Abstract
Recent work from our labs demonstrated that a metabolite(s) from the soil bacterium Streptomyces venezuelae caused dopaminergic neurodegeneration in Caenorhabditis elegans and human neuroblastoma cells. To evaluate the capacity for metabolite production by naturally occurring streptomycetes in Alabama soils, Streptomyces were isolated from soils under different land uses (agriculture, undeveloped, and urban). More isolates were obtained from agricultural than undeveloped soils; there was no significant difference in the number of isolates from urban soils. The genomic diversity of the isolates was extremely high, with only 112 of the 1509 isolates considered clones. A subset was examined for dopaminergic neurodegeneration in the previously established C. elegans model; 28.3% of the tested Streptomyces spp. caused dopaminergic neurons to degenerate. Notably, the Streptomyces spp. isolates from agricultural soils showed more individual neuron damage than isolates from undeveloped or urban soils. These results suggest a common environmental toxicant(s) within the Streptomyces genus that causes dopaminergic neurodegeneration. It could also provide a possible explanation for diseases such as Parkinson’s disease (PD), which is widely accepted to have both genetic and environmental factors.
Similar content being viewed by others
Introduction
Members of the filamentous bacterial genus Streptomyces are ubiquitous in soils and are well known producers of medically and agriculturally useful secondary metabolites. These nonessential metabolites are thought to promote the survival of the producing organism but are not directly involved in its growth, development, or reproduction, and include compounds such as antibiotics and toxins. Several classes of toxins produced by Streptomyces spp., including proteasome inhibitors (reviewed by)1 and mitochondrial complex I inhibitors2,3, have been recently receiving additional attention due to their potential role as environmental toxicants in human diseases4,5,6,7.
A metabolite(s) produced by Streptomyces venezuelae caused age- and dose-dependent degeneration of all neurons in the nematode model organism, Caenorhabditis elegans, with dopaminergic neurons selectively degenerating faster. Human SH-SY5Y neuroblastoma cells also degenerated in culture upon dose-dependent exposures5. The S. venezuelae metabolite caused cell death, in part, through decreased ATP production, modulation of mitochondrial complex I, and increased ROS7,8. We further determined that the metabolite induces disruptions in protein homeostasis, glutathione-tractable α-synuclein toxicity, and ubiquitin proteasome system activity. These activities are epistatically regulated by loss-of-function of the PARK6 homologue, pink-18. These phenotypes were not exclusive to neurons, but occurred in all cells of the worm. However, because of the high energy requirements of neurons and muscle cells, obvious phenotypes were observed in these cell types5,7,8.
These data suggest that mitochondrial dysfunction from environmental exposure to Streptomyces metabolites may be a factor in cellular toxicity. The previous studies focused on a single species of Streptomyces (S. venezuelae). Here, an evaluation of naturally occurring strains was performed to assess whether soils supported robust streptomycete communities with the capability to cause dopaminergic neurodegeneration.
The degeneration of dopaminergic neurons is a hallmark of Parkinson Disease (PD), the second most prevalent neurodegenerative disorder. PD affects more than 1% of the population over age 65, increasing to 4-5% in people over age 85. Familial PD represents only a small percentage (~10%) of known cases, with the remaining disease thought to be due to a combination of interactions among environmental and intrinsic genetic factors. Twin studies indicated that environmental influences are critical to disease onset and appear pivotal to sporadic causality9. A greater incidence of PD has been associated with a rural lifestyle and farming as a profession10,11. Exposure to insecticides (e.g., rotenone)12,13,14,15,16,17, fungicides (e.g., maneb)15,16,18 and herbicides (e.g., paraquat)15,16,17,18,19 have been linked to PD; however, these exposures do not account for all associations between environmental risk and disease10,20. We hypothesize that exposure to bacterial metabolites produced by soil streptomycetes could be an environmental factor associated with neurodegeneration.
Soils harbor large and diverse microbial communities that can be affected by a variety of environmental conditions. Numerous studies have shown that substantial changes to the chemical composition of soil, often due to human use of the land, impact the resident microbial communities (e.g.,)21,22,23,24. Because PD is more prevalent in individuals with rural lifestyles, we wanted to evaluate the impact of land use on the diversity of resident soil streptomycetes and assess their ability to produce neurodegenerative metabolite(s). Soil properties, including pH, texture, and organic content, were measured for each sample to determine their impact on the diversity of Streptomyces present. A subset of the 1509 Streptomyces isolates recovered from soils under three land uses (agricultural, undeveloped, urban) were evaluated for their ability to cause dopaminergic neurodegeneration in C. elegans. We have previously established the utility of C. elegans for evaluation of genetic and environmental factors that impact dopaminergic neurodegeneration25,26,27,28; these worm models have been correlative to results obtained in mammalian systems29. Additionally, BOX-PCR, a fingerprinting method targeting repetitive, intergenic bacterial sequences, was used to assess the genomic diversity of all isolates. Our data showed that in natural populations, 28.3% (51/180) of genomically unique soil Streptomyces spp. isolated from diverse land uses in Alabama caused C. elegans dopaminergic neurons to degenerate. These results suggest that there could be a common environmental toxicant(s) within the Streptomyces genus that causes dopaminergic neurodegeneration.
