Abstract
Recent advancements in computational chemistry utilize transformer-based models pre-trained on Simplified Molecular Input Line Entry System (SMILES) sequences to predict molecular properties. To improve upon existing methods, we propose MLM-FG, a molecular language model with a novel pre-training strategy that randomly masks subsequences corresponding to chemically significant functional groups. This technique compels the model to better infer molecular structures and properties by learning the context of these key units. Extensive evaluations across 11 benchmark tasks demonstrate the superiority of MLM-FG, outperforming existing SMILES- and graph-based models in 9 of the 11 tasks. Remarkably, MLM-FG surpasses even some 3D-graph-based models, highlighting its exceptional capacity for representation learning without explicit 3D structural information. These results indicate that MLM-FG effectively learns to interpret molecular properties from SMILES, offering a powerful new tool for computational chemistry and related disciplines.
Similar content being viewed by others
Introduction
While deep learning has been widely explored in cheminformatics with significant progress1,2, its potential is severely limited by the scale of labeled data. To reduce the cost of data annotation while enabling generalizable, transferable, and robust representation learning from unlabeled data, researchers extend the pre-training strategies3 from images and texts to molecular data4,5,6,7,8.
To work with machine learning algorithms, molecules can be represented by a chemical notation language named SMILES9, which explicitly represents meaningful substructures such as branches and cyclic structures. To enable pre-training, researchers use the variant of Transformers to pre-train on large-scale unlabeled SMILES strings8,10,11. To pre-train a molecular language model, given the SMILES string of every molecule in the training dataset, existing methods usually adopt an masked autoencoding strategy8,10,11,12. It first randomly selects a subsequence of the SMILES string. Then the strategy masks the selected part and trains models to predict the masked part. However, such random masking strategy would ignore the key chemical substructures of molecules, such as rings and functional groups4,7. For instance, consider aspirin, which is denoted by “O=C(C)Oc1ccccc1C(=O)O”. In this molecular structure, critical functional groups such as the carboxylic acid (“-COOH”) and the ester (“-COO-”) are at risk of being overlooked due to random masking. This oversight neglects their pivotal contributions to the molecular activity and properties. To the end, these methods may fail to learn the critical molecular properties, which are primarily relevant to the chemical substructures of a molecule, from SMILES strings13,14.
Previous investigations have highlighted the limitations of SMILES notation in terms of topology awareness, underscoring its inability to explicitly encode the structural information of molecules15. Several works have attempted to incorporate better molecular structural or grammatical information from SMILES strings16,17. To address this issue, structure-aware pre-training methods utilizing Graph Neural Networks (GNNs) have marked significant advancements. These approaches leverage graph-format representations of molecules—such as the topological arrangement of atoms in a 2D space—to enrich learning models with a deeper understanding of molecular structures4,6,7. More recently, 3D graph-based molecular data representations have been introduced in GNN pre-training, where the three-dimensional structures of molecules are used to boost the performance of pre-trained models by incorporating detailed structural and topological information18,19. However, while topological information (2D connectivity) is readily derived from SMILES, obtaining precise 3D structural information (conformations) presents challenges. While experimental methods can determine the 3D positions of atoms and the angles between bonds, these approaches can be costly and time-consuming, especially for the vast datasets typically used in pre-training. Alternatively, some studies directly convert SMILES strings or 2D topology graphs into 3D graphs using computational tools like the Merck Molecular Force Field (MMFF94)20 function in RDKit18. Yet, such computationally derived 3D structures may not always yield precise conformational information and can introduce inaccuracies, particularly for flexible molecules or across diverse chemical spaces without specific parameterization.
On the other hand, recent efforts on SMILES-based molecular models have explored fragment-based encoding techniques to enhance molecular representation learning. For example, Aksamit et al.21 introduced a hybrid fragment-SMILES encoding method focused on improving ADMET prediction22 by adjusting fragment frequency thresholds in tokenization. This approach modifies the input representation by incorporating frequent molecular fragments into the tokenization process. While this method improves the performance of SMILES-based molecular models for ADMET prediction, it relies on predefined fragment frequencies and fragment libraries, which may limit its generalizability and effectiveness across other diverse molecular property prediction tasks. Additionally, their approach is primarily optimized subject to ADMET prediction and does not address the challenges of capturing structural information without altering the input representation, potentially restricting its applicability to a broader range of molecular modeling tasks. Given these challenges, including the difficulties in obtaining consistently accurate 3D conformations at scale for pre-training and the limitations of some fragment-based methods, it is reasonable to question the added value of such automatic data format conversions and the precision of converted 3D graphs. Thus, it remains challenging to pre-train a structure-aware molecular model when precise structural information is not readily available.
