Abstract
The increasing prevalence of obesity-driven metabolic dysfunction-associated steatotic liver disease (MASLD) urges the development of new therapeutic strategies. Totum-448 is a patented blend of polyphenol-rich plant extracts designed to reduce hepatic steatosis, a key risk factor for steatohepatitis and type 2 diabetes. In a mouse model of MASLD, Totum-448 (1.5% w/w) significantly improved metabolic homeostasis by lowering hyperinsulinemia and systemic glucose intolerance, without affecting body weight, fat mass, or calorie intake. It also reduced hepatic steatosis and plasma alanine aminotransferase levels, as assessed by histological/biochemical assays and MS-based lipidomics. RNA sequencing and spectral flow cytometry approaches revealed that liver inflammation and pro-fibrotic gene expression were decreased, along with preservation of tissue-resident Kupffer cells and reduced recruitment of pro-inflammatory macrophages. These combined effects suggest that Totum-448 ameliorates both steatosis and hepatic inflammation, contributing to improved whole-body metabolic health. Totum-448 may represent a promising nutritional intervention for MASLD management in humans.
Similar content being viewed by others
Introduction
The development of metabolic dysfunction-associated steatotic liver disease (MASLD), formerly known as non-alcoholic fatty liver disease (NAFLD)1, and its progressive, more aggressive form, metabolic dysfunction-associated steatohepatitis (MASH), are closely intertwined with the current worldwide epidemic of obesity and type 2 diabetes2,3. Recent epidemiological studies have indeed highlighted the alarming rise of MASLD in both developing and developed countries, with a high prevalence rate (>50%) in overweight and obese adults that constitutes one of the health challenges of the 21st century4.
MASLD has a complex pathophysiology, characterized by hepatic lipid accumulation, insulin resistance, lipotoxicity, inflammation, and progressive fibrogenesis1, with an estimated annual cost exceeding 200 billion euros in the United States and Europe alone5. The disease spectrum is broad, ranging from isolated steatosis to MASH, fibrosis, and cirrhosis, ultimately increasing the risk of hepatocellular carcinoma6,7,8. Effective therapeutic treatments remain scarce as only Resmetirom, a modestly effective drug for fibrotic MASH, was recently approved by the FDA9.
Metaflammation, which refers to a state of chronic, low-grade inflammation arising from obesity-associated immunometabolic dysregulations in various organs, plays a pivotal role in MASLD initiation and progression10. Excessive hepatic influx of lipids and accumulation of triglycerides in the form of lipid droplets in hepatocytes triggers a cascade of inflammatory responses in the liver mediated by various immune cell types, notably those from the innate myeloid compartment11,12. In this context, macrophages are believed to play a central role13,14,15,16. During homeostasis, hepatic macrophages predominantly consist of self-replenishing, embryonically-derived tissue-resident Kupffer cells (resKCs)17,18. However, in response to obesity-associated lipotoxic stress and local inflammatory cues in their microenvironment, resKCs undergo cell death, leading to an influx of circulating bone-marrow-derived monocytes for replenishing the empty niche. These cells further differentiate into monocyte-derived macrophages (moMACS) and ultimately resKCs that display almost identical features to KCs of embryonic origin19,20. Collectively, these obesogenic-driven changes in the hepatic immunological landscape contribute to the chronic inflammatory milieu, leading to liver injury, fibrosis, and ultimately, the development of MASH and its complications21,22,23. Moreover, metaflammation extends beyond the liver, promoting systemic metabolic dysfunction and exacerbating both central and peripheral insulin resistance, dyslipidemia, and cardiovascular risk24.
The multifactorial nature of MASLD highlights the need for comprehensive strategies involving conventional lifestyle interventions and innovative preventive and/or therapeutic approaches to halt disease progression and reduce associated morbidity and mortality25. Recently, functional foods and nutraceuticals containing various bioactive compounds have received considerable attention due to their potential therapeutic benefits in the context of MASLD/MASH26,27. Indeed, their accessibility and relatively low risk of adverse effects make them attractive adjuncts to lifestyle modifications and/or pharmacological treatments. For instance, bioactive compounds such as omega-3 fatty acids, polyphenols, flavonoids, and vitamins, as well as probiotics, have been shown to mitigate lipid accumulation, inflammation, oxidative stress, and insulin resistance in the liver, potentially contributing to preventing MASLD/MASH progression and/or promoting disease regression28.
Totum-448 is a novel, polyphenol-rich plant-based active principle composed of a mixture of plant extracts designed to reduce obesity-induced hepatic steatosis, a risk factor for progression towards type 2 diabetes and MASH. In the present study, we aimed to investigate the effects of Totum-448 on hepatic steatosis, liver inflammation, and metabolic homeostasis in a dietary mouse model of MASLD.
Results
Totum-448 improves whole-body metabolic homeostasis in MASLD mice independently of body weight changes
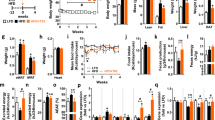
To select the optimal Totum-448 concentration to be administered, a pilot study was performed in MASLD mice (Supplementary Fig 1). For this purpose, C57BL/6 male mice were first fed a high-fat diet supplemented with sucrose in the drinking water (HFD/S) for 12 weeks, followed by HFD supplementation with or without Totum-448 at various concentrations (1.5, 2, and 2.5% w/w) for 4 additional weeks (Supplementary Fig 1a). We observed a substantial time- and dose-dependent decrease in body weight at the two highest Totum-448 concentrations (-3.8% and -9.8% at 2 and 2.5% Totum-448, respectively (Supplementary Fig 1b–c)). These effects were likely due to a dose-dependent decrease in food intake (Supplementary Fig 1d) even though concomitant increases in liquid energy intake were evidenced (Supplementary Fig 1e). Altogether, no significant impact on total energy intake (Supplementary Fig 1f) and feces production (Supplementary Fig 1g) were observed. To further study the effects of Totum-448 on metabolic homeostasis in insulin resistant obese MASLD mice, the concentration that did not affect body weight and food intake was selected, i.e. 1.5% w/w, and administered to HFD/S-fed mice using the same experimental settings as described above (Fig. 1a). In line with the pilot study, we did not observed any effect on body weight (Fig. 1b) and composition (Fig. 1c, d) in obese mice after a 4-week supplementation with 1.5% Totum-448. Similarly, this concentration of Totum-448 did not affect the total energy intake (14.8+/−0.5 Kcal/day versus 15.6+/−0.2 Kcal/day in HFD/S and HFD/S+Totum-448, respectively; p = 0.44). As expected, HFD/S feeding increased fasting plasma glucose and insulin levels (Fig. 1e, f), and HOMA-IR (Fig. 1g) when compared to low-fat diet (LFD)-fed mice. Furthermore, HFD/S impaired whole-body glucose homeostasis, as assessed by intraperitoneal glucose tolerance test (Fig. 1h). Although no effect was observed on fasting plasma glucose levels, Totum-448 supplementation in HFD/S-fed mice significantly reduced insulin levels, and, consequently, the calculated HOMA-IR (Fig. 1e–g). Congruent with HOMA-IR data, Totum-448 improved whole-body glucose homeostasis in obese mice (Fig. 1h), without increasing glucose-induced insulin levels (Fig. 1i). Of note, these weight change-independent effects of Totum-448 on insulin and HOMA-IR were already observed after 2 weeks of supplementation (Supplementary Fig. 2). In addition, although the HFD/S-induced increase in total blood leukocyte counts was unchanged, the circulating levels of monocytes was significantly reduced by Totum-448, while other myeloid (neutrophils, eosinophils) and lymphoid (B, NK, CD4+ and CD8+ T) subsets were not affected (Supplementary Fig 3).