Results
Differences in isolation of Streptomyces by land use and physiography
Samples were collected from soils under different land uses (e.g., agriculture, undeveloped, urban) across the state of Alabama (Fig. 1). For study purposes, agricultural soils were collected from lands used for cultivation of a plant species for consumption by humans or livestock, or retail. Land that supported substantial plant growth and appeared unused for any purpose was considered undeveloped. The presence of multiple tree species and significant undergrowth indicated that the land was not currently being utilized for agricultural purposes, nor had it been in recent years. Developed, or urban, lands included property near human populations, exclusive of lawns, which were likely to be impacted by pollution through exhaust and/or industrial applications. To represent the topographical and geologic diversity of the state, which affects the overlying soils, agricultural, undeveloped, and urban soil samples were collected and examined from each of the major physiographic provinces within Alabama (Fig. 2). Physiographic provinces are areas characterized by terrain texture, rock type and geologic structure with specific geomorphology or landforms different from adjacent provinces; Alabama lies at the confluence of five physiographic provinces (Coastal Plains, Piedmont Upland, Valley and Ridge, Cumberland Plateau, Interior Low Plateau). The Black Belt Prairies are a section of the Coastal Plains province. In total, 85 soil samples were obtained, with 27 from agricultural uses, 33 from undeveloped areas, and 25 from urban environments (Table 1).
(A). Agricultural soils were collected from between rows of crops, such as from this cotton field. (B). Soils from undeveloped lands were also collected; samples taken from these locations usually required the removal of leaf litter prior to soil sampling. (C). Urban soils originated from small areas of land likely to be impacted by human activities and pollution.
Cultivation of Streptomyces spp. from soils under different land uses varied, with a significant difference found in the number of isolates obtained between agricultural and undeveloped soils (one-way analysis of variance [ANOVA] with Tukey’s Honestly Significant Difference [HSD], p < 0.01; Fig. 3A). No significant differences in the isolation of Streptomyces spp. were found across physiographic provinces (p=0.905; Table 1). A total of 1509 isolates from 85 unique soil samples were characterized in this study, with an estimated range of 1.8 × 106 to 4.2 × 108 streptomycetes per gram of soil (Supplemental Table 1). On average, 24.1 ± 10 isolates were recovered from 0.25 g of soil used for agriculture compared to 15.1 ± 9.2 and 20.0 ± 9.1 isolates from 0.25 g of undeveloped and urban soils, respectively. There were no significant differences in isolation of streptomycetes among the 3 cultivation media employed (mannitol-soy, starch casein, and an environment specific agar; Chi-square, p = 0.43). The amount of organic matter (one-way ANOVA following a log conversion with Tukey’s HSD, p < 0.01; Fig. 3B) in the soils and soil pH (Kruskal-Wallis, p < 0.01) were significantly different across land use. A significant but weak positive correlation also existed between soil pH and isolation of Streptomyces (p < 0.05; r = 0.259). Soil pH differed significantly between agricultural and urban soils (Mann-Whitney, p < 0.01) and between undeveloped and urban soils (Mann-Whitney, p < 0.01). Unsurprisingly, across all land use patterns, soil pH significantly correlated with the amount of soil organic matter (SOM; Spearman’s correlation, p < 0.05). The greatest difference in SOM was found between agricultural and urban soils (Fig. 3B). No significant differences were found between land use and soil texture based on sand, silt or clay content (one-way ANOVA following a log conversion).
(A). A comparison of the average number of isolates obtained from dilutions of 0.25 g of agricultural (AG), undeveloped (UD) and urban (UR) soils. Statistical significance (p < 0.01) is shown between letters (ANOVA with Tukey’s HSD). Columns with the same letter are not significantly different from one another. Bars indicate the standard error of the mean (B). Organic matter content (mg/g) comparison among soils from AG, UD, and UR land uses. Statistical significance (p < 0.01) is shown between letters (ANOVA following a log conversion with Tukey’s HSD).
Genomic diversity of Streptomyces isolates
Within dendrograms based on the similarity of genomic banding patterns generated during BOX PCR, isolates from the various land uses did not cluster (data not shown). Analysis of composite fingerprint patterns for each soil also showed no significant clustering by land use (Analysis of Similarity [ANOSIM], p > 0.05; Fig. 4). Based on banding patterns, of the 1509 isolates, only 112 (7.4%) were considered clones of other isolates. Thus, Streptomyces spp. possessing diverse genomes were recovered from soils exhibiting a broad range of pH values, organic matter content, and textures.
C. elegans exposure to Streptomyces spp. supernatants
Streptomyces isolates cultured from soils under each land use (agricultural, undeveloped, urban) were selected for analysis in the C. elegans dopaminergic model, with the locations representing all of the physiographic provinces within the state. The isolates from these locations were inoculated into SYZ broth for growth, which is conducive for the production of secondary metabolites. However, approximately half of the Streptomyces spp. inoculated did not grow in this medium, possibly due to the salt concentration. This limitation impacted both the number of isolates that we were able to test in the neurodegeneration assay and the physiographic distribution of the tested isolates. However, sixty isolates per land use were successfully grown and the filtered supernatants used for assessing neurodegeneration in a C. elegans model system. Of the isolates tested, 28.3% (51 of the 180 strains) were capable of producing neurodegeneration in the dopaminergic neurons of C. elegans (one-way ANOVA followed by a Bonferroni correction, p < 0.05). There was variability in the distribution of compound-producing Streptomyces across land uses (Fig. 5). Specifically, there were more isolates with neurodegenerative activity from agricultural and undeveloped soils compared to urban soils (Fisher’s exact test, p < 0.01). Similar percentages of compound producing strains were observed in agricultural (38.3%; 23 of 60 isolates) and undeveloped (35.0%; 21 of 60) soils while urban soils yielded a lower percentage of compound-producing strains (11.6%; 7 of 60). While it would be intriguing to further differentiate the neurodegenerative potential of Streptomyces spp. isolated from different physiographic provinces, too few isolates per physiographic province were tested for reliable statistical analyses. However, questions regarding compound-producing isolates and soil texture, pH, and organic content were addressed. No significant correlations between the prevalence of compound production and these characteristics were identified.
