices (RIs) of the fibre and PS-b-P4VP layer were taken as constants (1.46 and 1.56, respectively). The permittivity of AgNPs in the visible region was calculated using the Lorentz-Drude model50. We used a normally distributed incident Gaussian beam characterised by its polarisation in the x-direction (transverse magnetic (TM) polarisation) or y-direction (transverse electric (TE) polarisation). Perfectly matched boundary conditions of the layers were used along the x and z axes, respectively, whilst the periodic boundary condition was used along the y axis. The diameter and period of AgNPs were set at 80 nm and 15 nm, respectively, and the polymer thickness was fixed at ~3 nm around the nanofibre. A mesh of 2 nm × 2 nm × 2 nm was deployed to ensure robust simulations (Supplementary Fig. 2a, b).
Bacterial cultivation conditions
In this study, two bacterial strains were tested against trimethoprim (TMP) to determine the MIC and antibiotic susceptibility patterns over time. The wild-type E. coli BL21 (DE3) is susceptible to TMP whilst the E. coli BL21 (DE3) strain transformed with a pET24 (Qiagen) plasmid harbouring the dfrB1 gene (UniProt ID P00383) is resistant to TMP owing to the dihydrofolate reductase activity of DfrB1. Expression of the DfrB1 enzyme is under the control of the lac repressor and is induced by the addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) to the growth medium. For convenience, the strains E. coli BL21 (DE3) and E. coli BL21 (DE3) (DfrB1) are hereafter named BL21 and DfrB1, respectively. BL21 and DfrB1 were pre-cultured three times on Luria-Bertani agar (LB-agar) at 37 oC and harvested after 24 h to obtain axenic bacterial cultures with stable phenotype51, prior to the AST experiments. Single colonies of each of the bacterial samples were separately inoculated in fresh 5 mL Luria-Bertani nutrient broth in cell culture tubes and incubated at 37 oC with shaking at 230 rpm for 24 h in a shaking incubator (New Brunswick Scientific, Edison, NJ, USA). Cell suspensions were collected after overnight growth and further processed as described below.
Susceptibility tests and determination of MIC
The OD600 for the overnight cultures were normalised to 0.1 and inoculated in fresh and sterile Luria-Bertani nutrient broth containing varying concentrations of TMP, 0.1 mM IPTG (ThermoFisher), with and without 50% D2O in triplicate on sterile 96-well plates and cultured under the same conditions as described above. The MIC for the susceptible strain (BL21) was determined by broth dilution on a 96-well plate4. The MIC for the resistant strain was previously determined by the broth dilution method to be 600 µg/mL, well above the TMP concentrations used in the present study38. For SERS-AST, OD600 (0.1)-normalised E. coli strains were cultured for 4 h under identical cultivation conditions to the broth method described above. The cell cultures were then used to inoculate nutrient broth cultures containing 4 µg/mL of TMP (2 × MIC of the susceptible strain) and 0.1 mM IPTG with and without D2O labelling to investigate the kinetics of isotope incorporation by bacteria at 0, 1, 2, and 4 h time intervals using the SERS-based nanofibre sensors.