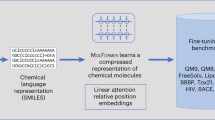
To tackle the aforementioned challenges, we propose a novel molecular representation framework MLM-FG – a SMILES-based Molecular Language Model, which randomly masking SMILES subsequences corresponding to specific molecular Functional Groups to incorporate structure information of atoms during the pre-training phase. Specifically, MLM-FG employs transformer-based models trained on a large corpus of SMILES strings for 100 million molecules. As shown in Fig. 1, given the SMILES string for every molecule in the training dataset, MLM-FG first parses the string and identify the subsequences corresponding to functional groups23 and key clusters of atoms in the molecules. Then MLM-FG randomly masks a certain proportion of subsequences and trains the model to predict the masked part as the pre-training task. Our work distinguishes itself from both fragment-based encoding methods and graph-based approaches utilizing GNNs. Compared to the hybrid fragment-SMILES encoding technique21, which modifies the input representation by incorporating frequent molecular fragments for ADMET prediction, our method maintains the standard SMILES syntax and introduces a novel pre-training strategy. By randomly masking functional groups within SMILES sequences during pre-training, our approach compels the model to learn to predict these substructures based on contextual information, enhancing its ability to capture structural relationships without altering the input representation. In contrast to graph-based methods4,6,7,18,19 that rely on explicit structural information represented in molecular graphs or 3D geometries—which may not always be precise or readily available–our approach effectively infers structural information implicitly from large-scale SMILES data, making it broadly applicable across various molecular property prediction tasks.
(1) MLM-FG adopts 12-layer multi-head transformer blocks (in either RoBERTa or MoLFormer architectures) with a hidden state dimension of Dh= 768 for pre-training and fine-tuning, (2) MLM-FG follows a functional group-aware random masking strategy to pre-train the model on a large corpus of 10 to 100 million SMILES sequences from PubChem, and (3) MLM-FG fine-tunes the pre-trained models to support a wide range of molecular machine learning applications.
Extensive experimental evaluations across 11 benchmark classification and regression tasks in the chemical domain demonstrate the robustness and superiority of MLM-FG. Our findings reveal that MLM-FG outperforms existing pre-training models, either based on SMILES or graphs, in 9 out of the 11 downstream tasks, ranking as a close second in the remaining ones. Remarkably, MLM-FG also surpasses 3D graph-based models, which explicitly incorporate molecular structures into their inputs, highlighting its exceptional capacity for representation learning even without explicit 3D structural information. These results show that pre-trained transformer encoders specialized in molecular SMILES demonstrate robust performance, matching or even exceeding existing supervised or unsupervised language models and GNN benchmarks in accurately forecasting a broad spectrum of molecular properties.
Results
In this section, we present a series of comprehensive experiments designed to illustrate the efficacy of MLM-FG. These experiments include performance comparisons across various downstream tasks and visual analysis of pre-trained representations. To assess the impact of model architecture and data size, we utilized two transformer-based models for pre-training on a corpus consisting of millions of molecules. These models are based on the MoLFormer8 and RoBERTa architectures.
Moreover, we conducted a comparative analysis of MLM-FG with models pre-trained using different strategies and methods derived from existing literature. These include models based on both molecular graphs, such as MolCLR4, GROVER24, and GEM18, and SMILES e.g., MoLFormer8. Notably, two recent works – GEM18, incorporating the explicit 3D structures of 20 million molecules in pre-training, and MoLFormer8, pre-trained using SMILES strings of 1.1 billion molecules, are two strong baselines in the line of research for molecular graph-based and SMILE-based solutions. Please note that the hybrid fragment-SMILES tokenization method by Aksamit et al.21 was not included in our experimental comparison. This exclusion is due to two key differences that make direct comparisons less meaningful in the context of our study: (1) their work focuses exclusively on ADMET prediction tasks, whereas our evaluation spans 11 diverse molecular property benchmarks, and (2) their hybrid fragment-SMILES encoding fundamentally alters the input sequence format by incorporating fragments into the tokenization process. In contrast, all approaches compared in our work, including MLM-FG, maintain standard SMILES notation as input, with our method applying random functional group masking directly to these standard SMILES strings. Given these differences in both evaluation scope and input representation, we focused our comparisons on existing models that align more closely with our methodology and objectives, which center on pre-training strategies for standard SMILES inputs.