10 week-old C57BL/6JOlaHsd male mice were fed either a low-fat diet (LFD, open squares/bars) or high-fat diet (HFD) supplemented with sucrose in the drinking water (10% w/v, HFD/S) for a period of 12 weeks after which the HFD was either supplemented with Totum-448 (T448, 1.5% g/g; purple squares/bars) or left without supplementation (control; black squares/bars) for 4 additional weeks (a). At week 4 of treatment, body weight (b) and body composition (c) were determined. Post-sacrifice, the weight of the liver, WAT, BAT and heart were determined (d). The fasting glucose and insulin levels (e, f) were determined at week 4 and used to calculate HOMA-IR (g). An intraperitoneal (i.p.) glucose tolerance test (GTT) was performed at week 4 in 6-hour fasted mice. Blood glucose levels were measured at baseline and 20, 40, 60 and 90 min post-injection, and the AUC was calculated (h). Insulin levels were measured at 20 min post-injection (i). Results are expressed as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. n = 10-12 mice per group from 2 independent experiments.
Totum-448 marginally impacts fecal microbiome composition
Given that obesity-induced changes in gut microbiota is associated with metabolic dysfunctions29, we next assessed the impact of Totum-448 on fecal microbiome composition by performing 16S ribosomal RNA sequencing on feces collected during the last week of the study. Importantly, Totum-448 supplementation had no effect on the length of the total intestine and colon nor the weight of the cecum content at sacrifice (Fig. 2a-c). At the phylum level, the Shannon index was decreased in the HFD/S groups when compared to the LFD group, indicating a reduction in bacterial species diversity, but no significant differences were observed in response to Totum-448 supplementation (Fig. 2d). Principal component analysis (PCA) of the relative abundance of intestinal microbial communities using the Bray-Curtis dissimilarity index also confirmed that microbial composition only differed significantly between the LFD and HFD/S groups (notably Actinobacteria and Tenericutes) but not in response to Totum-448 supplementation (Fig. 2e, f, Supplementary Table 1). At the genus level, the Shannon index was increased in both HFD/S and HFD/S+Totum-448 groups (Fig. 2g). The PCA analysis confirmed that the HFD/S strongly altered microbial composition when compared to LFD while Totum-448 supplementation had no significant effect (Fig. 2h). Similar PCA results were obtained using Jaccard distance (Fig. 2i), which is more sensitive to rare taxa by only taking into account the presence or absence of a dedicated taxon, independent of its abundance. Altogether, only a few bacterial taxa from the Firmicutes phylum were specifically affected by Totum-448 (Fig. 2j, Supplementary Table 2), indicating a marginal impact on fecal microbiota composition.
LFD- and HFD/S-fed mice were treated as described in Fig. 1. The intestine and colon lengths and the cecum weight were measured post-sacrifice (a–c). Fecal microbiome alpha and beta diversity: species richness and evenness were assessed at phylum (d) and genus (g) level by Shannon index. Principal coordinate analysis of microbial composition using Bray-Curtis (e, h) and Jaccard (i) distances at phylum (e) and genus (h, i) levels. Average relative microbiome composition of fecal samples at phylum (f) and genus (j) levels. For visual clarity, only the most abundant 5 phyla and 20 genera are presented individually, the rest being summed up into “Others”. Results are expressed as mean ± SEM. n = 10–12 mice per group from 2 independent experiments (for a–c). n = 4–6 mice per group from 1 experiment (d–j).
Totum-448 reduces hepatic steatosis and alters liver lipid composition
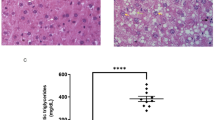
Ectopic lipid accumulation, especially in the liver, triggers immunometabolic dysfunctions contributing to insulin resistance and impaired nutrient homeostasis7. Therefore, we next investigated the impact of Totum-448 supplementation on hepatic steatosis in MASLD mice. Remarkably, Totum-448 almost completely reverted HFD/S-induced hepatic steatosis in MASLD mice, as assessed by H&E staining (Fig. 3a). This effect was mostly resulting from a reduction in macrovascular steatosis (Fig. 3b, c) and associated with a significant reduction of steatosis, inflammation, hepatocellular ballooning and MASLD activity scores (Fig. 3d). These findings were further supported by a potent decrease in both liver triglycerides (TG) and total cholesterol contents (−28% and −30% respectively; p < 0.05; Fig. 3e). Quantitative lipidomics further confirmed that Totum-448 significantly affected the hepatic lipid composition by reducing the liver content of a large numbers of TGs, diglycerides (DGs) and free fatty acid (FFA) species in MASLD mice (Fig. 3f, g).
LFD- and HFD/S-fed mice were treated as described in Fig.1. PFA-fixed, paraffin-embedded liver section were stained with Hematoxilin and Eosin (H&E, a) followed by computer-assisted determination of hepatic lipid droplet (LD) size distribution (b) and mean LD area (c). H&E-stained slides were also used to assess the hepatic steatosis, lobular inflammation, hepatocellular ballooning scores and overall MASLD activity score (d). Hepatic triglyceride (TG), total cholesterol (TC) and phospholipid (PL) contents (e) were determined post-sacrifice. The hepatic lipid composition was determined by targeted lipidomics using the Lipidyzer platform. The heatmap shows the relative abundance of the individual lipid species per class in each group (f). The relative increase and decrease of various lipid species per class in livers from HFD/S + T448 compared to HFD/S-fed mice are displayed on the volcano plot (g). CE Cholesterylester, CER ceramides, DG Diglycerides, FFA Free-fatty acids, LP Lipoprotein, PL Phospholipids, SM Sphingomyelin, TG Triglycerides. Results are expressed as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001. n = 10–12 mice per group from 2 independent experiments for a–e and n = 3-4 mice per group for lipidomics (f–g).
Totum-448 lowers inflammatory and pro-fibrotic transcriptomic signatures in the liver
To gain mechanistic insights into the beneficial metabolic effects of Totum-448, bulk RNA sequencing was performed in the livers from MASLD mice. Differential gene expression analysis showed that Totum-448 induced a significant up- and downregulation of 47 and 345 unique transcripts, respectively, in HFD/S-fed mice (Fig. 4a, b). Gene ontology and gene set enrichment analyses indicated an enrichment of downregulated genes involved in innate immune response, myeloid cell and platelet activation, pro-inflammatory cytokine production and extracellular matrix organization (Fig. 4c, d, Supplementary Fig 4a). A large number of genes encoding proteins involved in leukocyte chemotaxis (e.g. Ccl2), liver inflammation (e.g. Lcn2) and hepatic stellate cell pro-fibrotic activation (e.g. Acta2, Timp1, Col1a1 and Mmp12) were found to be among the most significantly downregulated by Totum-448. These findings were confirmed by targeted qPCR (Fig. 4e–f, Supplementary Fig 4b). In line with improvements in hepatic MASLD/MASH features, a decrease in circulating alanine aminotransferase (ALT) was observed in response to Totum-448 supplementation, indicating a reduction in hepatocyte injury and liver damage (Fig. 4g).
LFD- and HFD/S-fed mice were treated as described in Fig. 1. Bulk RNA sequencing was performed in liver samples to assess hepatic transcriptional changes in response to T448 supplementation. Hierarchically clustered heatmap displays differentially-expressed genes (DEGs) in each group (a). The volcano plot depicts significantly up- and down-regulated genes in livers from HFD/S + T448- compared to HFD/S-fed mice (b). A GO-term analysis on DEGs (c) and a gene set enrichment analysis (GSEA, d) were performed on the whole transcriptome. Targeted qPCR was performed to assess the expression of pan-inflammatory gene Lcn2 (e) and fibrosis-related genes (f). Plasma alanine transaminase (ALT) levels were determined at week 4 in 6-hour fasted mice (g). Results are expressed as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01. n = 3 mice per group for bulk RNA-seq (a–d) and n = 10–12 mice per group from 2 independent experiments for targeted qPCR (e–g).