Nanofibre nanosensor-based SERS spectral measurement
Bacterial cultures (1.5 mL) were transferred to 1.5 mL Eppendorf plastic tubes and washed by centrifugation at 5000 g for 3 min at 4 oC using a benchtop microcentrifuge (Brinkmann Instruments, Ontario, Canada). Bacterial cell supernatants were removed and discarded before the cell pellets were resuspended in an equal volume of distilled (DI) water. This process was repeated three times to ensure the removal of growth media components. For SERS analysis on nanofibre sensors, 30 µL of overnight cell samples (OD600 = 5) were incubated on nanofibre sensors in a sterile Petri dish and dried for 30 min at room temperature to allow for sufficient interactions between the sensor and cells, and the extraction of metabolites from bacteria. After the drying step, the nanofibre sensor was inserted into an adjustable magnetic holder and mounted onto a piezoelectric positioner attached to a confocal Raman microscope, as reported previously52. The optical nanofibre sensor was carefully adjusted to ~45o angle underneath a 40× air objective using a magnetic piezoelectric holder as the pivot to precisely focus the excitation laser beam onto the sensor through the objective lens. The position of the nanofibre sensor was adjusted either alone using a piezoelectric micromanipulator or synchronously with the microscope air objective in the x, y, and z coordinates53. With such well-controlled movements in the x-y and z axes, an excitation laser light spot was focused on bacterial cells adsorbed on the nanofibre (Supplementary Fig. 10) to measure SERS-AST spectral data. The sensors were not reused after bacterial sensing, as the protocol for cleaning bacteria and metabolites off the nanofibre sensor surface is not available at present. Hence, fresh sensors were used for each measurement condition.
Spectral data acquisition
SERS data were measured on the nanofibre sensors using a WITec alpha300 Raman system equipped with a confocal microscope and Raman spectrometer (Raman microspectroscopy). The excitation light beam with ~1 mW power was generated by a 632.8 nm HeNe gas laser focused on the sensor for 5 s via a 40x magnifying objective (NA = 0.65). The backscattered Stokes light was collected through the same optics and diffracted by a 300 grooves/mm grating to the CCD for spectral data acquisition. A total of five SERS spectra for each biological replicate were measured from individual studies on three separate nanofibre sensors. Data capture and instrument control were performed using the Control 5 software.
Spectral data processing and machine learning modelling
Machine learning exploration of the SERS-AST data was performed in Matlab software 2021b (The MathWorks Inc., Natwick, US). Spectral data preprocessing was performed sequentially; first, the data were subjected to baseline correction via asymmetric least squares54, followed by standardisation using the extended multiplicative signal correction (EMSC) scaling55. Multivariate analysis based on principal component analysis was employed to reduce the dimensionality of SERS spectral data, examine natural similarities and differences with respect to molecular diversity induced by TMP at different time points, and identify spectral outliers56. The first five principal components (PCs) retaining the highest explained variance were used as input data to build a discriminant function analysis (DFA). DFA is a supervised model that probes linear relationship(s) among spectral variables linked to sample classes (time points), which DFA then applies to probe bacterial AST based on characteristic molecular vibrations unique to each experimental condition57. In principle, a DFA algorithm minimises the within-class variance while simultaneously maximising the between-class variance to achieve sample classification. Importantly, a DFA model is interpretable by nature as it provides scores showing sample classes and identifies feature importance in classification via loadings plots58. A data splitting method (i.e., 70% train versus 30% test of randomised datasets) was applied to validate DFA, where an independent holdout test dataset (30%) was projected into subspaces generated by the model in the training set59. Statistical data were presented as averages of spectra, with the error bars representing the standard deviation of the mean of repeated spectral measurements.
Data availability
The data associated with this study are provided in this manuscript and accompanying Supplementary Information. Raw data are available from the corresponding authors upon reasonable request.
Code availability
The chemometric codes used in this study are available from the authors on request. Matlab functions used alongside our code are freely available in the cluster-toolbox-v2.0 courtesy of Biospec (Laboratory for Bioanalytical Spectroscopy) · GitHub.
References
Khan, Z. A., Siddiqui, M. F. & Park, S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics 9, 49 (2019).
Hassall, J., Coxon, C., Patel, V. C., Goldenberg, S. D. & Sergaki, C. Limitations of current techniques in clinical antimicrobial resistance diagnosis: examples and future prospects. NPJ Antimicrob. Resist. 2, 16 (2024).
Benfey, P. N. & Mitchell-Olds, T. From genotype to phenotype: systems biology meets natural variation. Science 320, 495–497 (2008).
Reller, L. B., Weinstein, M., Jorgensen, J. H. & Ferraro, M. J. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755 (2009).