Performance of MLM-FG on downstream tasks
Before fine-tuning MLM-FG to downstream tasks, we use 10 million, 20 million, and 100 million unlabeled molecules sampled from PubChem25, a public access database that contains purchasable drug-like compounds, to pre-train MLM-FG on two transformer-based models. Subsequently, we conduct experiments on multiple molecular benchmarks from the MoleculeNet26, including seven classification tasks and five regression tasks. Classification accuracy is reported as Area Under the Receiver Operating Characteristic Curve (AUC-ROC), and regression errors are reported as Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE). Following the previous work8,18,27, we adopt the scaffold split28 to split the datasets, which splits molecules based on their molecular substructure. By separating structurally distinct molecules into different subsets, scaffold splitting poses a more significant challenge and offers a robust test of model generalizability compared to random splitting methods. While conducting multiple runs with different random seeds or multiple random scaffold splits would be ideal for a more comprehensive robustness assessment, our current study adhered to the standard scaffold splits provided by MoleculeNet or utilized in the cited baseline papers. This approach ensures a fair and direct comparison with established reported results in the field.
We choose seven classification tasks from the MoleculeNet benchmark with six baseline models to evaluate and compare the performance of MLM-FG. Based on the experimental results shown in Table 1, MLM-FG, employing either MoLFormer or RoBERTa architectures, surpasses all of the baselines in five (BBBP, ClinTox, Tox21, HIV, and MUV) out of seven benchmarks and comes a close second in the other two (BACE and SIDER). For example, on ClinTox, MLM-FG (RoBERTa, 100M) achieves an AUC-ROC of 0.9606 compared to MoLFormer’s 0.9451, and on MUV, it achieves 0.7990 versus MoLFormer’s 0.7599. This result demonstrates the superiority of MLM-FG in dealing with the prediction of molecular properties; especially compared within SMILES-based solutions, MLM-FG delivers high classification accuracy. GEM outperforms MLM-FG in BACE and SIDER datasets, which could be attributed to its utilization of explicit 3D structural information of molecules.
We choose five regression tasks from the MoleculeNet benchmark with six baseline models to evaluate the performance of MLM-FG. Based on the experimental results shown in Table 2, we can conclude that MLM-FG exceeds the performance of all baselines in four out of five benchmarks (ESOL, FreeSolv, Lipo, and QM7) and attains comparable results on the QM8 dataset. Especially, MLM-FG showcases notable performance gains over the second-best model on ESOL dataset with 41.01% improvement. In particular, the QM8 dataset involve prediction of several quantum-chemical measures, which is considered challenging without 3D information8. While GEM and MoLFormer lead MLM-FG on the QM8 dataset, the performance of MLM-FG is still noteworthy given that GEM incorporates explicit 3D information and MoLFormer was pre-trained on a dataset ten times larger than ours (1.1 billion vs. 100 million molecules). The quantitative differences should be viewed in this context.
We hypothesize that MLM-FG’s advantage is more pronounced on datasets where the target molecular properties are strongly influenced by the presence, absence, or specific arrangement of well-defined functional groups. In such cases, explicitly training the model to recognize and understand the context of these FGs, as MLM-FG does, provides a more direct learning signal. For instance, properties like toxicity (ClinTox, Tox21 in Table 1) or biological activity (HIV, MUV in Table 1) are often linked to specific pharmacophoric FGs, potentially explaining MLM-FG’s strong performance on these tasks.
Ablation study
To comprehensively evaluate the effectiveness of MLM-FG, we conducted ablation studies to dissect the contribution of key treatments, including pre-training strategies, model architectures, and the size of pre-training datasets, to the overall performance.
The comparison between the functional group-based random masking, random subsequence masking, and training from scratch, underscores the effectiveness of the unique masking approach proposed by MLM-FG. Notably, MLM-FG demonstrated a significant performance improvement, for instance, achieving error reductions in the ESOL dataset to 0.3432 using functional group-based masking, compared to 0.4909 with random subsequence masking and 0.9721 when trained from scratch. Moreover, we can observe a clear performance improvement from the vanilla MoLFormer to the MoLFormer models pre-trained by MLM-FG in the most datasets for both classification and regression tasks. This observation confirms the advantage of the proposed functional group-aware random masking strategy, even with less molecules for pre-training. In addition, the comparisons between MoLFormer and RoBERTa models highlight a distinct advantage for the more extensive RoBERTa model in the most cases. For instance, in the regression tasks such as FreeSolv, RoBERTa models pre-trained by MLM-FG posted superior results (e.g., error of 1.7430 for the RoBERTa 10M) when compared to the MLM-FG pre-trained MoLFormer models under similar conditions (error of 5.5461 for MoLFormer 10M).
