Abstract
We developed a protocol of noninvasive prenatal testing (NIPT), employing a higher-resolution picodroplet digital PCR, to detect genetic imbalance in maternal plasma DNA (mpDNA) caused by cell-free fetal DNA (cffDNA). In the present study, this approach was applied to four families with autosomal recessive (AR) congenital sensorineural hearing loss. First, a fraction of the fetal DNA in mpDNA was calculated. Then, we made artificial DNA mixtures (positive and negative controls) to simulate mpDNA containing the fraction of cffDNA with or without mutations. Next, a fraction of mutant cluster signals over the total signals was measured from mpDNA, positive controls, and negative controls. We determined whether fetal DNA carried any paternal or maternal mutations by calculating and comparing the sum of the log-likelihood of the study samples. Of the four families, we made a successful prediction of the complete fetal genotype in two cases where a distinct cluster was identified for each genotype and the fraction of cffDNA in mpDNA was at least 6.4%. Genotyping of only paternal mutation was possible in one of the other two families. This is the first NIPT protocol potentially applicable to any AR monogenic disease with various genotypes, including point mutations.
Similar content being viewed by others
Introduction
The main benefit of prenatal diagnosis is the timely management of diseases before or after birth. To date, the two most commonly used methods for prenatal diagnosis have been chorionic villus sampling and amniocentesis, which carries a 1% risk of miscarriage1,2,3. Unless the benefit from diagnosing a certain disease outweighs the risk, it is difficult to justify using these two methods to perform prenatal diagnosis.
Recently, prenatal diagnosis using cell-free fetal DNA (cffDNA) has been developed4,5,6. This method, unlike the two aforementioned methods, provides genetic information of fetuses in a noninvasive manner, as cffDNA can be obtained from the maternal peripheral blood. Hence, this method is regarded as a noninvasive prenatal testing (NIPT). Several studies have demonstrated that fetal aneuploidies and chromosome abnormalities can be detected by NIPT using cffDNA7,8,9. Consequently, monogenic diseases, which may not be fatal but certainly beneficial to diagnose, can be detected. However, NIPT using cffDNA is a technically challenging procedure, because the fetal DNA is indistinguishable from the maternal genomic DNA (gDNA) in the maternal plasma10. This challenge can be overcome by measuring the genetic imbalance in the maternal plasma caused by cffDNA.
The genetic imbalance can be detected in two ways: Reconstruction and prediction of the fetal haplotype and direct genotyping of the residue of interest. The former technique is based on massive parallel sequencing (MPS)11. The latter is based on digital polymerase chain reaction (dPCR)4,6. NIPT using targeted MPS technology requires numerous informative single nucleotide polymorphisms (SNPs) around the residue of interest for reconstruction of the fetal haplotypes. This may not be possible in some cases and sometimes recombination of alleles of the fetus should be considered. The second technique mentioned may comparatively be simpler and more straightforward than the first with respect to direct genotyping of the residue of interest. The genetic imbalance in the maternal plasma is measured by dPCR. However, the previous chip-based dPCR does not have sufficient resolution to diagnose general monogenic diseases. Statistical correction, like the Poisson distribution, is required to measure the genetic imbalance4,5,6, reducing the accuracy of NIPT. Since the resolution of dPCR depends on the number and volume of partitions, a greater number of partitions with smaller volume can guarantee higher resolution12. Moreover, the previously suggested protocols that employ dPCR to diagnose autosomal recessive (AR) monogenic diseases cannot completely cover AR monogenic diseases with a compound heterozygous genotype4,5.
For the first time, we utilized picodroplet dPCR to perform NIPT, which generated millions of picoliter-sized droplets. Since there are no droplets with multiple copies of the target in picodroplet dPCR13,14, statistical compensation is not necessary to measure the genetic imbalance. It leads to a diagnosis with greater accuracy. To the best of our knowledge, no study to date has presented successful NIPT results from using dPCR to diagnose AR monogenic diseases with a compound heterozygous genotype. Although it is technically more challenging, a protocol of NIPT for these diseases is necessary.