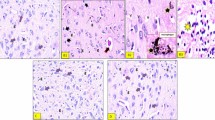
An unequivocal advantage of C. elegans is that detailed analyses of neurons are achievable. There are precisely six dopaminergic neurons within the anterior region of the worm that consistently display degenerative characteristics (Fig. 6A). We examined these six neurons in C. elegans for changes in the cell bodies as well as the neuronal processes for normal appearance vs. degenerative changes (Fig. 6B,C). If a dopaminergic neuron displayed an abnormality (cell body loss, blebbing or missing neuronal processes), it was scored as degenerating and the total number of damaged neurons was recorded and compared to controls.
(A–C). Representative images of C. elegans (strain BY200) expressing GFP specifically in the six anterior dopaminergic (DA) neurons. In all images, arrows show intact dopaminergic neuron cell bodies. Arrowheads indicate areas where dopaminergic neurons have degenerated. (A) An example of normal, intact, DA neurons that have not degenerated following 12 days exposure to negative control, E. coli. (B,C). C. elegans where all six DA neurons have degenerated following 12 days exposure to the S. venezuelae positive control (B) or a compound-producing Streptomyces sp. isolate (C). (D) Exposure to environmental isolates of Streptomyces spp. can cause significant neurodegeneration in populations of C. elegans.
Using the degeneration of C. elegans dopaminergic neurons from exposure to S. venezuelae supernatant as a positive control5, 28.5% of individual dopaminergic neurons were found to be degenerated (Fig. 6D). Similarly, exposure to the supernatants of compound-producing soil Streptomyces spp. resulted in the degeneration of 26.4% of dopaminergic neurons. In contrast, exposure to the supernatants of non-compound producing isolates did not cause significant neurodegeneration, as only 1.4% of neurons, on average, degenerated in these nematodes. These data were comparable to the 0.4% neuron degeneration observed when animals were exposed to E. coli supernatants, the negative control. Interestingly, the amount of individual dopaminergic neuron damage in C. elegans exposed to compound-producing strains varied between land uses. C. elegans exposed to supernatants of isolates from agricultural soils caused more individual neurons to degenerate compared to strains isolated from undeveloped (one-way ANOVA, p < 0.05; Fig. 7) and urban (p < 0.05; Fig. 7) soils. All dopaminergic neuron counts from compound-producing soil streptomycetes were non-significantly different from the compound-producing S. venezuelae positive control and significantly different from the E. coli negative control (p < 0.01).
To determine if strains capable of producing the neurodegenerative compound(s) were similar, the genomic banding patterns obtained from BOX PCR of the tested Streptomyces isolates (n=180) were evaluated using Gel Compar II. No clustering of species capable of producing the neurodegenerative compound(s) was apparent within the dendrograms (data not shown). Instead, compound-producing species were distributed throughout the dendrograms, suggesting that strains capable of producing the neurodegenerative compound(s) exhibit substantial genomic diversity.
Discussion
Exposure to the metabolites produced by Streptomyces venezuelae resulted in degeneration of all neuron classes in a C. elegans model and human neuroblastoma cells5. This led us to question the prevalence of neurodegenerative compound-producing Streptomyces spp. in natural environments. Based on the elevated incidence of PD, a neurodegenerative disease associated with rural living, drinking well water, and farming as a profession (e.g.,)10,11, we hypothesized that more neurodegenerative metabolite-producing organisms would be isolated from rural soils (e.g., agricultural, undeveloped) than urban soils.
C. elegans was ideal for these studies because it is a rapidly cultured transparent organism with an experimentally accommodating lifespan. Despite its evolutionary distance from humans, the 302 neurons of this genetically invariant hermaphroditic nematode retain many hallmarks of mammalian neuronal function including ion channels, neurotransmitters, vesicular transporters, receptors, and synaptic components30. The six anterior dopamine-producing neurons in this animal were readily evaluated for neurodegenerative changes using GFP as a fluorescent indicator. The use of C. elegans to rapidly evaluate the impact of environmental exposures on the survival of dopaminergic neurons is one of the strengths of this model system.
Notably, 28.3% of the Streptomyces isolates produced a compound(s) that caused dopaminergic neurodegeneration in C. elegans. The similarity in the percentages of neurodegenerative metabolite producing strains from agricultural (38.3%) and undeveloped (35.0%) soils compared to urban (11.6%) soils supported our hypothesis. Interestingly, significantly fewer Streptomyces spp. were isolated from undeveloped soils compared to agricultural soils, suggesting that land use alters the soil environment for Streptomyces. Additionally, strains isolated from agricultural soils resulted in more individual dopaminergic neuron damage than strains recovered from undeveloped or urban soils, indicating that the production of the neurodegenerative compound(s) is variable and may change with environment (Fig. 7). Although Basilio et al.31 reported differences in metabolite production from isolates obtained from environments with differing pH and salinity, the edaphic properties examined in this study (pH, organic matter content, and texture) did not appear to influence the distribution of producing strains. Alternately, the media used for isolation of the Streptomyces may select for strains with this capacity, as media composition affects bacterial recovery32 and metabolite production33,34,35. Obviously, much work remains to be done to address the potential impact of this neurodegenerative metabolite and its possible role as a bacterial toxicant in the etiology of PD.
This study utilized culture-dependent methods, which are generally thought to recover only a small portion (<1%) of the resident microorganisms, indicating that greater diversity of streptomycetes is likely in the soils examined. However, this approach was selected as it allowed us to specifically examine isolates for the production of the neurodegenerative compound, which, at present, cannot be assessed using culture-independent molecular techniques. It is possible that certain genera, including the aerobic, spore-forming Streptomyces with the ability to utilize a wide variety of carbon sources, are more amenable to cultivation than other genera. For example, a culture-dependent investigation of Antarctic soils indicated that the majority of actinobacteria isolated were Streptomyces (>80%), although they also found that the isolates were most closely related to culturable species while the phylotypes identified using cultivation-independent methods were related to uncultured species36. More work is needed to evaluate the full complement of resident Streptomyces spp. for the capacity to produce compounds that cause dopaminergic neurodegeneration, especially as this genus has been shown to account for ~6% of soil microbial libraries37.