Expanding the dataset size typically leads to improved performance for MLM-FG, as demonstrated across various benchmarks. For example, RoBERTa models pre-trained with MLM-FG achieve an accuracy of 0.6103 on the MUV dataset when utilizing 10 million molecules for pre-training. Meanwhile, this accuracy increases to 0.6428 with a dataset size of 20 million molecules and further rises to 0.7990 when leveraging the entire 100 million molecules for pre-training. Moreover, training with just 10 million molecules on the ESOL dataset yields an error of 0.3432, which represents a significant improvement over MoLFormer models trained from scratch (0.9721). However, as seen with the MLM-FG (MoLFormer) variants on ESOL, on expanding the training set to 20 million and 100 million molecules, we see a performance dip (0.4407 and 0.5135, respectively). This phenomenon, where more data does not monotonically improve performance, can be attributed to factors such as those discussed by Nakkiran et al.29, including potential overfitting to idiosyncrasies in larger pre-training corpora or fixed model capacity. It might also indicate inefficiencies in handling larger datasets without fine-tuning the methodology accordingly. Thus, while increased dataset sizes generally improve the accuracy of MLM-FG, the specific nature of the data, the model architecture, the pre-training techniques, and their interactions also play a critical role in extracting the maximal benefit from larger datasets.
Pre-trained representations visualization
The pre-training representation visualization results provide comprehensive insights into the learned molecular representations by MLM-FG.
Our first visualization analysis intends to connect the weights of the molecules and the distribution of their learned representations. These representations, extracted from the downstream datasets without fine-tuning, encompass 312,879 unique molecules. As shown in Fig. 2, our experiment maps these presentations onto a 2D space using UMAP30, where each point in the visualization is color-coded based on its corresponding molecular weight (g/mol). It can be seen in Fig. 2 that even without task-specific fine-tuning, MLM-FG is capable of distinguishing between light-weighted and heavily-weighted molecules, indicating that the pre-training representation of MLM-FG has successfully captured molecular property information. While other pre-training methods also show some separation by molecular weight in their raw pre-trained embeddings, the key impact of MLM-FG’s functional group-aware masking may lie in learning more nuanced structural relationships relevant for specific downstream tasks, which become more evident after fine-tuning. The improved performance on these tasks (Tables 1 and 2) is the primary validation of this benefit.
Yet another visualization study has been conducted to analyze the 2D graph and 3D geometric information content in the pre-training representation of MLM-FG. This study specifically focused on the comparisons between SMILES-based transformer models, including the standard MoLFormer as well as its variants and RoBERTa pre-trained using MLM-FG. For each model, given a SMILES string as the input, we extracted attention vectors for every atomic token, analyzed attentions across different atomic token pairs, and constructed attention matrices for these atoms. We subsequently compare these attention matrices with the corresponding matrices representing covalent bond connectivity and 3D distances between atom pairs. Figure 3 showcases the adjacency matrix, 3D distance matrix, and attention matrices for the molecule “CP(Br)C=O”. It shows that the attention matrices obtained by MLM-FG show a notable correlation with the 3D distance matrix of the molecule, especially when comparing to MoLFormer and MoLFormer/RoBERTa’s variants pre-trained by other strategies. This suggests an enhanced structural awareness. The domain-aware nature of MLM-FG, stemming from its training to predict functional groups, likely guides it to learn attention patterns that better reflect these spatial relationships. Table 3 presents the results for the cosine similarity between the 3D distance matrices and attention matrices, averaged over 50,000 molecules, showcasing the performance of various models. Compared to MoLFormer, MoLFormer trained from scratch, and RoBERTa trained from scratch, MLM-FG with both MoLFormer and RoBERTa architectures pre-trained for 100M steps exhibits superior correlation in capturing the relationship between attention matrices and 3D distance matrices, an empirical finding further supporting its ability to learn structurally relevant representations.
Discussion
This work proposes the MLM-FG framework that incorporates functional groups – as prior information on molecular structures – and leverages a functional group-aware random masking strategy to pre-train molecular language models on large-scale SMILES databases, yielding enhanced performance and generalization capability for downstream tasks. Our model has been evaluated across 12 datasets, including 7 classification and 5 regression tasks, outperforming the existing state-of-the-art models in 9 of these datasets. Notably, 5 of these datasets involve predictions on multiple sub-tasks of molecular properties, with up to 27 sub-tasks. We also validate the impact of data scale and model type, pre-training on MoLFormer and RoBERTa models across datasets of 10 million, 20 million, and 100 million molecules, followed by an analysis of downstream task performance. We employe AUC, RMSE, and MAE metrics to ensure a fair comparison among molecule analysis methods. Currently, there is limited research on leveraging Transformer architectures with added external knowledge for molecule data analysis. As a publicly available tool, MLM-FG offers a powerful resource for molecule analysis and further advanced applications.
Several key findings of this work could be summarized as follows.
-
Compared to 2D/3D molecular graphs, SMILES strings lack of explicit structural information and are with limited topological awareness. In the meanwhile, the proposed MLM-FG framework overcomes these limitations by incorporating functional group-aware random masking during pre-training, which enables implicit learning of structural features and functional group interactions from SMILES data, ultimately leading to more accurate predictions of molecular properties.