Prelingual sensorineural hearing loss (SNHL) usually occurs in an AR fashion. Diverse combinations of genotypes have been documented for prelingual, hereditary SNHL. SNHL requires a timely auditory rehabilitation for the proper development of language skills. Any delay in auditory rehabilitation will hinder language development in subjects with hearing loss15. Given this, prelingual SNHL is a good target disease to test a novel comprehensive NIPT approach for various AR genotype combinations. For this purpose, we recruited four families segregating genetically diagnosed AR type prelingual SNHL and expecting a new baby. We developed a protocol of NIPT employing picodroplet dPCR, with an expectation for it to be applicable to diagnose AR diseases with any combination of genotypes.
Methods
Subjects and Ethical Considerations
The institutional review boards of both Seoul National University Hospital (IRBY-H-0905–041–281) and Seoul National University Bundang Hospital (IRB-B-1007-105-402 and IRB-B-1508-312-304) approved all procedures used in this study. All subjects provided written informed consent. All methods were performed in accordance with the relevant guidelines and regulations. Four families with the first baby already confirmed to have SNHL due to AR mutations of known deafness genes and an unborn baby (fetus) were included in this study. The causative mutations of SNHL from these four families have previously been documented (the first family (SH123): SLC26A4 c.1529 T > A (p.V510D)/c.2168 A > G (p.H723R), the second family (SB191): GJB2 c.299_300delAT (p.H100RfsX14)/c.123 G > A (p.G45E), the third family (SB170): GJB2 c.235delC homozygote, the fourth family (SB251): GJB2 c.508_511dupAACG (p.A171EfsX40)/c.257 C > G (p.T86R) (Supplementary Figure S1). NIPT was performed for genotyping of the causative deafness gene from the unborn baby of each family.
Plasma DNA extraction protocol
Blood samples were collected from all pregnant mothers. At the time of this procedure, the maternal gestational age of the first, second, third, and fourth families was 15, 20, 18, and 10 weeks, respectively. The maternal body weight was 60.0, 58.2, 68.5 and 55.2 kg, respectively. Plasma was centrifuged for 10 min at 2000 g using MACHEREY-NAGEL, NucleoSpin Plasma XS (Germany) kit. We strictly followed the manufacturer’s guidelines using the manual, which involved the extraction of circulating DNA in 1–2 days or freezing of plasma at −20 °C. We used the ‘high sensitivity protocol’, with the exception of the first step. Based on our repeated trials that resulted in severe loss of yield, we added 1.5 ml plasma—not 240 μl plasma, as instructed—to a microcentrifuge tube. The buffer volume was adjusted accordingly.
gDNA preparation
The gDNA previously obtained from the father, mother, and first baby in each family was fragmented to mimic the plasma DNA using Covaris S220 (Covaris, MA, USA). This fragmented gDNA was used as a constituent of positive and negative control DNAs so that the control DNAs were close to pDNA at least in terms of size. The fragment size of 150 base pair length was confirmed by Bioanalyzer High Sensitivity DNA Chips (Agilent Technologies, CA, USA). DNA concentration was determined using a fluorescence assay of Picogreen (Invitrogen, Grand Island, NY, USA).