Although BOX PCR was found to be effective in discriminating between closely related Streptomyces species in another study38, evaluation of our BOX banding patterns indicated that there was no uniformity within producing and non-producing strains (as determined by our C. elegans bioassay). However, Davelos et al.39 reported poor correspondence between 16 S rRNA gene grouping and BOX PCR fingerprints, suggesting that the fingerprinting method may reflect the instability of the Streptomyces genome, especially in the divergent ‘arms’ (reviewed by)40. In our samples, isolates producing a compound that caused dopaminergic neurodegeneration were widespread, implying that the genes for compound production may have been moved between hosts via lateral gene transfer (LGT). Much debate exists regarding the occurrence and ecological significance of LGT in streptomycetes, with some biosynthetic gene clusters suggested to be transferred between species laterally41,42,43,44 while others are more likely vertically transmitted45.
Of the 1509 Streptomyces isolates recovered in this study, 92.5% (1397 isolates) exhibited unique genomic banding patterns by BOX-PCR methods. As many important biosynthetic and metabolic genes have been found to be plasmid-encoded within this genus, the entire genetic information for an isolate was examined, including any plasmids. All colonies demonstrating the distinctive wrinkled, powdery morphology of Streptomyces were designated for isolation, eliminating potential selection bias by the investigators. Additionally, the predicted densities of streptomycetes per gram of soil (roughly 106–108 cells) are equivalent to, or in excess of, previous estimates derived from cultivation based analyses39,46,47,48,49, suggesting that the genus was adequately sampled and highlighting the extensive genomic diversity. As many natural products research programs were discontinued due to the high rediscovery rate of known compounds, these data support the prediction that less than 10% of bioactive metabolites have been discovered from this genus50,51 and indicate that members of Streptomyces possess extremely diverse genomes that likely include many novel biosynthetic gene clusters overlooked in previous phenotypic studies.
In this study, genomically diverse Streptomyces isolates were obtained from all samples but similar communities were not detected within soils under the same land use (Fig. 4) or from the same physiographic province (data not shown). We isolated more Streptomyces strains from agricultural soils compared to undeveloped soils but found no significant difference in streptomycete isolation from urban and undeveloped soils. However, as discussed by Lauber et al.52 soil microbial communities often reflect the characteristics of the soil in which they reside, indicating that land use alone cannot be used to predict community composition, as edaphic properties are not necessarily consistent within soils from a specific land use. Due to the exceedingly high level of genomic diversity discovered within the cultivated Streptomyces strains, it was not possible to evaluate what environmental factors affected their distribution. However, similar to results published by Lauber et al.53 more Streptomyces strains were isolated from soils with neutral to slightly alkaline pH. This result was somewhat unexpected, as a number of previous studies had portrayed Streptomyces as an acidophilic genus commonly recovered from soils exhibiting lower pH (e.g.,)54,55,56. Regardless, it is interesting that none of the strains appeared to have substantial localized populations, minimizing the possibility for cell-to-cell communication via quorum sensing approaches. However, the small amount of soil used for cultivation and the known spatial heterogeneity of soil populations may have influenced the recovery of similar strains. Even with these limitations, it appears that substantial genetic variation exists within members of the Streptomyces, reinforcing the utility of the genus for continued discovery of novel natural products.
Conclusions
The role of bacterial metabolites, particularly toxins, in human health has been well established (reviewed by)57,58. However, considerably less is known about the role of environmental toxins in the establishment of PD6,59. Although exposure to herbicides, fungicides, and pesticides have been shown to elicit Parkinsonian-like symptoms in various model systems, these environmental factors are not thought to account for all associations between environmental risk and disease10,20. This study demonstrated that a substantial portion of the cultivable Streptomyces community from soil samples has the ability to produce a neurodegenerative compound(s) and may be another mechanism for environmental exposure.
Methods
Soil Collection
Approximately 10 g of soil was collected from ~3–5 cm below the surface using a clean garden trowel and placed into a small resealable plastic bag. Each soil sample was thoroughly homogenized. From agricultural soils, samples were obtained between crop rows to avoid damage to the crop and collection of root rhizosphere communities (Fig. 1A). Undeveloped samples were collected from unused land, often requiring that significant leaf litter and debris be removed to access the soil surface, while urban samples were taken from soil near roadways, parking lots and large businesses (Fig. 1B,C). Additional soil (~1.5 liters) was collected from each site and placed into a resealable plastic bag for preparation of an environmental-specific medium and measurement of edaphic properties. Following collection, soil was stored at 4 °C until use. Soil samples were analyzed for particle size distribution, organic content, and pH. Particle size distribution within soils was assessed using a dried sieving method. Approximately 2 g of air-dried soil samples were sieved using U.S. standard mesh sizes #230,#60,#35, and #10. Sieves were shaken at a constant rate of 1100 rpm for 20 minutes using a titer plate shaker (Lab Line Instruments). Based on retention on the sieves, particles were classified as large grain sand, medium grain sand, fine grain sand, or clay/silt. A Bouyoucos hydrometer (VWR International) was employed to further differentiate between clay and silt. For this analysis, 50 g of air-dried soil samples were shaken at 270 rpm overnight on a C1 platform shaker (New Brunswick Scientific) in 50 mL of 5% sodium hexametaphosphate in water solution. Prior to analysis, the samples were diluted to 1 L with deionized water, mixed for 30 seconds, and readings of specific gravity were taken at 40 seconds and 2 hours. A control solution, consisting of 50 mL 5% sodium hexametaphosphate diluted to 1 L, was used to account for variation in specific gravity resulting from the dispersing solution. Following drying of soil at 100 °C overnight, soil organic content was determined by ashing 1 g of soil at 500 °C overnight in a muffle furnace (Thermo Scientific). Samples were weighed to 10−4 gram to determine carbon loss and measured in triplicate. Soil pH was measured on 10 g soil in 10 mL deionized water using a waterproof handheld pH meter (Oakton).