Totum-448 prevents loss of tissue-resident Kupffer cells and reduces both hepatic monocyte infiltration and accumulation of pro-inflammatory monocyte-derived macrophages
To further investigate the inhibitory effect of Totum-448 on hepatic inflammation, we performed an in-depth immunophenotyping of liver leukocytes by spectral flow cytometry (see Supplementary Fig 5a for gating strategy). The total number of CD45+ hepatic leukocytes tended to be higher in HFD/S-fed mice when compared to LFD mice but was not affected by Totum-448 (Fig. 5a). Using Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) to visualize global changes in the major hepatic immune cell subsets (Fig. 5b), we observed that while Totum-448 treatment had no impact on neutrophils, NK cells, dendritic cells, and T and B cells subsets (Supplementary Fig 5b), it led to significant reduction of both eosinophil (Fig. 5c) and Ly6Chi monocytes (Fig. 5d) in the liver from MASLD mice. Remarkably, although the total macrophage abundance was not affected in any of the groups (Fig. 5e), the proportion of CD11c+ and TREM2+ expressing macrophages was increased in HFD/S-fed mice and significantly lowered by Totum-448 (Fig. 5f), indicating a reduction in pro-inflammatory and lipid-associated macrophages, respectively. The total macrophage pool was further divided into monocyte-derived CD11b+CLEC2- macrophages (moMACS) and CD11blowCLEC2+ Kupffer cells (KCs), the latter being further divided into CLEC2+TIM4- monocyte-derived Kupffer cells (moKCs) and resident CLEC2+TIM4+ Kupffer cells (resKCs). As expected, HFD/S induced a potent loss of resKCs and a concomitant increase in moMACS in order to repopulate the KCs niche when compared to LFD-fed mice (Fig. 5g, h). Remarkably, Totum-448 supplementation significantly decreased both KC loss and increased accumulation of moMACS in the livers from MASLD mice (Fig. 5g, h), strongly suggesting a reduction in HFD/S-induced KC activation and death. Of note, immunophenotyping was also performed in eWAT from a subset of the mice (Supplementary Fig 6). Totum-448, while not significantly affecting tissue leukocyte content and relative abundances of eosinophils, monocyte and T cells (Supplementary Fig 6b–f), may also dampen tissue inflammation by reducing tissue accumulation of both total adipose tissue macrophages (ATMs), obesity-associated pro-inflammatory CD11c+ATMs (Supplementary Fig 6g, h) and neutrophils (Supplementary Fig 6i).
LFD- and HFD/S-fed mice were treated as described in the legends of Fig. 1. The total number of CD45+ hepatic leukocytes was determined after isolation (a). Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) was used to assess global changes in the major hepatic immune cell subsets (b). The proportion of eosinophils (c) monocytes (d) and total hepatic macrophages (e) expressed as frequency of total CD45+ leukocytes and the proportion of CD11c+ and TREM2+ expressing macrophages were determined (f). The abundance of resident Kupffer cells (g) and monocyte-derived macrophages (moMACS, h) expressed as frequency of the total hepatic macrophage pool was determined. Results are expressed as mean ± SEM. *p ≤ 0.05, **p ≤ 0.01. n = 10-12 mice per group from 2 independent experiments.
Discussion
In this study, we report and dissect the beneficial effects of Totum-448, a polyphenol-rich plant extract, on hepatic steatosis, liver inflammation and whole-body metabolic homeostasis in a dietary mouse model of MASLD.
Previous studies have shown the potential of nutraceuticals in improving cardiometabolic health, particularly in the context of insulin resistance, type 2 diabetes, and MASLD26,27,28,30. Various plant-derived bioactive compounds, such as polyphenols, flavonoids, and specific fiber blends, have been shown to improve insulin sensitivity and glucose/lipid homeostasis through mechanisms independent of weight loss, notably through modulation of gut microbiota28.
Our results indicate that Totum-448 has a limited impact on microbiota composition in MASLD mice. While HFD/S feeding significantly altered fecal microbial diversity and composition, the marginal differences only observed in some Firmicutes taxa in response to Totum-448 supplementation suggest a negligible impact rather than a broad restructuring of the gut microbiome. While numerous animal studies have reported major effect of polyphenols on microbiota in the context of MASLD31, the relatively marginal impact of Totum-448 on microbiome composition observed in this work may be partly related to the study design. Indeed, the majority of the preclinical studies were actually carried out with the administration of polyphenols starting simultaneously with the initiation of the dietary regimen, i.e., assessing the impact on disease progression rather than on its regression32. A few studies did report significant changes in microbiota in a context of pre-established dysbiosis, but the duration of supplementation was significantly longer compared to this study33,34, suggesting that 4-week supplementation with Totum-448 might have been insufficient to counteract the deep-seated microbial changes induced by HFD/S.
Despite this, significant effects on hepatic steatosis were observed in Totum-448-supplemented mice. While this study was not designed to determine which specific compound was responsible for these benefits, previous research has demonstrated direct actions of certain isolated polyphenols found in Totum-448 on the liver, leading to reduced lipid accumulation. Among the most extensively studied polyphenols in HFD-fed mice, oleuropein has been shown to inhibit Wnt10b- and FGFR1-mediated signaling pathways involved in hepatic lipogenesis, while also suppressing TLR2- and TLR4-mediated pro-inflammatory signaling implicated in hepatic steatosis35. Additionally, it was shown to regulate lipid oxidation, lipogenesis, and inflammation via PPAR-α36 and activates autophagy pathways through AMPK37. Moreover, chlorogenic acid alleviated steatosis by inhibiting ALKBH5 activity, which in turn suppressed the ERK signaling pathway and regulated autophagy38. Similarly, luteolin has been reported to enhance mitochondrial biogenesis via the AMPK/PGC1α pathway, promoting fatty acid oxidation39. It also inhibits IL-1 and IL-18 pro-inflammatory pathways40 and reduces lipid accumulation by preventing LXR-mediated sterol regulatory element-binding protein-1 (SREBP-1c) activation41. Finally, caffeic acid has demonstrated promising hepatoprotective effects both in vivo and in vitro, reducing hepatocyte lipid accumulation and increasing autophagy, possibly through modulation of either fibroblast growth factor 21 (FGF21), FGF receptor 1 (FGFR1), β-Klotho (KLB), and/or the AMPK-SREBP-1c axis42,43,44. Totum-448 also contains choline, a nutrient known to exert hepatoprotective effects in various models of liver diseases. In both dietary and genetic models of MASLD, choline supplementation has been shown to reduce hepatic steatosis, normalize cholesterol metabolism, and attenuate inflammation and fibrosis45,46,47,48. For instance, choline restored cholesterol homeostasis and markedly improved liver function in HFD-fed Pemt/Ldlr double knockout mice, thereby preventing the onset of steatohepatitis48. It was also reported to improve steatosis and liver damage via enhanced fatty acid oxidation and reduced VLDL secretion in APOE*3-Leiden.CETP mice46. Furthermore, long-term choline supplementation in HFD-fed mice decreased hepatic steatosis and reduced hepatocellular carcinoma burden47. Of note, the choline concentrations used in these preclinical studies ranged from 0.4% to 1.2% w/w, which are higher than the 0.2% w/w included in the present study; however, we cannot exclude the possibility that choline contributed, at least in part, to the beneficial hepatic and metabolic effects observed in our study. Overall, polyphenols and choline are believed to exert anti-steatotic effects in the liver through a combination of anti-inflammatory and antioxidant mechanisms, which collectively could help alleviating insulin resistance, along with the activation of PPAR-α-mediated fatty acid oxidation and the inhibition of lipogenesis via the AMPK/SREBP-1c pathway49,50. In the present work, however, our liver transcriptomic analysis did not fully reflect all these pathways in the metabolic signature. Instead, the most pronounced effects were related to anti-inflammatory and anti-fibrotic responses, suggesting the participation of extrahepatic mechanisms. For instance, increased adipose tissue lipolysis due to insulin resistance is a well-recognized contributor to excessive free fatty acid delivery to the liver, resulting in steatosis. Although we did not specifically assess insulin resistance or inflammation in adipose tissue, the fact that WAT weight tended to be higher in Totum-448-supplemented mice raises intriguing possibilities for future investigations into its role in the observed metabolic effects.