Picodroplet digital PCR (dPCR) methods
RainDrop Digital PCR System (RainDance Technologies Inc., Billerica, MA, USA) was used to assess picodroplet dPCR. In a pre–polymerase chain reaction environment, PCR reaction mixes were combined with primers and probes (the sequences and concentrations of primers and probes are given in Supplementary Table S1) along with 1.25 μl Drop Stabilizer (RainDance Technologies), 12.5 μl TaqMan Genotyping Master Mix (Life Technologies), DNase/RNase-free sterile water, and template DNA (either the minimum 2 ng of plasma DNA or 30 ng of the fragmented gDNA), which made up a total reaction volume of 25 μl. All probes were validated (Supplementary Figure S2). To emulsify the PCR reaction mix, it was loaded onto the RainDrop Source instrument (RainDance Technologies), carefully following the guidelines. Each 25 μl PCR mix was emulsified into 5 pl droplet volumes, partitioning a single molecule of DNA into approximately 5 million droplets. After emulsion, the PCR mixes were placed in a C1000 with deep-well (Bio-Rad) to be amplified, following the protocol outlined in Supplementary Table S2. The thermal cycled samples were loaded onto the RainDrop Sense instrument (RainDance Technologies), identifying the fluorescent intensity of each droplet for two fluorophores (FAM and VIC) simultaneously using a 488 nm laser. After evaluating all the samples, data from the cluster plots were spectrally-compensated and analyzed using the RainDrop Analyst data analysis software, in accordance with the standard procedures. The sample containing the highest mutant titration was used as the control sample to define the gates around the cluster of droplet events. These gates were applied across all evaluated samples within each assay. The same mutant gate was set within all wildtype-only samples, in which the droplets with mutant signals (droplet events that are counted within the mutant gate) were considered false-positive. These false-positive events were subtracted from the total mutant signal when counting the true-positive-mutant events across the samples.
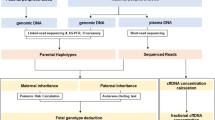
Noninvasive prenatal testing (NIPT) protocol
The known causative mutations were confirmed by Sanger sequencing from gDNAs of the father, mother, and first baby in each family. Then, NIPT was performed by two-track approach, depending on the homozygosity of the causative deafness mutations in each family.
In case of compound heterozygosity for the causative mutation
Preparatory step (Validation of our methodology)
The precision and applicability of our protocol were tested prior to genotyping of the fetal DNA. We attempted to evaluate whether consistent values were obtained throughout the repeated measurements of three different samples that were supposed to have mutant and wildtype residues at a ratio of 1:1.1368 (first family) or 1:1 (second family) (Supplementary Table S3). The result was expressed as the mean fraction of mutant sequence over the total reads at the mutated residue with standard deviations (SD). The values obtained by two measurements from the three samples were 0.4667 ± 0.0003, 0.4957 ± 0.0068, and 0.4794 ± 0.0008. SD below 0.0068 and detection of slightly low fraction from the first family ensured that our system can be applicable to genotyping of the fetal DNA (Supplementary Table S3).
Phase I (Genotyping of a paternal mutation): A fraction of the fetal DNA of unborn baby in the maternal plasma DNA (mpDNA) was calculated using either a paternal mutation of our interest or other known SNP exclusively from the paternal gDNA. The paternal mutation or the previously chosen SNP exclusively from paternal gDNA would not exist theoretically in mpDNA unless it had been inherited to the fetus. Firstly, the fraction of signals from the mutant cluster over the total signals from both the wildtype and mutant clusters in mpDNA was measured at the paternal mutant residue by picodroplet dPCR. If the mutant signals from the paternal causative mutant residue was close to nil, then we designed the primers and probes for detection of the paternal gDNA-specific homozygous SNP and calculated the fetal DNA fraction using signals from this SNP. Based on the calculated fraction, we made an artificial DNA mixture simulating the composition of positive and negative controls for the paternal causative mutation: The positive and negative controls account for maternal gDNA artificially containing the gDNA components with and without paternal mutation in the calculated fetal DNA fraction, respectively. The positive control was a mixture of gDNAs from the mother and first baby, represented as a ratio of the fetal DNA to the total mpDNA; the negative control comprised of the plasma DNA from any subjects without paternal mutation. Next, a fraction of signals from the mutant cluster over the total signals from both the wildtype and mutant clusters in positive and negative controls was measured at the paternal mutant residues, using picodroplet dPCR.
We determined whether the fetal DNA carried a paternal mutation by analyzing the direction of genetic imbalance caused by cffDNA between the wildtype and mutant alleles at the paternal mutant residues in mpDNA—either toward the positive controls or negative controls. This was quantified by calculating and comparing the sum of the log-likelihood of the study samples under the assumption that they followed a normal distribution of the positive and negative controls, respectively. The fetus was diagnosed as having a paternal mutation if the sum of the log-likelihood of the study samples following a normal distribution of the positive control was greater than that of the negative control. Otherwise, the fetus was diagnosed as containing no paternal mutation. A calculation of the test statistic was not necessary as the acquired data were well discriminated enough to make the uncertainty of likelihood negligible.