Bacterial Cultivation
Soil suspensions were prepared by adding 0.25 g of soil from the small plastic bag to 1.0 mL of sterile deionized water, mixing thoroughly by vortexing, and serially diluting the suspension to 10−2 and 10−3. From each dilution, 75 μL was plated in duplicate onto actinobacterial-specific and environment-specific media. Mannitol-soy agar60 supplemented with 1% sodium pyruvate and starch casein agar (1.5% bacteriological agar, 1% soluble starch, 1% sodium pyruvate, 0.2% potassium phosphate dibasic, 0.2% potassium nitrate, 0.2% sodium chloride, 0.03% casein, 0.005% MgSO4+7 H2O, 0.002% CaCO3, 0.002% FeSO4+7H2O in 1 L deionized water) were used for their high selectivity for Actinobacteria. To make environment specific media, 1 liter of each soil sample was mixed with 1 liter of deionized water, incubated at 4 °C overnight, and the supernatant was filtered with cheesecloth to remove debris and large sand particles. Bacteriological agar (1.5%) and sodium pyruvate (1%) were added to the filtered supernatant prior to autoclaving. All media were supplemented with cycloheximide and naladixic acid at concentrations of 25 μg/ml to inhibit the growth of fungi and Gram-negative bacteria, respectively, as well as sodium pyruvate to reduce the occurrence and impact of reactive oxygen species61. Over a six-week incubation at room temperature, the distinctive colonial morphology (e.g., dry, wrinkled, powdery) for which Streptomyces are known was used to select colonies for isolation.
Molecular Methods
DNA was extracted by suspending cells from isolates in 100 μL of a sterile 5% Chelex (analytical grade 100 resin; Bio-Rad Laboratories) in deionized water solution followed by 4 freeze/thaw cycles with periodic vigorous vortexing. Isolates were verified as members of the genus Streptomyces through PCR amplification using Streptomyces-specific primers, StrepB (5′-ACAAGCCCTGGAAACGGGGT-3′) and StrepF (5′-ACGTGTGCAGCCCAAGACA-3′)62. Each reaction contained 25 pmol of each primer, 10 μL PerfectTaq buffer (5 Prime), 0.125 μg MgCl2, 100 mM of each deoxynucleoside triphosphate, 2 U PerfectTaq DNA Polymerase (5 Prime), 1 μL template DNA, and sterile deionized water to a final volume of 50 μL. Reaction conditions were 5 min at 95 °C, followed by 30 cycles of 45 sec at 95 °C, 40 sec at 50 °C, and 120 sec at 72 °C, with a final 10 min incubation at 72 °C. PCR products were analyzed by electrophoresis at 80 V on 1.5% agarose gels. Gels were stained with GelRed™ (Biotium) and visualized under UV transillumination with a gel imaging system (Fotodyne Inc.).
Isolates confirmed as streptomycetes by a positive amplification with Streptomyces-specific primers were subjected to BOX PCR38 using the BOXA1R primer. The PCR reaction mixtures consisted of 0.3 μg BoxA1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′), 10 μL 5x Green Go Taq Buffer (Promega), 0.125 μg MgCl2, 4 μL DMSO, 100 mM of each deoxynucleoside triphosphate, 5 U Go Taq DNA Polymerase (Promega), 3 μL template DNA, and sterile deionized water to a final volume of 50 μL. Reaction conditions were 7 min at 95 °C, followed by 30 cycles of 90 °C for 30 sec, 53 °C for 60 sec and 65 °C for 8 min, with a final 16 min incubation at 65 °C. PCR products were analyzed by electrophoresis on 1.5% agarose gels at 80 V for 85 minutes. Gels were stained with GelRed™ (Biotium) and visualized under UV transillumination with a gel imaging system (Fotodyne Inc.).
Growth and isolation of environmental samples for neurodegenerative testing
Streptomyces isolates from collection locations representing each soil classification (agricultural, undeveloped, and urban) and all of the major physiographic provinces within the state were selected haphazardly. Each isolate was inoculated into 10 mL of SYZ broth63 and incubated at 30o C with shaking at 220 rpm for 17–21 days. In total, 60 isolates from each land use were grown for testing. Samples were harvested approximately 14 days after maximum cell density had been reached. Cell debris was removed by centrifugation at 10,000 x g for 10 minutes and supernatants were sequentially passed through three PES filter membranes with pore sizes of 1 μm, 0.45 μm and 0.22 μm. After the final filtration, the supernatant of each sample was dispensed into five equal aliquots and frozen for subsequent experiments. Each aliquot was thawed and used only once.
Caenorhabditis elegans strains
Nematodes were maintained using standard procedures64. Strain BY200 vtIs1 [Pdat-1::GFP, pRF4(rol-6(su1006))] (an integrated Pdat-1::GFP strain), a generous gift from Randy Blakely (Vanderbilt University), was used in all experiments.