AlMasoud, N. et al. Discrimination of bacteria using whole organism fingerprinting: the utility of modern physicochemical techniques for bacterial typing. Analyst 146, 770–788 (2021).
Ma, H. et al. Surface-enhanced Raman spectroscopy: current understanding, challenges, and opportunities. ACS Nano 18, 14000–14019 (2024).
Huang, H.-K. et al. Bacteria encapsulation and rapid antibiotic susceptibility test using a microfluidic microwell device integrating surface-enhanced Raman scattering. Lab Chip 20, 2520–2528 (2020).
Cialla-May, D. et al. Recent advances of surface enhanced Raman spectroscopy (SERS) in optical biosensing. TrAC Trends Anal. Chem. 181, 117990 (2024).
Liu, C.-Y. et al. Rapid bacterial antibiotic susceptibility test based on simple surface-enhanced Raman spectroscopic biomarkers. Sci. Rep. 6, 23375 (2016).
Han, Y. Y. et al. SERS-based rapid susceptibility testing of commonly administered antibiotics on clinically important bacteria species directly from blood culture of bacteremia patients. World J. Microbiol. Biotechnol. 39, 16 (2023).
Dina, N. E. et al. SERS-based antibiotic susceptibility testing: towards point-of-care clinical diagnosis. Biosens. Bioelectron. 219, 21 (2023).
Samek, O., Bernatová, S. & Dohnal, F. The potential of SERS as an AST methodology in clinical settings. Nanophotonics 10, 2537–2561 (2021).
Chang, K. W. et al. Antibiotic susceptibility test with surface-enhanced Raman scattering in a microfluidic system. Anal. Chem. 91, 10988–10995 (2019).
Pérez-Jiménez, A. I., Lyu, D., Lu, Z., Liu, G. & Ren, B. Surface-enhanced Raman spectroscopy: benefits, trade-offs and future developments. Chem. Sci. 11, 4563–4577 (2020).
Chisanga, M. et al. Metabolism in action: stable isotope probing using vibrational spectroscopy and SIMS reveals kinetic and metabolic flux of key substrates. Analyst 146, 1734–1746 (2021).
Berry, D. et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl Acad. Sci. USA 112, E194–E203 (2015).
Song, Y. et al. Raman-deuterium isotope probing for in-situ identification of antimicrobial resistant bacteria in Thames river. Sci. Rep. 7, 16648 (2017).
Shams, S. et al. Optical photothermal infrared spectroscopy: a novel solution for rapid identification of antimicrobial resistance at the single-cell level via deuterium isotope labeling. Front. Microbiol. 14, 1077106 (2023).
Matanfack, G. A. et al. Influence of carbon sources on quantification of deuterium incorporation in heterotrophic bacteria: a Raman-stable isotope labeling approach. Anal. Chem. 92, 11429–11437 (2020).
Wang, Y. et al. Reverse and multiple stable isotope probing to study bacterial metabolism and interactions at the single cell level. Anal. Chem. 88, 9443–9450 (2016).
Masters, P. A., O’Bryan, T. A., Zurlo, J., Miller, D. Q. & Joshi, N. Trimethoprim-sulfamethoxazole revisited. Arch. Intern. Med. 163, 402–410 (2003).
Premasiri, W. R. et al. The biochemical origins of the surface-enhanced Raman spectra of bacteria: a metabolomics profiling by SERS. Anal. Bioanal. Chem. 408, 4631–4647 (2016).
Chisanga, M. et al. Rapid differentiation of Campylobacter jejuni cell wall mutants using Raman spectroscopy, SERS and mass spectrometry combined with chemometrics. Analyst 145, 1236–1249 (2020).
Hassanain, W. A., Johnson, C. L., Faulds, K., Graham, D. & Keegan, N. Recent advances in antibiotic resistance diagnosis using SERS: focus on the “Big 5” challenges. Analyst 147, 4674–4700 (2022).