Phase II (Genotyping of a maternal mutation): Next, we checked for whether the fetal DNA had a maternal mutation. Theoretically, the ratio between the signals from the wildtype and mutant clusters at the maternal mutant residues is expected to be 1:1 without the fetal DNA. Given that the fetal DNA is contained in mpDNA, we aimed to detect any deviation from the expected ratio of 1:1. Both positive and negative controls for maternal mutation were also generated as calculated above, considering the fraction of fetal DNA in mpDNA. The positive control was a mixture of maternal gDNA harboring the mutation in a heterozygous state and first baby’s gDNA as a ratio of the fetal DNA to the total plasma DNA. The negative control included maternal gDNA mixed with gDNA not harboring the maternal mutation as a ratio of the fetal DNA proportion. Next, a fraction of signals from the mutant cluster over the total signals from both the wildtype and mutant clusters in mpDNA with unknown fetal DNA genotypes, positive controls, and negative controls was measured at the maternal mutant residues, using picodroplet dPCR.
The same discriminant analysis was done as in the first phase. The fetus was diagnosed as containing a maternal mutation if the sum of the log-likelihood of the study samples following a normal distribution of the positive control was greater than that of the negative control. Otherwise, the fetus was diagnosed as containing no maternal mutation (Fig. 1).
In case of homozygosity for the causative mutation
If the first baby carried a homozygous mutation, genotyping was tried in a single stage. In this situation, a fraction of the fetal DNA in mpDNA was calculated from the fraction of a previously chosen SNP, which was documented to exist exclusively from the paternal gDNA. Based on this result, positive and negative control samples for the homozygous mutation were generated with consideration to the calculated fraction of the fetal DNA. The positive control was a mixture between maternal gDNA with a mutation in the heterozygous state and the first baby’s gDNA harboring a mutation in the homozygous state as a ratio of the fetal DNA to the total plasma DNA. The negative control was a mixture of maternal gDNA and gDNA with a mutation in the heterozygous state as a ratio of the fetal DNA proportion. A fraction of signals from the mutant cluster over the total signals from both the wildtype and mutant clusters in mpDNA with unknown fetal DNA genotypes, positive controls, and negative controls was measured, using picodroplet dPCR.
The same discriminant analysis was performed as above. The fetus was diagnosed with having a homozygous mutation if the sum of the log-likelihood of the study samples following a normal distribution of the positive controls was greater than that of the negative controls. Otherwise, the fetus was diagnosed as containing no mutation or carrying a mutation in the heterozygous state. Clinically, the latter was expected to be unaffected (Fig. 1).
Genetic study for confirmation of fetal genotype
Sanger sequencing of gDNA from buccal mucosa of the second baby for targeted gene after birth served as a gold standard for the genotyping and the predicted fetal genotypes were checked against these Sanger sequencing results.
Results
Prediction of fetal genotypes by our NIPT protocol
In the first family, the father was a carrier of SLC26A4 c.1529 T > A (p.V510D), mother was a carrier of SLC26A4 c.2168 A > G (p.H723R), and the first baby was a compound heterozygote of SLC26A4 c.1529 T > A (p.V510D)/c.2168 A > G (p.H723R). The mean fraction of paternal mutation in mpDNA was 0.0319. A fraction of the fetal DNA in mpDNA was 6.4% ((64 × 2)/(1845 + 64) + (60 × 2)/(1921 + 60))/2). The mean fractions of paternal mutation in positive and negative controls were 0.0267 and 0.0015, respectively (Table 1 and Figs 2A and 3). Sequentially, the fetus was diagnosed as having a paternal mutation by discriminant analysis (sum of the log-likelihood: positive control, 6.8677; negative control, -infinity) (Table 2). The mean fractions of maternal mutation in mpDNA, as well as positive and negative controls were calculated as 0.4557, 0.4939, and 0.4685, respectively (Table 1, Fig. 2B and Supplementary Figure S3). Sequentially, the fetus was diagnosed as having no maternal mutation by discriminant analysis (sum of the log-likelihood: positive control, −36.5065; negative control, −12.2726) (Table 2). The fetus was diagnosed as unaffected.