C. elegans neurodegeneration assay
Each filtered sample was added to the surface of nematode growth medium (NGM) petri plates at a final concentration of 25 μl/ml along with E. coli (strain OP50)5. Adult Pdat-1::GFP animals (strain BY200) were placed on the environmental sample plates and allowed to lay eggs for ~4 hours before the adults were removed. The worm embryos were grown under constant exposure to Streptomyces supernatants until analysis. Worms were transferred to freshly made plates every other day and scored for neurodegeneration after 12 days of exposure. Similarly, Pdat-1::GFP worms exposed to S. venezueulae supernatant or E. coli supernatant were used as positive and negative controls, respectively, and were analyzed along with worms exposed to the metabolites from the environmental Streptomyces isolates. A total of 20-30 worms were analyzed for each environmental isolate. Worms were considered normal when all six anterior dopaminergic neurons were intact and no visible signs of degeneration were observed. If a worm displayed a neuron with a missing or shortened neuronal process, rounding or cell body loss, or blebbing process, the neuron was scored as exhibiting a degenerative change5. Further, the number of abnormal neurons/worm was scored on a scale of 0 to 6, with 0 representing a worm with no visible dopamine neuron degeneration, and 6 signifying that a worm exhibited degeneration in all six anterior dopamine neurons. For example, if a worm lacked one dopaminergic cell body and also had two degenerating dendrites, the worm was assigned a score of 3, signifying that 3 out of 6 neurons showed degeneration. This scale allowed for an assessment of the severity of neuronal damage within a population of neurons. After assigning each worm a score, the scores for all worms exposed to each supernatant were added together to yield the total number of degenerating neurons.
Statistical Analysis
The densiometric curves of all Streptomyces isolates and the 180 tested isolates were statistically evaluated using the software package Gel Compar II (BioSystematica). Each densiometric curve was based on the BOX banding pattern and the intensity of each band in relation to other bands within the sample38. To prevent ambiguities associated with size determination, only bands between 300 and 3000 base pairs were used for analyses. Isolates with greater than 90% similarity of densiometric curves were considered clones39. To produce a similarity matrix, Pearson’s correlations were calculated using 2% optimization and 1% curve smoothing with negative similarities unchanged from the default value65. The cluster analysis was calculated using unweighted pair group-method arithmetic average (UPGMA) with advanced clustering. Similarly, to compare banding patterns for all isolates recovered from a particular sample, composite profiles were generated using the create_averaged_fingerprints script provided by Gel Compar II staff. Comparisons of the profiles were performed using both curve-based (e.g., Pearson) and band-based (e.g., Jaccard) coefficients to generate similarity matrices. These matrices were imported into Primer66 and one-way ANOSIM, non-metric multidimensional scaling analyses, and cluster analyses were performed to evaluate if Streptomyces communities from particular land uses were similar.
SPSS statistical software (version 19) was used to analyze differences in isolation of Streptomyces spp. and neurodegenerative compound-producing species across land use patterns and soil characteristics. Measurements of streptomycete isolation followed a normal distribution; parametric tests including a one-way ANOVA and Pearson correlation were used to measure significance. Measurements of sand, silt, clay, and SOM were log transformed to achieve assumptions of normal distribution and parametric tests such as one-way ANOVA with Tukey’s HSD and Pearson correlation were used to measure significance. Nonparametric tests including the Kruskal-Wallis, Mann Whitney and Spearman correlation were used in analyses involving soil pH. Examination of compound-producing species by land use exhibited a normal distribution and parametric tests including ANOVA and Pearson’s correlation were used in analyses.
To determine which Streptomyces isolates caused significant dopaminergic neurodegeneration in C. elegans (in comparison to the negative E. coli control), a one-way ANOVA followed by a Bonferroni correction was employed (Prism 3.0 software; GraphPad). For analysis of the Streptomyces isolates that caused significant neurodegeneration with varying land uses, total numbers of damaged neurons were compared using a one-way ANOVA followed by Tukey’s HSD.
Additional Information
How to cite this article: Watkins, A. L. et al. The Prevalence and Distribution of Neurodegenerative Compound-Producing Soil Streptomyces spp. Sci. Rep. 6, 22566; doi: 10.1038/srep22566 (2016).
References
Genin, E., Reboud-Ravaux, M. & Vidal, J. Proteasome inhibitors: recent advances and new perspectives in medicinal chemistry. Curr Top Medicinal Chem . 10, 232–256 (2010).
Lümmen, P. Complex I inhibitors as insecticides and acaricides. Biochim. Biophys. Acta. 1364, 287–296 (1998).
Surup, F., Shojaei, H., von Zezschwitz, P., Kunze, B. & Grond, S. Iromycins from Streptomyces sp. and from synthesis: new inhibitors of the mitochondrial electron transport chain. Bioorg. Medicinal Chem . 16, 1738–1746 (2008).
Betarbet, R., Sherer, T. B., Di Monte, D. A. & Greenamyre, J. T. Mechanistic approaches to Parkinson’s disease pathogenesis. Brain Pathol. 12, 499–510 (2002).
Caldwell, K. A. et al. Investigating bacterial sources of toxicity as an environmental contributor to dopaminergic neurodegeneration. PLoS ONE. 4, e7227 (2009).
Reichmann, H. View point: Etiology in Parkinson’s disease. Dual hit or spreading intoxication. J. Neurol. Sci. 310, 9–11 (2011).
Ray, A., Martinez, B. A., Berkowitz, L. A., Caldwell, G. A. & Caldwell, K. A. Mitochondrial dysfunction, oxidative stress, and neurodegeneration elicited by a bacterial metabolite in a C. elegans Parkinson’s model. Cell Death Dis . 5, e984 (2014).
Martinez, B. A., Kim, H., Ray, A., Caldwell, G. A. & Caldwell, K. A. A bacterial metabolite induces glutathione-tractable proteostatic damage, proteasomal disturbances, and PINK1-dependent autophagy in C. elegans . Cell Death Dis . 6, e1908 (2015).