Zhu, H. et al. Block copolymer brush layer-templated gold nanoparticles on nanofibers for surface-enhanced Raman scattering optophysiology. ACS Appl. Mater. Interfaces 11, 4373–4384 (2019).
Masson, J. F. et al. Optoplasmonic effects in highly curved surfaces for catalysis, photothermal heating, and SERS. ACS Appl. Mater. Interfaces 15, 46181–46194 (2023).
Zhao, X. J. et al. Branched Au nanoparticles on nanofibers for surface-enhanced Raman scattering sensing of intracellular pH and extracellular pH gradients. ACS Sens. 5, 2155–2167 (2020).
Zhu, H., Masson, J.-F. & Bazuin, C. G. Templating gold nanoparticles on nanofibers coated with a block copolymer brush for nanosensor applications. ACS Appl. Nano Mater. 3, 516–529 (2020).
Zhu, H., Masson, J. F. & Bazuin, C. G. Monolayer arrays of Au nanoparticles on block copolymer brush films for optical devices and biosensors. ACS Appl. Nano Mater. 7, 5637–5651 (2024).
Mistark, P. A. et al. Block-copolymer-based plasmonic nanostructures. ACS Nano 3, 3987–3992 (2009).
Allison, G. et al. A fabry-perot cavity coupled surface plasmon photodiode for electrical biomolecular sensing. Nat. Commun. 12, 7 (2021).
Jia, Y., Tang S., Wang Z. & Y., L. Plasmonic resonant cavity provides a flexible substrate for surface-enhanced Raman spectroscopy detection in a wide band range. ACS Appl. Opt. Mater. 2, 761–766 (2024).
Grys, D.-B. et al. Eliminating irreproducibility in SERS substrates. J. Raman Spectrosc. 52, 412–419 (2021).
Tahir, M. A., Dina, N. E., Cheng, H. Y., Valev, V. K. & Zhang, L. W. Surface-enhanced Raman spectroscopy for bioanalysis and diagnosis. Nanoscale 13, 11593–11634 (2021).
Lemay-St-Denis, C. & Pelletier, J. N. From a binding module to essential catalytic activity: how nature stumbled on a good thing. Chem. Commun. 59, 12560–12572 (2023).
van Duijkeren, E., Schink, A.-K., Roberts, M. C., Wang, Y. & Schwarz, S. Mechanisms of bacterial resistance to antimicrobial agents. Microbiol. Spectrum. https://doi.org/10.1128/microbiolspec.arba-0019-2017 (2018).
Bhosle, A. et al. A strategic target rescues trimethoprim sensitivity in Escherichia coli. iScience 23, 100986 (2020).
Lemay-St-Denis, C. et al. A conserved SH3-like fold in diverse putative proteins tetramerizes into an oxidoreductase providing an antimicrobial resistance phenotype. Philos. Trans. R. Soc., B 378, 20220040 (2023).
Chisanga, M., Muhamadali, H., Ellis, D. I. & Goodacre, R. Surface-enhanced raman scattering (SERS) in microbiology: illumination and enhancement of the microbial world. Appl. Spectrosc. 72, 987–1000 (2018).
Su, X. et al. Estuarine plastisphere as an overlooked source of N2O production. Nat. Commun. 13, 3884 (2022).
Muhamadali, H., Chisanga, M., Subaihi, A. & Goodacre, R. Combining Raman and FT-IR spectroscopy with quantitative isotopic labeling for differentiation of E. coli cells at community and single cell levels. Anal. Chem. 87, 4578–4586 (2015).
Fornasaro, S. et al. Surface enhanced Raman spectroscopy for quantitative analysis: results of a large-scale European multi-instrument interlaboratory study. Anal. Chem. 92, 4053–4064 (2020).
Allen, D. M., Einarsson, G. G., Tunney, M. M. & Bell, S. E. J. Characterization of bacteria using surface-enhanced Raman spectroscopy (SERS): influence of microbiological factors on the SERS spectra. Anal. Chem. 94, 9327–933 (2022).