The mean fractions of paternal and maternal mutations.
(A) The mean fractions of a paternal mutation in the maternal plasma DNA, and the positive and negative controls of the first family were 0.0319, 0.0267 and 0.0015, respectively. The mean fractions of a paternal mutation in maternal plasma DNA, and the positive and negative controls of the second family were 0.0725, 0.0804 and 0.0002, respectively. The mean fractions of a paternal mutation in maternal plasma DNA, and the positive and negative controls of the fourth family were 0.0423, 0.0490 and 0.0000, respectively. (B) The mean fractions of a maternal mutation in maternal plasma DNA, and the positive and negative controls of the first family were 0.4557, 0.4939 and 0.4685, respectively. The mean fractions of a maternal mutation in maternal plasma DNA, and the positive and negative controls of the fourth family were 0.4662, 0.5070 and 0.4655, respectively.
In the second family, the father was a carrier of GJB2 c.299_300delAT (p.H100Rfs*14), mother was a carrier of GJB2 c.123 G > A (p.G45E), and the first baby was a compound heterozygote of GJB2 c.299_300delAT (p.H100Rfs*14)/c.123 G > A (p.G45E). The mean fraction of paternal mutation in mpDNA was 0.0725. The fraction of fetal DNA was 14.5% ((93 * 2)/(1190 + 93)). The mean fraction of paternal mutation in positive and negative controls were 0.0804, and 0.0002, respectively (Table 1, Fig. 2A and Supplementary Figure S4). Sequentially, the fetus was diagnosed as having a paternal mutation by discriminant analysis (sum of the log-likelihood: positive control, −89.6183; negative control, -infinity) (Table 2). The mean fractions of maternal mutation in mpDNA, as well as positive and negative controls were calculated as 0.3957, 0.4710, and 0.3904, respectively (Table 1 and Supplementary Figure S5). However, from the result of picodroplet dPCR using the probe for maternal mutation, mpDNA did not show a distinct cluster for the wildtype. The tail around the cluster resulted in an ambiguous wildtype count (Supplementary Figure S5A and B). Although the fraction of signals from the maternal mutant cluster over the total signals from both the wildtype and mutant clusters in mpDNA were achieved through a data analysis software, we were unable to determine whether the fetus had a maternal mutation (Table 1 and Supplementary Figure S5). Therefore, only a partial prenatal diagnosis of the fetus was made.
In the third family, both the father and mother were carriers of GJB2 c.235delC, and the first baby was a GJB2 c.235delC homozygote. The mean fraction of SNP, detected exclusively from the paternal gDNA, in mpDNA was 0.014 (Table 1 and Supplementary Figure S6). The fraction of the fetal DNA in mpDNA was only 2.7% (((24 * 2)/(2507 + 24) + (10 * 2)/(560 + 10))/2). The genotype status of the fetus could not be predicted due to a low fraction of cffDNA.
In the fourth family, the father was a carrier of GJB2 c.508_511dupAACG (p.A171Efs*40), the mother was a carrier of GJB2 c.257 C > G (p.T86R), and the first baby was a compound heterozygote of GJB2 c.508_511dupAACG (p.A171Efs*40)/c.257 C > G (p.T86R). The mean fraction of paternal mutation in mpDNA was 0.0423. The fraction of the fetal DNA in mpDNA was 8.5% ((41 × 2)/(899 + 41) + (35 × 2)/(820 + 35))/2). The mean fraction of the paternal mutation in positive and negative controls was 0.0490 and 0.0000, respectively (Table 1, Fig. 2A and Supplementary Figure S8). Sequentially, the fetus was diagnosed as having a paternal mutation by discriminant analysis (sum of the log-likelihood: positive control, −16.3308; negative control, −1050.1540) (Table 2). The mean fraction of maternal mutation in mpDNA, as well as positive and negative controls were calculated as 0.4662, 0.5070, and 0.4655, respectively (Table 1, Fig. 2B and Supplementary Figure S9). Sequentially, the fetus was diagnosed as having no maternal mutation by discriminant analysis (sum of the log-likelihood: positive control, -infinity; negative control, 10.4021) (Table 2). The fetus was diagnosed as unaffected.