-
The functional group-aware random masking strategy proposed by MLM-FG demonstrates a significant performance improvement compared to masking strategies used in MoLFormer, random subsequence masking, and training from scratch. Furthermore, our analysis indicates that the attention patterns between atomic tokens extracted from MLM-FG show a notable correlation with the actual 3D distances between atoms, suggesting a more accurate representation of molecular structures.
-
It has been observed that leveraging a larger model like RoBERTa or pre-training with a larger volume of data typically results in enhanced performance in downstream tasks, especially in the experiments for classification tasks. However, it is important to note that in many scenarios, employing larger models with more data may actually hurt the performance29.
The MLM-FG model represents a significant advancement in molecular modeling by capturing essential structural information through functional group-aware masking within SMILES strings. This capability enhances the prediction of molecular properties and may aid in understanding of structure-activity relationships, making it a valuable tool across drug discovery and metabolomics studies. In drug discovery, MLM-FG can be applied to virtual screening of large compound libraries to identify potential drug candidates, help prioritize compounds that are more likely to interact with specific biological targets, and aid in optimizing design of lead compounds. Additionally, MLM-FG may facilitate drug repurposing by screening existing drugs for new therapeutic targets, broadening the utility of known compounds. In metabolomics studies, MLM-FG may help identify unknown metabolites, providing valuable insights into metabolic processes and potential therapeutic targets. Overall, MLM-FG emerges as a transformative tool in computational chemistry with broad implications and applications across the life sciences. Integrating MLM-FG into various research workflows can accelerate innovation and yield more efficient and targeted outcomes in their respective fields.
The MLM-FG model, despite its innovations in molecule analysis, confronts several challenges. Firstly, it cannot model very long SMILES sequences. Those exceeding 512 tokens are truncated (less than 0.01% in the pre-training datasets), potentially leading to loss of information. While recent decoder-only language models have addressed long-context training and inference31,32, applying these techniques to molecular language models remains a key area for our future work. However, please note that earlier works, such as MoLFormer8, set 202 as the token limit, which still delivered reasonable performance. Secondly, our focus is on molecular modeling, limiting our ability to extend to predicting chemical reactions between molecules and molecular generation. Additionally, the performance of MLM-FG could be further improved through incorporating 3D information of molecules in pre-training, fine-tuning, and testing. Moreover, data re-sampling of pre-training datasets and advanced fine-tuning strategies could enhance MLM-FG in downstream tasks. The specific masking ratios used in our functional group-aware strategy are hyperparameters, and their optimization could lead to further performance gains; this remains an avenue for future work. Another potential challenge relates to the definition and identification of functional groups; while RDKit provides a standardized approach, ambiguities in FG definitions (e.g., in complex conjugated systems or overlapping SMARTS patterns) could introduce noise or complexity for the model. Our future work would address these issues for potential performance enhancements.
Methods
This section provides a comprehensive overview of the design features associated with each component of MLM-FG. We introduce the model architecture of MLM-FG and the pre-training strategy.
Model architectures
In this work, we present MLM-FG, an approach for large-scale pre-training of molecules based on the Transformer blocks, which incorporates multi-layer and multi-head transformer blocks. Specifically, MLM-FG offers the same architectural configuration as the one shared by MoLFormer and RoBERTa, employing a 12-layer transformer and a hidden state dimension of Dh = 768. Consider an input SMILES sequence denoted as s = (s1, s2, …, sL), where L represents the length of the sequence. MLM-FG first tokenizes the sequence and subsequently feeds them into the transformer. This process enables us to extract token embeddings \({\boldsymbol{h}}=({h}_{1},{h}_{2},\ldots ,{h}_{l})\in {{\mathcal{R}}}^{L\times {D}_{h}}\), where Dh represents the dimension of the hidden representations for the tokens. Then the model takes the series of token embeddings as input and transform them into a lower-dimentional vector to output the embedding of the SMILES sequence. The total number of trainable parameters in MLM-FG (MoLFormer) is approximately 48.1M and MLM-FG (RoBERTa) is approximately 93.8M.
Pre-training datasets
Following many other pre-training based approaches, ours MLM-FG is structured into two main phases: pre-training and fine-tuning. During the pre-training phase, which is not tailored to any particular task, MLM-FG is trained on a vast corpus of range from 10 million to 100 million SMILES sequences sampled from PubChem25. These molecules were obtained via random sampling from the PubChem database and were not explicitly stratified by property or structure, beyond standard filtering common for creating such pre-training sets. Summary statistics of these pre-training datasets (e.g., molecular weight distributions, atom type frequencies) and FG coverage statistics have been provided in the Supplementary Tables. The self-supervised training phase enables MLM-FG to discern sequential distributions and substructures in molecule sequences, thereby gaining a holistic grasp of their structural and functional insights.