Tanner, C. M. Is the cause of Parkinson’s disease environmental or hereditary? Evidence from twin studies. Adv. Neurol. 91, 133–142 (2003).
Gorell, J. M., Johnson, C. C., Rybicki, B. A., Peterson, E. L. & Richardson, R. J. The risk of Parkinson’s disease with exposure to pesticides, farming, well water, and rural living. Neurol. 50, 1346–1350 (1998)
Priyadarshi, A., Khuder, S. A., Schaub, E. A. & Priyadarshi, S. S. Environmental risk factors and Parkinson’s Disease: a metaanalysis. Environ. Res. 86, 122–127 (2001).
Betarbet, R. et al. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nature Neurosci. 3, 1301–1306 (2000).
Sherer, T. B. et al. Mechanism of toxicity in rotenone models of Parkinson’s disease. J. Neurosci. 23, 10756–10764 (2003).
Sherer, T. B., Kim, J. H., Betarbet, R. & Greenamyre, J. T. Subcutaneous rotenone exposure causes highly selective dopaminergic degeneration and α-synuclein aggregation. Exp. Neurol. 179, 9–16 (2003).
Uversky, V. N. Neurotoxicant-induced animal models of Parkinson’s disease: understanding the role of rotenone, maneb and paraquat in neurodegeneration. Cell Tissue Res. 318, 225–241 (2004).
Landrigan, P. J. et al. Early environmental origins of neurodegenerative disease in later life. Environ. Health Perspect. 113, 1230–1233 (2005).
Tanner, C. M. et al. Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect. 119, 866–872 (2011).
Costello, S., Cockburn, M., Bronstein, J., Zhang, X. & Ritz, B. Parkinson’s disease and residential exposure to maneb and paraquat from agricultural applications in the Central Valley of California. Amer. J. Epidemiol. 169, 919–926 (2009).
Goldman, S. M. et al. Genetic modification of the association of paraquat and Parkinson’s disease. Movement Disord. 27, 1652–1658 (2012).
Thiruchelvam, M., Richfield, E. K., Baggs, R. B., Tank, A. W. & Cory-Slechta, D. A. The nigrostriatal dopaminergic system as a preferential target of repeated exposures to combined paraquat and maneb: implications for Parkinson’s disease. J. Neurosci. 20, 9207–9214 (2000).
Steenwerth, K. L., Jackson, L. E., Calderon, F. J., Stromberg, M. R. & Scow, K. M. Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biol. Biochem. 34, 1599–1611 (2002).
Bossio, D. A. et al. Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microb. Ecol. 49, 50–62 (2005).
Jenkins, S. N. et al. Actinobacterial community dynamics in long term managed grasslands. Antonie Van Leeuwenhoek. 95, 319–334 (2009).
Zhang, H., Lu, Y. & Cao, L. Effect of fertilization of paddy soil bacterial diversity and spatial distribution. Fresenius Environ. Bull . 20, 1558–1563 (2011).
Cooper, A. A. et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 313, 324–328 (2006).
Gilter, A. D. et al. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nature Genet. 41, 308–315 (2009).
Ruan, Q., Harrington, A. J., Caldwell, K. A., Caldwell, G. A. & Standaert, D. G. VPS41, a protein involved in lysosomal trafficking, is protective in Caenorhabditis elegans and mammalian cellular models of Parkinson’s disease. Neurobiol. Dis. 37, 330–338 (2010).
Tardiff, D. F. et al. Yeast reveal a “druggable” Rsp5/Nedd4 network that ameliorates α-synuclein toxicity in neurons. Science. 342, 979–983 (2013).
Dexter, P. M., Caldwell, K. A. & Caldwell, G. A. A predictable worm: application of Caenorhabditis elegans for mechanistic investigation of movement disorders. Neurotherapeutics. 9, 393–404 (2012).
Chalfie, M. & White, J. The nervous system. In The Nematode Caenorhabditis elegans, (ed Wood, W. B. ) pp. 337–391 (Cold Spring Harbor Laboratory Press, 1988).
Basilio, A. et al. Patterns of antimicrobial activities from soil actinomycetes isolated under different conditions of pH and salinity. J. Appl. Microbiol. 95, 814–823 (2003).
Wawrik, B., Kerkhof, L., Kukor, J. & Zylstra, G. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl. Environ. Microbiol. 71, 6776–6783 (2005).
Duffy, B. K. & Défago, G. Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438 (1999).
Rigali, S. et al. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces . EMBO Rep. 9, 670–675 (2008).
Ruiz, B. et al. Production of microbial secondary metabolites: regulation by the carbon source. Crit. Rev. Microbiol. 36, 146–167 (2010).
Babalola, O. O. et al. Phylogenetic analysis of actinobacterial populations associated with Antarctic Dry Valley mineral soils. Environ. Microbiol. 11, 566–576 (2009).
Janssen, P. H. Identifying the dominant soil bacterial taxa in libraries of 16 S rRNA and 16 S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728 (2006).
Lanoot, B. et al. BOX-pCR Fingerprinting as a powerful tool to reveal synonymous names in the genus Streptomyces. Emended descriptions are proposed for the species Streptomyces cinereorectus, S. fradiae, S. tricolor, S. colombiensis, S. filamentosus, S. vinaceus and S. phaeopurpureus . Syst. Appl. Microbiol. 27, 84–92 (2004).
Davelos, A. L., Xiao, K., Samac, D. A., Martin, A. P. & Kinkel, L. L. Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microb. Ecol. 70, 1051–1058 (2004).
Hopwood, D. A. Soil to genomics: the Streptomyces chromosome. Annu. Rev. Genet. 40, 1–23 (2006).