Confirmation of fetal genotype by Sanger sequencing
Sanger sequencing from the second baby in the first, second, third, and fourth families confirmed the following: a single heterozygote of SLC26A4 c.1529 T > A (p.V510D), compound heterozygote of GJB2 c.299_300delAT (p.H100Rfs*14) and c.123 G > A (p.G45E), a single heterozygote of GJB2 c.235delC, and a single heterozygote GJB2 c.508_511dupAACG (p.A171Efs*40) (Fig. 4A–D). The prenatal diagnosis of paternal and maternal mutations for the second baby in the first and fourth family and paternal mutation for the second baby in the second family was correct (Table 3).
Genetic study for confirmation of fetal genotype.
(A) Sanger sequencing traces of the second-born baby of the first family: SLC26A4 c.1529 T > A (p.V510D) single heterozygote. (B) Sanger sequencing traces of the second-born baby of the second family: GJB2 c.299_300delAT (p.H100Rfs*14)/c.123 G > A (p.G45E). (C) Sanger sequencing traces of the second-born baby of the third family: GJB2 c.235delC single heterozygote. (D) Sanger sequencing traces of the second-born baby of the fourth family: GJB2 c.508_511dupAACG (p.A171Efs*40) single heterozygote.
Discussion
Up until recently, prenatal diagnosis, despite having many advantages for certain diseases, has never been performed on a regular basis. This was the case because prior methods—chorionic villus sampling and amniocentesis—were highly invasive with associated risks. However, with the discovery of cffDNA in the peripheral blood of pregnant women, which provides genetic information of the fetus, prenatal testing became more feasible16.
NIPT using cffDNA started from determination of fetal sex16 and RhD status17,18. The indication of NIPT was extended to aneuploidies, which is relatively an easier target. In 2007, it was detected that genetic material derived from chromosome 21 increased in the plasma of pregnant woman carrying a fetus with trisomy 2119,20. Prenatal diagnosis of trisomy 21 was performed through measuring the genomic representation of chromosome 21 with random MPS21,22. With the improvement in technology and strategy, the indication criteria for NIPT have expanded. Monogenic diseases have been diagnosed using NIPT4,5,23,24.
Our method using picodroplet dPCR, compared with previous methods of NIPT—MPS and chip-based dPCR, has several merits in diagnosing monogenic diseases. First, the biggest difference is that MPS deducts the fetal haplotypes through sequencing numerous SNPs around the residue of interest25,26,27,28,29. Conversely, our method sequences a residue of interest directly even if a mutation exists in a homozygous fashion. Sequentially, the method using MPS requires a lot of time and labor, and it poses concerns about chromosomal recombination inevitably, while our method offers greater clarity and simplicity. Additionally, if an efficient probe for a certain founder allele has been constructed previously, early diagnosis can be facilitated in many cases. Second, to date, several researchers have attempted NIPT employing chip-based dPCR for the following diseases: beta-thalasemia, hemophilia, and sickle cell anemia. Lun et al. detected fetal alleles of the beta-thalasemia mutation that were inherited from the mother30. Tsui et al. successfully reported the applicability of NIPT in detecting a female carrier of hemophilia with male fetuses6. Moreover, Barrett et al. successfully reported the applicability of NIPT for sickle cell anemia4. Although these studies showed successful results, they had limitations. As chip-based dPCR has less than 800 partitions, there must be a chamber containing multiple copies of a target. Containing one or no target copy in one partition is the major premise of dPCR to achieve high resolution. Although the Poisson distribution has been used to overcome this weakness, it can only compensate to a limited extent, reducing the accuracy of NIPT. To overcome this, we employed picodroplet dPCR instead of chip-based dPCR. To the best of our knowledge, this is the first study to use picodroplet dPCR in NIPT. Picodroplet dPCR generates millions of picoliter droplets. One droplet contains one or no target copy, and there is no droplet containing multiple copies of a target13,14. Consequently, statistical compensation, such as the Poisson distribution, is not needed to get the actual number of target molecules. This contributes to a more accurate prenatal diagnosis.