In addition to its impact on metabolic homeostasis, this study provides an overview of the effects of Totum-448 on hepatic immune cell composition in HFD/S-fed mice, supporting the growing evidence that nutraceuticals, especially those of polyphenol-rich nature, can exert immunomodulatory effects which may contribute to improved metabolic outcomes51. In the liver, couple of key features were also associated with Totum-448 supplementation, namely reductions in HFD/S-induced eosinophilia, KC loss, and moMACS accumulation. Eosinophilic inflammation has been linked to progressive MASLD and suggested to be a potential contributor to fibrotic remodeling observed in later-stage MASH52,53. Interestingly, we found that Totum-448 supplementation almost completely reversed HFD/S induced eosinophil accumulation, indicating a potential protective effect against progressive MASH. Remarkably, Totum-448 supplementation also decreased the loss of embryonically-derived resKCs and reduced the recruitment and/or differentiation of moMACS, highlighting a beneficial remodeling of the hepatic macrophage compartment associated with reduced inflammation and MASLD progression. In addition to changes in hepatic macrophage ontogeny, modulation of their activation states has also been linked to MASLD/MASH progression, with CD11c expression being a hallmark of pro-inflammatory macrophage activation54,55. In our study, the expression of CD11c among the total macrophage pool was significantly reduced in response to Totum-448 supplementation, indicating an overall decrease in the inflammatory activation of the hepatic macrophage compartment. Furthermore, one of the key macrophage subsets recently identified during MASLD development, called lipid-associated macrophages (LAMs), was shown to arise in both WAT and the liver during obesity and to display a unique ability to store and oxidize lipids when compared to resKCs56. LAMs are intimately linked with fibrotic areas in the liver during MASH development and have been shown to play an essential role in the regression of fibrosis12,57. One of the key defining markers of LAMs is the expression of Triggering receptor expressed on myeloid cells-2 (TREM2), which is strongly associated with steatohepatitis in different diet-induced murine models of MASH58,59. Although the exact role of TREM2+ macrophages in the MASLD/MASH pathophysiology still remains to be clarified, they are intimately linked with disease progression of MASLD to MASH and the rise of fibrosis and serve as an indicator of disease severity. It is however, tempting to speculate that the observed decrease in TREM2+ macrophages induced by Totum-488 supplementation may result from a reduction in their hepatic recruitment secondary to dampening of pro-inflammatory and pro-fibrotic signaling.
In addition to the potential anti-fibrotic and immunomodulatory effects of Totum-448, the RNA sequencing data highlighted a potential lowering of platelet activation in response to Totum-448 supplementation. MASLD/MASH have been associated with a pro-thrombotic state and increased intrahepatic platelet accumulation and activation has previously been associated with various stages in MASLD and MASH pathophysiology58,59,60. For example, one study reported that platelet-derived growth factor B (PDGF-B) could activate hepatic stellate cells (HSCs) and promote liver fibrosis60, whilst other studies demonstrated the anti-steatotic/fibrotic effects of aspirin use58 and various anti-platelet drugs61. Platelets also directly interact with KCs, a feature that has been demonstrated in early steatosis and shown to contributes to MASH development through increased immune cell recruitment62. One may therefore speculate that part of the beneficial effects of Totum-448 could be related to a direct effect on intrahepatic platelet dynamics and activation, an interesting aspect to investigate that would require further studies.
Several limitations of this study ought to be acknowledged, one of them being the lack of a clear underlying mechanism explaining the observed immunometabolic effects of Totum-448. Given its polyphenol-rich composition, its benefits are likely mediated through the pleiotropic action of various bioactive molecules, influencing multiple cell types and organs both directly and indirectly. Although the current study does not investigate the bioavailability of Totum-448, a dedicated clinical study is currently ongoing to assess the bioavailability of its major metabolites in humans (ClinicalTrials.gov Identifier: NCT06047847). However, tissue-specific changes in insulin sensitivity were not assessed in our study, which limits our understanding of whether peripheral organs, such as the liver, adipose tissue, or skeletal muscle, were specifically affected by Totum-448 supplementation. Furthermore, it is worth mentioning that the dietary model used in the current study induces a rather mild form of MASLD/MASH, with hepatic steatosis and some degree of inflammation, but without detectable fibrosis as assessed by collagen accumulation using Sirius red staining or hydroxyproline assay (data not shown). Similarly, the majority of studies describing immunological changes in the liver during MASH also rely on the use of more advanced models of MASH. One might therefore speculate that Totum-448 may eventually exert even more beneficial effects in advanced MASH stages, or could reveal stronger immunomodulatory and anti-fibrotic properties than the ones observed with the current experimental settings. It should also be noted that this study did not include Totum-448 supplementation in LFD conditions, which might provide insights into the potential effects of Totum-448 in the absence of HFD/S induced MASLD. Another limitation is that the study was conducted exclusively in male mice. It is well established that metabolic responses to dietary interventions, including to HFD exposure63 or polyphenol supplementation64, are exhibiting sexual dimorphism, with female mice displaying different adaptations due to hormonal variations, gut microbiota composition, immune cell profiles, and intrinsic metabolic flexibility. Hence, the absence of female subjects prevents a comprehensive evaluation of whether the observed effects would be similar in both sexes. Finally, while rodent models provide valuable insights into metabolic disorders such as MASLD, their relevance to human physiology remains a key consideration. Therefore, further clinical trials are necessary to determine whether these findings translate to human populations. As such, Totum-448 is currently being evaluated in a randomized, double-blind clinical trial involving MASLD patients (ClinicalTrials.gov Identifier: NCT06704321).
In summary, we show that Totum-448 supplementation reduces both hepatic steatosis and liver inflammation, and improves whole-body metabolic homeostasis in a diet-induced MASLD mouse model. Although the underlying mechanism(s) of Totum-448 remain to be elucidated, its beneficial immunometabolic properties likely result from pleiotropic actions on various cell types and/or organs driven by a variety of plant-derived polyphenolic molecules. Altogether, supplementation with Totum-448 may constitute a promising novel nutritional approach for MASLD patients.
Methods
Totum-448
Totum-448 is a patented blend of 5 plant extracts and choline designed to act in combination to target the risk factors of developing MASLD. The mixture contains extracts from olive leaf (Olea europaea), bilberry (Vaccinium myrtillus), artichoke leaf (Cynara scolymus), chrysanthellum (Chrysanthellum indicum subsp. afroamericanum B.L. Turner), black pepper (Piper nigrum), and choline.
The total levels of phenolic compounds (in gallic acid equivalent), sugars (in glucose equivalent), and fat (in sunflower oil equivalent) were determined in Totum-448 using Folin-Ciocalteu, Dubois, and sulfo-phospho-vanillin (SPV) colorimetric assays, respectively, as previously reported65. A fluorometric method using fluoraldehyde o-phthaldialdehyde reagent (OPA) was used to quantify the protein content66, and the Lee method was used to quantify insoluble dietary fibers67. A more precise characterization of phytochemical compounds was performed by HPLC-UV/Visible/RID-MS using a 1260 LC system and a 1200 LC system with a 6110 Single Quad MS-ESI detector (Agilent Technologies, Santa Clara, CA, USA) with a C18 Prodigy reversed-phase column (250 mm × 4.6 mm, 5 μm; Phenomenex, Torrance, CA, USA) and an Atlantis HILIC Silica column (150 × 4.6 mm, 5 μm, Waters, The Netherlands). Supplementary Table 3 shows the chemical characterization of Totum-448.