Functional group-aware pre-training strategy
Pre-training strategies in molecular representation learning highly correlate with molecule formats. For pre-training with unlabeled data, the prevalent approach involves reconstructing randomly masked tokens in SMILES strings. Given that molecules with similar structures may have vastly different properties, this method might overlook the complex interrelations among molecular features and potentially distort molecular semantics. Our objective is to weave chemical domain knowledge, specifically regarding substructures, into the pre-training process.
Rather than randomly masking subsequences or tokens in SMILES, we mask the cluster of tokens in SMILES that represent these substructures. During the pre-training phase, we start by identifying the substructures that correspond to specific molecular functional groups with RDKit18, which uses a predefined, rule-based set of SMARTS patterns for this identification. A list of commonly encountered functional groups or references to RDKit’s definitions have been provided in the Supplementary Information. Then we randomly select a subset of these identified substructures, followed by masking the associated tokens within these substructures. If multiple functional groups are candidates for masking based on the rules below, the specific group(s) to be masked are chosen randomly, without a predefined priority. Based on the count of these functional groups within a molecule, our masking strategy adaptively adjusts. If a molecule does not contain any functional groups, atom masking is employed as the default strategy. This ensures that the model still learns general structural aspects of molecules lacking specific functional groups. For molecules with fewer than 10 functional groups, we mask only one functional group. This approach is designed to preserve the overall structural integrity of the molecule while still introducing the model to the complexity of functional groups. In cases where a molecule contains more than 10 functional groups, we randomly mask 10% of these groups. This strategy introduces a higher level of complexity and variability, challenging the model to better generalize its learning across a more diverse set of molecular substructures. This adaptive strategy, based on functional group counts, aims to provide a consistent yet challenging learning signal across diverse molecules. It leverages general chemical knowledge (the importance of FGs) rather than introducing detrimental dataset-specific biases, with the randomness in selection promoting generalization.
The model leverages self-supervised learning to predict masked atoms, thereby acquiring structural information about molecules. This methodical selection and masking process is instrumental in guiding the model to understand and predict the underlying structural characteristics of molecules, enhancing its ability to infer molecular properties and functionalities based on structural cues. By integrating domain knowledge about molecular substructures into our pre-training strategy, we enable the model to develop a more nuanced and accurate representation of molecular structures, paving the way for more effective learning and prediction in downstream tasks.
Setups of MLM-FG in experiments
MLM-FG approach gives rise to several model variants distinguished primarily by their underlying architecture and the size of the pre-training dataset. This section introduces the key variants leveraged in our experiments, which include MLM-FG (MoLFormer) and MLM-FG (RoBERTa). Specifically, MLM-FG (MoLFormer) utilizes the MoLFormer architecture designed to capture complex molecular representations using rotary positional embeddings and an efficient linear attention mechanism. The MoLFormer models were pre-trained using three different dataset sizes: 10 million, 20 million, and 100 million molecules. These variations allow for an understanding of how the scale of the pre-training data impacts the effectiveness of model embeddings on downstream tasks, such as molecular property prediction. In addition, MLM-FG (RoBERTa) builds upon the RoBERTa architecture, renowned for its robustness in handling masked language modeling tasks due to its bidirectional encoder representations. Similarly to MoLFormer, RoBERTa models were pre-trained on datasets of 10 million, 20 million, and 100 million molecules. These multiple pre-training data scales enable evaluations of the RoBERTa model’s performance adaptability and efficiency when applied to different molecular prediction applications.
Both variants of MLM-FG, based MoLFormer and RoBERTa transformers, are instrumental in drawing comparative insights between different transformer-based architectures and dataset sizes. The insights gained from these variants help delineate the potential benefits and limitations inherent in each architecture, fostering an advanced understanding of their applicability within molecular informatics.
Setups of baseline methods for comparisons
The models we are comparing against are based on cutting-edge methodologies derived from contemporary literature. These methods are as follows. MolCLR-gin and MolCLR-gcn are two 2D molecular graph-based models designed to leverage molecular graphs. They are equipped with distinct features focusing on graph-based learning paradigms and trained on 10 million unlabeled molecules. The total number of trainable parameters in MolCLR-gin is approximately 2.2M and in MolCLR-gcn is approximately 0.8M. In addition, GEM is A 3D molecular graph-based model built upon a geometry-based approach. It incorporates innovative strategies based on molecular geometry in a 3D space for pre-training on 20 million unlabeled molecules. The total number of trainable parameters in GEM is approximately 0.107M. Furthermore, GROVER-base and GROVER-large are two methods that integrate Message Passing Networks into the Transformer-style architecture. They predict contextual properties based on atomic embeddings, encoding contextual information into node embeddings. The dataset for GROVER pre-training includes 10 million molecules. The total number of trainable parameters in GROVER-base is approximately 48M and in GROVER-large is approximately 100M. Finally, MoLFormer is another model based on SMILES representations. This model employs pre-trained representations to capture molecular information encoded as SMILES strings. The pre-training dataset contains 1.1 billion molecules, and the total number of trainable parameters in MoLFormer is approximately 48.1M.