Wiener, P., Egan, S. & Wellington, E. M. H. Evidence for transfer of antibiotic-resistance genes in populations of streptomycetes. Mol. Ecol. 7, 1205–1216 (1998).
Egan, S., Wiener, P., Kallifidas, D. & Wellington, E. M. H. Phylogeny of Streptomyces species and evidence for horizontal transfer of entire and partial antibiotic gene clusters. Antonie van Leeuwenhoek. 79, 127–133 (2001).
Widenbrant, E. M., Tsai, H.-H., Chen, C. W. & Kao, C. M. Spontaneous amplification of the actinorhodin gene cluster in Streptomyces coelicolor involving native insertion sequence IS466. J. Bacteriol. 190, 4754–4758 (2008).
Deng, M.-R., Guo, J., Li, X., Zhu, C.-H. & Zhu, H.-H. Granaticins and their biosynthetic gene cluster from Streptomyces vietnamensis: evidence of horizontal gene transfer. Antonie van Leeuwenhoek. 100, 607–617 (2011).
Matter, A. M., Hoot, S. B., Anderson, P. D., Neves, S. S. & Chen, Y.-Q. Valinomycin biosynthetic gene cluster in Streptomyces: conservation, ecology and evolution. PLoS ONE. 4, e7194 (2009).
Porter, J. N., Wilhelm, J. J. & Tresner, H. D. Method for the preferential isolation of actinomycetes from soils. Appl. Environ. Microbiol. 8, 174–178 (1960).
Huddleston, A. S. et al. Molecular detection of streptomycin-producing streptomycetes in Brazilian soils. Appl. Environ. Microbiol. 63, 1288–1297 (1997).
Lee, J. Y. & Hwang, B. K. Diversity of antifungal actinomycetes in various vegetative soils of Korea. Can. J. Microbiol. 48, 407–417 (2002).
Wiggins, B. E. & Kinkel, L. L. Green manures and crop sequences influence alfalfa root rot and pathogen inhibitory activity among soil-borne streptomycetes. Plant Soil. 268, 271–283 (2005).
Watve, M. G., Tckoo, R., Jog, M. M. & Bhole, B. D. How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol. 176, 386–390 (2001).
Clardy, J., Fishbach, M. A. & Walsh, C. T. New antibiotics from bacterial natural products. Nature Biotechnol. 24, 1541–1550 (2006).
Lauber, C. L., Strickland, M. S., Bradford, M. A. & Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415 (2008).
Lauber, C. L., Hamady, M., Knight, R. & Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Xu, C. et al. Neutrotolerant acidophilic Streptomyces species isolated from acidic soils in China: Streptomyces guanduensis sp. nov., Streptomyces paucisporeus sp. nov., Streptomyces rubidus sp. nov. and Streptomyces yanglinensis sp. nov. Int. J. Syst. Evol. Microbiol. 56, 1109–1115 (2006).
Jenkins, S. N. et al. Actinobacterial community dynamics in long term managed grasslands. Antonie van Leeuwenhoek. 95, 319–334 (2009).
Rousk, J. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351 (2010).
Codd, G. A. et al. Cyanobacterial toxins, exposure routes and human health. European J. Phycol. 34, 405–415 (1999).
Schmitt, C. K., Meysick, K. C. & O’Brien, A. D. Bacterial toxins: friends or foes? Emerg. Infect. Dis. 5, 224–234 (1999).
Cicchetti, F., Drouin-Ouellet, J. & Gross, R. E. Enviromental toxins and Parkinson’s disease: what have we learned from pesticide-induced animal models? Trends Pharmacol. Sci. 30, 475–483 (2009).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. In Practical Streptomyces genetics (John Innes Foundation, 2000).
Olson, J. B., Lord, C. C. & McCarthy, P. J. Improved recoverability of microbial colonies from marine sponge samples. Microb. Ecol. 40, 139–147 (2000).
Rintala, H., Nevalainen, A., Rönkä, E. & Suutari, M. PCR primers targeting the 16 S rRNA gene for the specific detection of streptomycetes. Molec. Cell Probes . 15, 337–347 (2001).
Park, Y. C., Gunasekera, S. P., Lopez, J. V., McCarthy, P. J. & Wright, A. E. Metabolites from the marine-derived fungus Chromocleista sp. isolated from a deep-water sediment sample collected in the Gulf of Mexico. J. Nat. Prod. 69, 580–584 (2006).
Brenner, S. The Genetics of Caenorhabditis Elegans. Genetics. 77, 71–94 (1973).
Häne, B. G., Jäger, K. & Drexler, H. G. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis. 14, 967–972 (1993).
Clarke, K. R. & Warwick, R. M. In Change in marine communities: an approach to statistical analysis and interpretation 2nd edition (Plymouth, 2001).
Acknowledgements
Funding for this study was provided by NIH grant 1R15ES019310-01 to J.B.O. Thanks to A. Richardson, C. Marsh, O. Grubbs, P. Mullen, L. Hotard, and J. Porter for assistance with cultivation of soil Streptomyces spp. and measurement of edaphic properties.
Author information
Authors and Affiliations
Contributions
A.L.W. performed the microbial analyses, wrote the manuscript, and prepared several figures. A.R. performed the neurodegeneration experiments and analysed the resulting data. L.R.R. assisted with neurodegeneration experiments. K.A.C. and J.B.O. conceived and designed the experiments, prepared several figures and jointly supervised the work. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Watkins, A., Ray, A., R. Roberts, L. et al. The Prevalence and Distribution of Neurodegenerative Compound-Producing Soil Streptomyces spp.. Sci Rep 6, 22566 (2016). https://doi.org/10.1038/srep22566
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22566
This article is cited by
-
Caenorhabditis elegans: a model to understand host–microbe interactions
Cellular and Molecular Life Sciences (2020)