Animals and diet
All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Institute for Laboratory Animal Research and were approved by the Dutch ethical committee on animal experiments (Centrale Commissie Dierproeven; AVD1060020174364). An a priori power calculation was performed, and the reporting of the research on animals was done according to the ARRIVE guidelines. Ten-week-old C57BL/6JOlaHsd male mice were purchased from Envigo (Horst, The Netherlands) and housed in a temperature-controlled room with a 12-hour light-dark cycle and ad libitum access to food and drink. To reduce variation due to sex hormone cycles on whole-body metabolism, male mice were used for all in vivo experiments. Mice were fed a low-fat diet (LFD, 10% energy derived from fat, D12450H, Research Diets, New Brunswick, NJ, USA) or high fat diet (HFD, 45% energy derived from fat, D12451, Research Diets, New Brunswick, NJ, USA) supplemented with sucrose in the drinking water (10% w/v, HFD/S) for 12 weeks. The experimental groups were randomized after removal of HFD/S low responders (~5%; body weight gain <6 g), after which HFD was supplemented or not with Totum-448 (Valbiotis SA, Perginy, France) by incorporating the compound in the diet for an additional 4 weeks (n = 10–12 mice per experimental group). The experimenters were not blinded to the diet supplementation on the metabolic test days, however, most of the subsequent analyses were performed in blind conditions.
Body composition, energy intake, and feces production
Body weight was frequently measured during the 4 weeks of supplementation (every day during the first week and at least twice a week for the rest of the study) using a conventional weighing scale. Body composition was measured by MRI (Echo Medical Systems, Houston, TX, USA) in conscious, unrestrained mice. At the end of the experiment, mice were euthanized with an overdose of ketamine/xylazine/fentanyl (100/10/0.07 mg/kg body weight), then blood was collected and visceral white adipose tissue (epidydimal; eWAT), supraclavicular brown adipose tissue (BAT), heart and liver were dissected, weighed and either stored or frozen in liquid nitrogen for further processing, as described in details below. The intestines were collected and measured (total and colon separately), and the weight of the cecum was determined using a precision scale. Food and sucrose intake were assessed at least twice a week throughout the study by weighing food pellets and measuring liquid volume in the drinking bottle for every cage (2–3 mice per cage). At week 4, a couple of days before study termination, feces produced over 24 h (starting from 9 am) were carefully collected in cage bedding and weighed.
Glucose tolerance test
Whole-body intraperitoneal (i.p.) glucose tolerance (ipGTT) test was performed at week 4 of Totum-448 supplementation, as previously reported68. In short, a bolus of glucose (2 g D-glucose/kg body weight; Sigma-Aldrich, St. Louis, MO, USA) was administered i.p. in 6h-fasted mice and blood glucose was measured at t = 0, 20, 40, 60, and 90 min post glucose injection using a Glucometer (Accu-Check; Roche Diagnostics, Basel, Switzerland).
Plasma analysis
Blood samples were collected from the tail vein of 4h-fasted mice using paraoxon-coated glass capillaries. Plasma insulin was determined using a commercially available ELISA kit (Chrystal Chem, Elk Grove Village, IL, USA) according to the manufacturer’s instructions. The homeostatic model assessment of insulin resistance (HOMA-IR) adjusted for mice69 was calculated as followed ([glucose (mg/dl)*0.055]*[insulin (ng/ml)*172.1])/3875. Plasma alanine aminotransferase (ALAT) was measured using a Reflotron® kit (Roche diagnostics, Basel, Switzerland).
Fecal microbiota analyses
DNA was extracted from fecal samples using the FastDNATM Spin Kit for Feces and a FastPrep-24 5 G (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions. Microbial 16S library preparation was performed at the PGTB (Plateforme Génome Transcriptome de Bordeaux, Bordeaux, France) by amplification and sequencing of the V3-V4 region of the 16S rRNA gene on an Illumina MiSeq using the 2x250bp Illumina v2 kit (Illumina, San Diego, CA, USA). Quality filtering and processing of the raw sequencing data were done as previously described70 using cutadapt v4.071 with Python v3.9.9, usearch v11.0.66772, and mothur v1.48.073. Classification of quality filtered reads was performed by comparison with the ribosomal database project (RDP trainset 14)74,75. Abundance tables were imported and analyzed in R v4.1. Alpha and beta diversity were estimated using the package vegan 2.6-276 and tested using Kruskal-Wallis and MANOVA (adonis2) tests, respectively. Indicator taxa analysis was performed using Kruskal-Wallis test. False Discovery Rate (FDR) correction for multiple testing was used were applicable. Indicator taxa were identified by selecting those with a significant overall association with the experimental group (p < 0.05), and that had at least one pairwise comparison between groups that reached significance (p < 0.05).
Hepatic lipid composition
Liver triglycerides (TG), total cholesterol (TC), and phospholipid (PL) contents were measured using commercial kits (Instruchemie, Delfzijl, The Netherlands) #2913, #10015, and #3009, respectively) and expressed as nmol per mg of total protein content using the Bradford protein assay kit (Sigma-Aldrich, St. Louis, MO, USA), as previously reported77,78. For lipidomics, lipids were extracted from 10 mg of liver by the methyl-tert-butylether method and analyzed using the Lipidyzer™, a direct infusion-tandem mass spectrometry-based platform (Sciex, Redwood City, CA, USA), as previously described78. Lipid concentrations were expressed as pmol/mg of liver.
Histological analysis
Pieces of liver (~30 mg) were fixed in 4% formaldehyde (Sigma-Aldrich, St. Louis, MO, USA), paraffin-embedded, sectioned at 4 μm, and stained with Hematoxilin and Eosin (H&E). After scanning, 5 fields at 40x or 20x magnification were used for the determination of lipid droplet (LD) size distribution and mean area, and MASLD activity score (NAS), respectively, as previously reported79.
Isolation of blood, liver, and adipose tissue leukocytes
At the final day of the experiment, blood was collected retro-orbitally in heparin-coated tubes for leukocyte isolation, as described previously68. Samples were diluted 1:1 in PBS (Fresenius Kabi, Bad Homburg, Germany, with calcium and magnesium), and erythrocytes were lysed for 15 min at room temperature using an erythrocyte lysis/fixation solution (BD Biosciences, Franklin Lakes, NJ, USA). Blood leukocytes were then centrifuged at 500 x g for 5 min at 4 °C and then subsequently washed three times in PBS. After washing, cell pellets were resuspended in PBS supplemented with 1% heat inactivated fetal calf serum (hiFSC; Serana, Pessin, Germany) and 2.5 mM ethylenediaminetetraacetic acid (EDTA; Sigma-Aldrich, St. Louis, MO, USA), counted using a hemocytometer and 1*106 cells per sample were further processed for flow cytometry.