Cheng, H.-W., Tsai, H.-M. & Wang, Y.-L. Exploiting purine as an internal standard for SERS quantification of purine derivative molecules released by bacteria. Anal. Chem. 95, 16967–16975 (2023).
Kneis, D. et al. Trimethoprim resistance in surface and wastewater is mediated by contrasting variants of the dfrB gene. ISME J. 17, 1455–1466 (2023).
Chisanga, M., Muhamadali, H., Ellis, D. I. & Goodacre, R. Enhancing disease diagnosis: biomedical applications of surface-enhanced Raman scattering. Appl. Sci. Basel 9, 1163–1186 (2019).
Kubryk, P., Niessner, R. & Ivleva, N. P. The origin of the band at around 730 cm-1 in the SERS spectra of bacteria: a stable isotope approach. Analyst 141, 2874–2878 (2016).
Lee, P. C. & Meisel, D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J. Phys. Chem. 86, 3391–3395 (1982).
Chuang, Y.-C. et al. Bacterial adenine cross-feeding stems from a purine salvage bottleneck. ISME J. 18, wrae034 (2024).
Rodrigo, S. G., García-Vidal, F. J. & Martín-Moreno, L. Influence of material properties on extraordinary optical transmission through hole arrays. Phys. Rev. B 77, 8 (2008).
Muhamadali, H. et al. Chicken, beams, and campylobacter: rapid differentiation of foodborne bacteria via vibrational spectroscopy and MALDI-mass spectrometry. Analyst 141, 111–122 (2016).
Lussier, F. et al. Dynamic-SERS optophysiology: a nanosensor for monitoring cell secretion events. Nano Lett. 16, 3866–3871 (2016).
Chisanga, M. & Masson, J. F. Machine learning-driven SERS nanoendoscopy and optophysiology. Annu. Rev. Anal. Chem. 17, 313–338 (2024).
Eilers, P. H. C. Parametric time warping. Anal. Chem. 76, 404–411 (2004).
Afseth, N. K. & Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemometrics Intell. Lab. Syst. 117, 92–99 (2012).
Chisanga, M. et al. SERS-based assay for multiplexed detection of cross-reactivity and persistence of antibodies against the spike of the native, P.1 and B.1.617.2 SARS-CoV-2 in non-hospitalised adults. Sens. Diagn. 1, 851–866 (2022).
Gromski, P. S. et al. A tutorial review: metabolomics and partial least squares-discriminant analysis—a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 879, 10–23 (2015).
Goodacre, R. et al. Proposed minimum reporting standards for data analysis in metabolomics. Metabolomics 3, 231–241 (2007).
Chisanga, M. et al. Label-free SERS for rapid differentiation of SARS-CoV-2-induced serum metabolic profiles in non-hospitalized adults. Anal. Chem. 95, 3638–3646 (2023).
Acknowledgements
We acknowledge financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Foundation for Innovation (CFI).
Author information
Authors and Affiliations
Contributions
Conceptualisation: M.C. and J.-F.M. Methodology: M.C., C.L.S.D. and J.-F.M. Investigation: M.C. and C.L.S.D. FDTD simulations and analysis: X.W., Y.L., M.L., W.P., and M.C. Data acquisition and interpretation: M.C., C.L.S.D., J.N.P., and J.-F.M. Code development and data analysis: M.C. Writing the original draft: M.C. Supervision: J.N.P. and J.-F.M. Funding: J.-F.M. All the authors have read and approved the final article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chisanga, M., Lemay-St-Denis, C., Wei, X. et al. Metabolite microextraction on surface-enhanced Raman scattering nanofibres and D2O probing accelerate antibiotic susceptibility testing. npj Biosensing 2, 21 (2025). https://doi.org/10.1038/s44328-025-00039-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44328-025-00039-w