These methods are included for comparison due to their representation of state-of-the-art molecular modeling techniques, each offering distinct advantages. MolCLR-gin and MolCLR-gcn focus on 2D graph representations, GEM provides a 3D approach, GROVER integrates Message Passing Networks with Transformer architectures for contextual analysis, and MoLFormer utilizes SMILES representations with extensive pre-training. Comparing against these varied advanced models allows a comprehensive evaluation of our proposed models’ effectiveness and improvements in predictive accuracy.
Hyper-parameters and training details
In the MLM-FG, both MoLFormer and RoBERTa comprise 12 layers, each equipped with 12 attention heads. For pre-training, we initialized with a learning rate of 3 × 10−5, gradually reducing it using a LambdaLR scheduler, and utilized the AdamW optimizer with a batch size of 1024 across 16 NVIDIA V100 GPUs. We conducted 50 epochs for datasets of 10M and 20M molecules and reduced the epoch count to 20 for the 100M dataset to balance computational demands and training depth. For the 100M dataset, one epoch of pre-training requires 6 hours and utilizes 16 GPU. In comparison, the 20M dataset takes 1.5 h, and the 10M dataset takes 0.7 hours under the same settings. In the fine-tuning phase, we maintained the learning rate at 3 × 10−5 but switched to the FusedLAMB optimizer for better efficiency, with a smaller batch size of 64 to ensure precise model adjustments tailored to specific tasks.
Data availability
The datasets used for pre-training and fine-tuning are derived from previous studies. These datasets are publicly available via download links as follows. - PubChem: https://pubchem.ncbi.nlm.nih.gov/- QM8: https://moleculenet.org/datasets-1- ESOL: https://moleculenet.org/datasets-1- FreeSolv: https://moleculenet.org/datasets-1- MUV: https://moleculenet.org/datasets-1- BBBP: https://moleculenet.org/datasets-1- BACE: https://moleculenet.org/datasets-1- ClinTox: https://moleculenet.org/datasets-1- Tox21: https://moleculenet.org/datasets-1- SIDER: https://moleculenet.org/datasets-1- HIV: https://moleculenet.org/datasets-1.
Code availability
We built MLM-FG using Python and PyTorch. The code repository of MLM-FG, readme files and tutorials are all available at https://github.com/Tianhao-Peng/MLM-FG. The checkpoints of pre-trained models are available for download at https://drive.google.com/drive/folders/16vOW0rzMJJAC0iNFbzb6E_40yQ3lbLaF.
References
Huang, B. & Von Lilienfeld, O. A. Communication: understanding molecular representations in machine learning: the role of uniqueness and target similarity. J. Chem. Phys. 145, 161102-1–161102-6 (2016).
David, L., Thakkar, A., Mercado, R. & Engkvist, O. Molecular representations in ai-driven drug discovery: a review and practical guide. J. Cheminform. 12, 56 (2020).
Devlin, J., Chang, M., Lee, K. & Toutanova, K. BERT: pre-training of deep bidirectional transformers for language understanding. In: Burstein, J., Doran, C. & Solorio, T. (eds.) Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, NAACL-HLT 2019, Minneapolis, MN, USA, June 2-7, 2019, Volume 1 (Long and Short Papers), 4171–4186 https://doi.org/10.18653/v1/n19-1423 (Association for Computational Linguistics, 2019).
Wang, Y., Wang, J., Cao, Z. & Farimani, A. B. Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4, 279–287 (2022).
Zhu, J. et al. Dual-view molecule pre-training. In KDD '23: Proc. 29th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, 3615–3627, https://arxiv.org/abs/2106.10234 (2023).
Li, P. et al. Learn molecular representations from large-scale unlabeled molecules for drug discovery. CoRR abs/2012.11175 https://arxiv.org/abs/2012.11175 (2020).
Rong, Y. et al. Self-supervised graph transformer on large-scale molecular data. In: Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M. & Lin, H. (eds.) Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems 2020, NeurIPS 2020, December 6-12, 2020, virtual https://proceedings.neurips.cc/paper/2020/hash/94aef38441efa3380a3bed3faf1f9d5d-Abstract.html (2020).
Ross, J. et al. Large-scale chemical language representations capture molecular structure and properties. Nat. Mach. Intell. 4, 1256–1264 (2022).
Weininger, D. Smiles, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inform. Comput. Sci. 28, 31–36 (1988).
Wang, S., Guo, Y., Wang, Y., Sun, H. & Huang, J. Smiles-bert: large scale unsupervised pre-training for molecular property prediction. In Proc. 10th ACM International conference on bioinformatics, computational biology and health informatics, 429–436 (Association for Computing Machinery (ACM), 2019).