For liver samples, the organs were collected after a 1 min post-sacrifice transcardial perfusion with PBS and further digested for isolation of leukocytes, as previously reported80. Livers were first collected in 10 mL ice-cold RPMI 1640 + Glutamax (Thermo Fisher Scientific, Waltham, MA, USA), minced and digested for 25 min at 37˚C under agitation (200 RPM) in 5 mL RPMI 1640 + Glutamax supplemented with 1 mg/ml Collagenase Type IV from Clostridium histolyticum (Sigma-Aldrich, St. Louis, MO, USA, 125 CDU/ml), 1 mg/ml Dispase II (Sigma-Aldrich, St. Louis, MO, USA, 1.4 U/ml), 1 mg/ml Collagenase D from C. histolyticum (Roche, Basel, Switzerland, 250 Mandl U/ml) and 2000 U/mL DNase I (Sigma-Aldrich, St. Louis, MO, USA). After digestion, samples were filtered (100 µM cell strainer; Corning, NY, USA) and pelleted at 300 x g for 10 min at 4 °C after which the pellets were washed twice with 40 mL PBS/hiFSC/EDTA. After washing, the pellets were treated with 3 mL erythrocyte lysis buffer consisting of 0.15 M NH4Cl (Merck, Rahway, NJ, USA), 1 mM KHCO3 (Merck, Rahway, NJ, USA), and 0.1 mM EDTA in ddH2O for 2 min at room temperature. Next, total leukocytes were isolated by MACS-sorting using CD45 positive selection MicroBeads and LS columns (Mitenyi Biotec, Bergisch Gladback, USA) according to the manufacturer’s instructions. Post-isolation, total leukocytes were counted using a hemacytometer and 1*106 cells per sample were further processed for flow cytometry.
For adipose tissue samples, epidydimal white adipose tissues (eWAT) were collected after a 1 min post-sacrifice transcardial perfusion with PBS and further digested for isolation of stromal vascular fraction (SVF), as previously reported80. Briefly, eWAT samples were minced and digested for 1 hour at 37 °C under agitation (60 RPM) in 2 mL HEPES-buffered Krebs solution containing 1 mg/mL Collagenase Type I from C. histolyticum (Sigma-Aldrich, St. Louis, MO, USA, 125 CDU/ml), 20 mg/mL Bovine Serum Albumin (Fraction V; Sigma-Aldrich, St. Louis, MO, USA) and 6 mM D-Glucose Sigma-Aldrich, St. Louis, MO, USA). After digestion, samples were filtered (100 µM cell strainer; Corning, NY, USA) and washed with 30 mL PBS/hiFCS/EDTA, as described above. After allowing the adipocytes to settle for 10 min, the infranatant consisting of the SVF was collected and pelleted at 350 x g for 10 min at room temperature. The pellet was treated with 1 mL erythrocyte lysis buffer, washed with 10 mL PBS/hiFCS/EDTA, and counted using a hemacytometer. As for the liver, 1*106 cells per sample were further processed for flow cytometry.
Flow cytometry
For blood leukocytes, isolated cells were washed with PBS/hiFSC/EDTA, pelleted at 500 x g for 5 min at 4 °C and incubated with a cocktail of antibodies directed against CD3, CD4, CD8, CD11b, CD19, CD45, Ly6C, NK1.1 and Siglec-F (see Supplementary Table 4 for details) in PBS/hiFSC/EDTA supplemented with Brilliant Stain Buffer Plus (BD Biosciences, Franklin Lakes, NJ, USA) and True Stain Monocyte Blocker (Biolegend, San Diego, CA, USA) for 30 min at room temperature.
For liver leukocytes, isolated cells were pelleted at 500 x g for 5 min at 4 °C and subsequently incubated with Zombie-NIR viability dye in PBS supplemented with True Stain Monocyte Blocker for 20 min at room temperature. After washing with PBS and pelleting as described above, the cells were fixed using a 2% paraformaldehyde solution (PFA; Sigma-Aldrich, St. Louis, MO, USA) in PBS for 10 min at room temperature. Post-fixation, the cells were washed with PBS/hiFSC/EDTA and incubated with a cocktail of antibodies directed against CD3, CD11b, CD11c, CD19, CD45, CD64, CD90.2, CLEC2, F4/80, Ly6C, Ly6G, NK1.1, Siglec-F, TIM4 and TREM2 (see Supplementary Table 4 for details) in PBS/hiFCS/EDTA supplemented with Brilliant Stain Buffer Plus and True Stain Monocyte Blocker for 30 min at 4 °C.
For eWAT leukocytes, cells were pelleted, incubated with Zombie-NIR viability dye, and fixed with PFA similarly to liver leukocytes. Post-fixation, the cells were washed with PBS/hiFSC/EDTA and incubated with a cocktail of antibodies directed against CD3, CD11b, CD11c, CD19, CD45, CD64, F4/80, Ly6C, Ly6G, NK1.1, and Siglec-F in PBS/hiFCS/EDTA supplemented with Brilliant Stain Buffer Plus (BD Biosciences, Franklin Lakes, NJ, USA) and True Stain Monocyte Blocker (Biolegend, San Diego, CA, USA) for 30 min at 4 °C.
After washing, blood, liver, and eWAT cells were resuspended in PBS/FSC/EDTA and acquired on a 5-laser Cytek Aurora(Cytek Biosciences, Fremont, CA, USA). SpectroFlo v3.0(Cytek Biosciences, Fremont, CA, USA) was used for spectral unmixing and FlowJoTM v10.8 was used to gate the flow cytometry data for all samples. Representative gating strategies used to gate the blood, liver, and eWAT cells can be found in Supplementary Fig. 3a, Supplementary Fig. 5a, and Supplementary Fig. 6a, respectively.
RNA isolation and RNA sequencing analyses
RNA was extracted from snap-frozen liver samples (~10-20 mg) using an RNA purification kit (NucleoSpin RNA Midi, Macherey-Nagel, Düren, Germany) followed by an on-filter DNAse treatment, according to the instructions provided by the manufacturer. RNA concentration and purity were measured using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Further RNA sample preparation, sequencing, and data pre-processing were done at Biomarker Technologies (BMK GmbH Münster, Germany).
For library preparation, a total amount of 1 μg RNA per sample was used as input material for the RNA sample preparation. Sequencing libraries were generated using NEBNext UltraTM RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) following manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer. First-strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase. Second-strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 240 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Brea, CA, USA). Then 3 μl USER Enzyme (New England Biolabs, Ipswich, MA, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min followed by 5 min at 95 °C before PCR. PCR was then performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers, and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system (Agilent Technolgies, Santa Clara, CA, USA).
Clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v4-cBot-HS (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform, and paired-end reads were generated.
For quality control, raw sequences (raw reads) in fastq format were first processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing low-quality reads and reads containing adapter or ploy-N from raw data. At the same time, Q20, Q30, GC-content, and sequence duplication level of the clean data were calculated. Clean reads were next mapped to the reference genome sequence (Mus musculus, NCBI GRCm39) using the HISAT2 alignment program81. Only reads with a perfect match or one mismatch were further analyzed and annotated based on the reference genome. Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KO (KEGG Ortholog database) and GO (Gene Ontology).
Gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped (FPKM). The formula is shown as follows: FPKM = cDNA Fragments/(Mapped Fragments (Millions)) ∗ Transcript Length (kb)). Differential expression analysis was performed using the DESeq2 bioconductor pipeline82. The resulting P-values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 were assigned as differentially expressed. Finally, gene Ontology (GO) enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages based Wallenius non-central hyper-geometric distribution, which can adjust for gene length bias in DEGs83. KOBAS software to test the statistical enrichment of differential expression genes in KEGG pathways84.
RNA isolation and RT-qPCR
RNA was extracted from snap-frozen liver (~10-20 mg) using TriPure RNA Isolation reagent (Roche Diagnostics, Basel, Switzerland). Total RNA (1–2 μg) was reverse transcribed using the M-MLV Reverse Transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA). Real-time qPCR runs were performed on a CFX96 Real-time C1000 thermal cycler (Biorad, Hercules, CA, USA) using the GoTaq qPCR Master Mix kit (Promega, Madison, WI, USA). Gene expression was normalized using housekeeping gene RplP0 and expressed as fold change compared to LFD-fed mice. Primer sequences can be found in Supplementary Table 5.
Statistical analysis
All data are presented as mean ± standard error of the mean (SEM). Statistical analysis was performed using GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA) with one-way analysis of variance (ANOVA) followed by Fisher’s post-hoc test unless indicated otherwise. Visualization of statistical significance was limited to the comparison between HFD/S and HFD/S + T448. Differences between groups were considered statistically significant at p < 0.05. Outliers were identified according to the two-standard deviation method (GraphPad Software, La Jolla, CA, USA).
Data availability
The original datasets generated during the current study are available from the corresponding author on reasonable request. Raw 16S rRNA sequences are available in the NCBI database under the bioproject number PRJNA1071341, with SRA numbers SRR27797200 to SRR27797215 and biosamples number SAMN39684451 to SAMN39684466.
References
Rinella, M. E. et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. Hepatology 78, 1966–1986 (2023).
Quek, J. et al. Global prevalence of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in the overweight and obese population: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 8, 20–30 (2023).
En Li Cho, E. et al. Global prevalence of non-alcoholic fatty liver disease in type 2 diabetes mellitus: an updated systematic review and meta-analysis. Gut 72, 2138–2148 (2023).
Liu, J. et al. Estimating global prevalence of metabolic dysfunction-associated fatty liver disease in overweight or obese adults. Clin. Gastroenterol. Hepatol. 20, e573–e582 (2022).
Younossi, Z. M. et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64, 1577–1586 (2016).
Kisseleva, T. & Brenner, D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat. Rev. Gastroenterol. Hepatol. 18, 151–166 (2021).
Loomba, R., Friedman, S. L. & Shulman, G. I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021).
Tilg, H., Adolph, T. E., Dudek, M. & Knolle, P. Non-alcoholic fatty liver disease: the interplay between metabolism, microbes and immunity. Nat. Metab. 3, 1596–1607 (2021).
Harrison, S. A. et al. A Phase 3, randomized, controlled trial of Resmetirom in NASH with liver fibrosis. N. Engl. J. Med 390, 497–509 (2024).
Sawada, K., Chung, H., Softic, S., Moreno-Fernandez, M. E. & Divanovic, S. The bidirectional immune crosstalk in metabolic dysfunction-associated steatotic liver disease. Cell Metab. 35, 1852–1871 (2023).
Barreby, E., Chen, P. & Aouadi, M. Macrophage functional diversity in NAFLD - more than inflammation. Nat. Rev. Endocrinol. 18, 461–472 (2022).
Daemen, S. et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 34, 108626 (2021).
Park, M. D., Silvin, A., Ginhoux, F. & Merad, M. Macrophages in health and disease. Cell 185, 4259–4279 (2022).
Guilliams, M. & Scott, C. L. Liver macrophages in health and disease. Immunity 55, 1515–1529 (2022).
Shapouri-Moghaddam, A. et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 233, 6425–6440 (2018).
Lazarov, T., Juarez-Carreno, S., Cox, N. & Geissmann, F. Physiology and diseases of tissue-resident macrophages. Nature 618, 698–707 (2023).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Liu, Z. et al. Fate Mapping via Ms4a3-Expression history traces monocyte-derived cells. Cell 178, 1509–1525 e1519 (2019).
Bleriot, C. et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity 54, 2101–2116 e2106 (2021).
Scott, C. L. et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 7, 10321 (2016).
Hammerich, L. & Tacke, F. Hepatic inflammatory responses in liver fibrosis. Nat. Rev. Gastroenterol. Hepatol. 20, 633–646 (2023).
Hoogerland, J. A., Staels, B. & Dombrowicz, D. Immune-metabolic interactions in homeostasis and the progression to NASH. Trends Endocrinol. Metab. 33, 690–709 (2022).
Huby, T. & Gautier, E. L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat. Rev. Immunol. 22, 429–443 (2022).
Hotamisligil, G. S. Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185 (2017).
European Association for the Study of the, L. European Association for the Study of, D. & European Association for the Study of, O. EASL-EASD-EASO Clinical Practice Guidelines on the management of metabolic dysfunction-associated steatotic liver disease (MASLD). J. Hepatol. 81, 492–542 (2024).
Rizzo, M. et al. Nutraceutical approaches to non-alcoholic fatty liver disease (NAFLD): A position paper from the International Lipid Expert Panel (ILEP). Pharm. Res. 189, 106679 (2023).
Cicero, A. F. G., Colletti, A. & Bellentani, S. Nutraceutical Approach to Non-Alcoholic Fatty Liver Disease (NAFLD): The available clinical evidence. Nutrients 10, https://doi.org/10.3390/nu10091153 (2018).
Cogorno, L. et al. Non-alcoholic fatty liver disease: Dietary and nutraceutical approaches. Liver Res. 7, 216–227 (2023).
Hou, K. et al. Microbiota in health and diseases. Signal Transduct. Target Ther. 7, 135 (2022).
Meegaswatte, H., Speer, K., McKune, A. J. & Naumovski, N. Functional foods and nutraceuticals for the management of cardiovascular disease risk in postmenopausal women. Rev. Cardiovasc Med 25, 460 (2024).
Mohammadhasani, K., Vahedi Fard, M., Mottaghi Moghaddam Shahri, A. & Khorasanchi, Z. Polyphenols improve non-alcoholic fatty liver disease via gut microbiota: A comprehensive review. Food Sci. Nutr. 12, 5341–5356 (2024).
Molinari, R., Merendino, N. & Costantini, L. Polyphenols as modulators of pre-established gut microbiota dysbiosis: State-of-the-art. Biofactors 48, 255–273 (2022).
Zhao, L. et al. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 8, 4644–4656 (2017).
Cheng, M. et al. A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food Funct. 9, 1079–1087 (2018).
Park, S., Choi, Y., Um, S. J., Yoon, S. K. & Park, T. Oleuropein attenuates hepatic steatosis induced by high-fat diet in mice. J. Hepatol. 54, 984–993 (2011).
Zhou, W. & Du, Z. Oleuropein mitigates non-alcoholic fatty liver disease (NAFLD) and modulates liver metabolites in high-fat diet-induced obese mice via activating PPARalpha. J. Sci. Food Agric. 104, 8634–8645 (2024).
Porcu, C. et al. Oleuropein induces AMPK-dependent autophagy in NAFLD Mice, regardless of the Gender. Int. J. Mol. Sci. 19, https://doi.org/10.3390/ijms19123948 (2018).
Meng, F. et al. Chlorogenic acid modulates autophagy by inhibiting the activity of ALKBH5 Demethylase, thereby ameliorating hepatic steatosis. J. Agric Food Chem. 71, 15073–15086 (2023).
Wang, T. et al. Luteolin ameliorates hepatic Steatosis and enhances mitochondrial biogenesis via AMPK/PGC-1alpha pathway in Western diet-fed mice. J. Nutr. Sci. Vitaminol.69, 259–267 (2023).
Abu-Elsaad, N. & El-Karef, A. Protection against nonalcoholic steatohepatitis through targeting IL-18 and IL-1alpha by luteolin. Pharm. Rep. 71, 688–694 (2019).
Yin, Y. et al. Luteolin improves non-alcoholic fatty liver disease in db/db mice by inhibition of liver X receptor activation to down-regulate expression of sterol regulatory element binding protein 1c. Biochem Biophys. Res Commun. 482, 720–726 (2017).
Zhang, J. et al. Caffeic acid ameliorates metabolic dysfunction-associated steatotic liver disease via alleviating oxidative damage and lipid accumulation in hepatocytes through activating Nrf2 via targeting Keap1. Free Radic. Biol. Med. 224, 352–365 (2024).
Liao, C. C., Ou, T. T., Huang, H. P. & Wang, C. J. The inhibition of oleic acid induced hepatic lipogenesis and the promotion of lipolysis by caffeic acid via up-regulation of AMP-activated kinase. J. Sci. Food Agric. 94, 1154–1162 (2014).