Broberg, J., Bånkestad, M. M. & Hellqvist, E. Y. Pre-training transformers for molecular property prediction using reaction prediction. In ICML 2022 2nd AI for Science Workshop (2022).
Irwin, R., Dimitriadis, S., He, J. & Bjerrum, E. J. Chemformer: a pre-trained transformer for computational chemistry. Machine Learning: Science and Technology 3, 015022 (2022).
Zhang, Z., Liu, Q., Wang, H., Lu, C. & Lee, C. Motif-based graph self-supervised learning for molecular property prediction. In Advances in Neural Information Processing Systems 34: Annual Conference on Neural Information Processing Systems 2021, NeurIPS 2021, (eds Ranzato, M. et al.) December 6-14, 2021, virtual, 15870–15882 (Neural Information Processing Systems Foundation, Inc, 2021).
Sun, M., Xing, J., Wang, H., Chen, B. & Zhou, J. Mocl: Contrastive learning on molecular graphs with multi-level domain knowledge. CoRR https://arxiv.org/abs/2106.04509 (2021).
Zhang, S. et al. Applications of transformer-based language models in bioinformatics: a survey. Bioinform. Adv. 3, vbad001 (2023).
Zhang, K., Mann, V. & Venkatasubramanian, V. G-matt: single-step retrosynthesis prediction using molecular grammar tree transformer. AIChE J. 70, e18244 (2024).
Mann, V. & Venkatasubramanian, V. Predicting chemical reaction outcomes: a grammar ontology-based transformer framework. AIChE J. 67, e17190 (2021).
Fang, X. et al. Geometry-enhanced molecular representation learning for property prediction. Nat. Mach. Intell. 4, 127–134 (2022).
Atz, K., Grisoni, F. & Schneider, G. Geometric deep learning on molecular representations. Nat. Mach. Intell. 3, 1023–1032 (2021).
Halgren, T. A. Merck molecular force field. i. basis, form, scope, parameterization, and performance of mmff94. J. Comput. Chem. 17, 490–519 (1996).
Aksamit, N., Tchagang, A., Li, Y. & Ombuki-Berman, B. Hybrid fragment-smiles tokenization for admet prediction in drug discovery. BMC Bioinform. 25, 255 (2024).
Norinder, U. & Bergström, C. A. Prediction of admet properties. ChemMedChem: Chemistry Enabling Drug Discovery 1, 920–937 (2006).
Nic, M. et al. International union of pure and applied chemistry compendium of chemical terminology (the Gold Book). Research Triangle Park, NC: IUPAC. https://doi.org/10.1351/goldbook (2019).
Rong, Y. et al. Grover: self-supervised message passing transformer on large-scale molecular data. arXiv preprint arXiv:2007.02835 2, 17 (2020).
Kim, S. et al. Pubchem 2019 update: improved access to chemical data. Nucleic Acids Res. 47, D1102–D1109 (2019).
Wu, Z. et al. Moleculenet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018).
Hu, W. et al. Strategies for pre-training graph neural networks. In International Conference on Learning Representations (ICLR 2020).
Ramsundar, B., Eastman, P., Walters, P. & Pande, V. Deep learning for the life sciences: applying deep learning to genomics, microscopy, drug discovery, and more (“O’Reilly Media, Inc.”, 2019).
Nakkiran, P. et al. Deep double descent: Where bigger models and more data hurt. J. Stat. Mech. Theory Exp. 2021, 124003 (2021).
McInnes, L., Healy, J., Saul, N. & Großberger, L. Umap: uniform manifold approximation and projection. J. Open Sour. Softw. 3 (2018).
An, S. et al. Make your llm fully utilize the context. Adv. Neural Inform. Process. Syst. 37, 62160–62188 (2024).
Tang, J. et al. Quest: query-aware sparsity for efficient long-context llm inference. In International Conference on Machine Learning, 47901–47911 (PMLR, 2024).
Acknowledgements
The authors would like to thank the anonymous reviewers for their constructive feedback which helped improve the manuscript. The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors have made contributions in this paper. T.P. designed studies conducted experiments and wrote part of the manuscript. Y.L., X.L., J.B., Z.X., N.S., S.M., and Y.X. involved in the discussion and wrote part of the manuscript. L. K. oversaw the research progress, involved in the discussion and wrote part of the manuscript. H. X. oversaw the research progress, designed the study and experiments, involved in the discussion, and wrote the manuscript. T.P. and H.X. made the equal technical contributions to this work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Peng, T., Li, Y., Li, X. et al. Pre-trained molecular language models with random functional group masking. npj Artif. Intell. 1, 28 (2025). https://doi.org/10.1038/s44387-025-00029-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44387-025-00029-3