Kim, H. M., Kim, Y., Lee, E. S., Huh, J. H. & Chung, C. H. Caffeic acid ameliorates hepatic steatosis and reduces ER stress in high fat diet-induced obese mice by regulating autophagy. Nutrition 55-56, 63–70 (2018).
Schenkel, L. C. et al. Choline supplementation restores substrate balance and alleviates complications of Pcyt2 deficiency. J. Nutr. Biochem 26, 1221–1234 (2015).
Liu, C. et al. Dietary choline increases brown adipose tissue activation markers and improves cholesterol metabolism in female APOE*3-Leiden.CETP mice. Int J. Obes.47, 236–243 (2023).
Brown, A. L. et al. Dietary Choline supplementation attenuates high-fat-diet-induced hepatocellular carcinoma in mice. J. Nutr. 150, 775–783 (2020).
Al Rajabi, A. et al. Choline supplementation protects against liver damage by normalizing cholesterol metabolism in Pemt/Ldlr knockout mice fed a high-fat diet. J. Nutr. 144, 252–257 (2014).
Rodriguez-Ramiro, I., Vauzour, D. & Minihane, A. M. Polyphenols and non-alcoholic fatty liver disease: impact and mechanisms. Proc. Nutr. Soc. 75, 47–60 (2016).
Li, S. et al. The potential and action mechanism of polyphenols in the treatment of liver diseases. Oxid. Med Cell Longev. 2018, 8394818 (2018).
Medoro, A. et al. Nutraceuticals as modulators of immune function: a review of potential therapeutic effects. Prev. Nutr. Food Sci. 28, 89–107 (2023).
Hart, K. M. et al. Type 2 immunity is protective in metabolic disease but exacerbates NAFLD collaboratively with TGF-beta. Sci. Transl. Med. 9, https://doi.org/10.1126/scitranslmed.aal3694 (2017).
Gieseck, R. L. et al. Interleukin-13 activates distinct cellular pathways leading to ductular reaction, steatosis, and fibrosis. Immunity 45, 145–158 (2016).
Wentworth, J. M. et al. Pro-inflammatory CD11c+CD206+ adipose tissue macrophages are associated with insulin resistance in human obesity. Diabetes 59, 1648–1656 (2010).
Corbin, A. L. et al. IRF5 guides monocytes toward an inflammatory CD11c(+) macrophage phenotype and promotes intestinal inflammation. Sci. Immunol. 5, https://doi.org/10.1126/sciimmunol.aax6085 (2020).
Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698.e614 (2019).
Ganguly, S. et al. Lipid-associated macrophages’ promotion of fibrosis resolution during MASH regression requires TREM2. Proc. Natl. Acad. Sci. USA 121, e2405746121 (2024).
Simon, T. G. et al. Daily Aspirin use associated with reduced risk for fibrosis progression in patients with nonalcoholic fatty liver disease. Clin. Gastroenterol. Hepatol. 17, 2776–2784.e2774 (2019).
Miele, L. et al. Nonalcoholic fatty liver disease (NAFLD) severity is associated to a nonhemostatic contribution and proinflammatory phenotype of platelets. Transl. Res. 231, 24–38 (2021).
Yoshida, S. et al. Extrahepatic platelet-derived growth factor-beta, delivered by platelets, promotes activation of hepatic stellate cells and biliary fibrosis in mice. Gastroenterology 147, 1378–1392 (2014).
Fujita, K. et al. Effectiveness of antiplatelet drugs against experimental non-alcoholic fatty liver disease. Gut 57, 1583–1591 (2008).
Malehmir, M. et al. Platelet GPIbalpha is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 25, 641–655 (2019).
Zhu, Q. et al. Sexual dimorphism in lipid metabolism and gut microbiota in mice fed a high-fat diet. Nutrients 15, https://doi.org/10.3390/nu15092175 (2023).
Santangelo, C. et al. Insights into the sex-related effects of dietary polyphenols and metabolic disruptors on inflammatory and (neuro) endocrine pathways in obesity: The HEAL Project. Nutrients 16, https://doi.org/10.3390/nu16213595 (2024).
Tobiasz-Adamczyk, B., Flak, E. & Jedrychowski, W. A. Impact of psychosocial factors on the prevalence of headaches in the industrial setting. A case control approach. Neuroepidemiology 4, 86–95 (1985).
Benson, J. R. & Hare, P. E. O-phthalaldehyde: fluorogenic detection of primary amines in the picomole range. Comparison with fluorescamine and ninhydrin. Proc. Natl. Acad. Sci. USA 72, 619–622 (1975).
Lee, S. C. & Prosky, L. International survey on dietary fiber: definition, analysis, and reference materials. J. AOAC Int 78, 22–36 (1995).
van der Zande, H. J. et al. Dendritic cell-intrinsic LKB1-AMPK/SIK signaling controls metabolic homeostasis by limiting the hepatic Th17 response during obesity. JCI Insight 8, https://doi.org/10.1172/jci.insight.157948 (2023).
Lee, S. et al. Comparison between surrogate indexes of insulin sensitivity and resistance and hyperinsulinemic euglycemic clamp estimates in mice. Am. J. Physiol. Endocrinol. Metab. 294, E261–E270 (2008).
Vallier, M. et al. Pathometagenomics reveals susceptibility to intestinal infection by Morganella to be mediated by the blood group-related B4galnt2 gene in wild mice. Gut Microbes 15, 2164448 (2023).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet j. 17, https://doi.org/10.14806/ej.17.1.200 (2011).
Edgar, R. C. & Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ. Microbiol. 75, 7537–7541 (2009).
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Thomas, A. et al. Hypoxia-inducible factor prolyl hydroxylase 1 (PHD1) deficiency promotes hepatic steatosis and liver-specific insulin resistance in mice. Sci. Rep. 6, 24618 (2016).
Zinsou, J. F. et al. Schistosoma haematobium infection is associated with lower serum cholesterol levels and improved lipid profile in overweight/obese individuals. PLoS Negl. Trop. Dis. 14, e0008464 (2020).
van der Zande, H. J. P. et al. Effects of a novel polyphenol-rich plant extract on body composition, inflammation, insulin sensitivity, and glucose homeostasis in obese mice. Int J. Obes.45, 2016–2027 (2021).
Lambooij, J. M., Tak, T., Zaldumbide, A. & Guigas, B. OMIP-104: A. 30-color spectral flow cytometry panel for comprehensive analysis of immune cell composition and macrophage subsets in mouse metabolic organs. Cytom. A 105, 493–500 (2024).
Siren, J., Valimaki, N. & Makinen, V. Indexing Graphs for Path Queries with Applications in Genome Research. IEEE/ACM Trans. Comput Biol. Bioinform. 11, 375–388 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793 (2005).
Acknowledgements
This study was partly funded by Valbiotis (to B.G.). The funder played no role in study design, data collection, analysis, and interpretation of data, or the writing and decision to submit this manuscript.
Author information
Authors and Affiliations
Contributions
V.C., Y.F.O., P.S., and B.G. conceptualized research; J.L., M.V., H.J.P.vd.Z., F.O., R.S., and F.L.J. performed research; J.L., M.V., F.O., R.S., F.L.J., and B.G. analyzed data; T.M., M.G., S.L.P., A.Z., P.S., and B.G. supervised the study; J.L., V.C., and B.G. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
V.C., Y.F.O., M.V., S.L.P., and P.S. are/were all employees of Valbiotis. S.L.P. and P.S. are listed as co-inventors on Totum-448 patent and possess company stocks. None of the other authors has any potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lambooij, J.M., Chavanelle, V., Vallier, M. et al. The polyphenol-rich plant extract Totum-448 decreases hepatic steatosis and inflammation in diet-induced MASLD mice. npj Gut Liver 2, 20 (2025). https://doi.org/10.1038/s44355-025-00033-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44355-025-00033-z