Additionally, we utilized positive and negative control samples. The results of the tested samples were compared with that of the control samples; prenatal diagnosis was made based on a discriminant analysis. In previous studies, however, they calculated the theoretically expected proportion of the mutant and wildtype alleles, comparing the value obtained from the tested sample against the calculated value4,6. The calculated value cannot reflect the effect of intrinsic variables of the experiment. There must be a difference between the calculated value and the value from a real sample, like the control sample in our study. Considering that dPCR requires a highly sensitive technique on a molecular-level, simulation with control samples could contribute to greater accuracy. As such, an accurate prenatal diagnosis can be made for point mutations in a compound heterozygous fashion using our novel protocol.
It is, however, worth noting that in our second and third families, a complete prenatal diagnosis was not achieved. The failure of NIPT in the third family was attributed to low cffDNA fraction (2.7%). A previous study using dPCR also reported that a higher cffDNA fraction was required to make a correct prenatal diagnosis30. Barrett et al. reported that 100% accuracy of NIPT was achieved at a cffDNA fraction of greater than 7%4. The effect of cffDNA fraction showed a similar tendency in NIPT using MPS. Yoo et al. reported that the lowest cffDNA fraction allowing successful NIPT with MPS was 5.8%29. Although New et al. reported a successful prenatal diagnosis with cffDNA fraction of 1.4%28, it was for a paternal mutation. In our results, the presence of SNP exclusively for a paternal DNA was detected in mpDNA under the cffDNA fraction of 2.7% (third family), which suggests that prenatal prediction of a paternal mutation is much more feasible compared with a maternal mutation even under a low fraction of cffDNA among mpDNA. NIPT using cffDNA is based on the measurement of imbalance in mpDNA caused by cffDNA. Consequently, if the fraction of cffDNA is too small to make a detectable imbalance in mpDNA, a prenatal diagnosis would not be possible. High enough cffDNA fraction seems to be one of the most important prerequisites for NIPT that uses dPCR or MPS. A cffDNA fraction is suggested to be influenced by gestational age and maternal body weight31,32. As cffDNA fraction tends to rise with increasing gestational age, this should be considered when performing NIPT at early gestational ages. In addition, as mother’s weight increases, maternal blood volume increases, which lowers the cffDNA fraction. In this present study, the mother of the third family, who was unable to successfully complete NIPT, was heavier that the other three mothers. As speculated, higher maternal body weight may indeed negatively influence the accuracy of NIPT.
Although we modified the probes for maternal mutation several times in the second family, the tail around the cluster of the wildtype was not removed (Supplementary Figure S5A and B), and a prenatal diagnosis for maternal mutation was not achieved. This might be attributed to the characteristics of the individual plasma DNA or a peculiar reaction between the individual plasma DNA and the probe. However, a successful prenatal diagnosis was achieved for a paternal mutation. In this case, NIPT using MPS could be an alternative. As NIPT utilizing dPCR or MPS has its own advantages and disadvantages, they can be complementary to one another. Therefore, an appropriate use of dPCR or MPS will contribute to a more accurate prenatal diagnosis.
In summary, we were able to make a successful prediction of the fetal genotype for AR monogenic diseases that result from point mutations by utilizing picodroplet dPCR in NIPT. This was possible particularly in cases where we were able to identify a distinct cluster for each genotype and where the fractions of cffDNA in mpDNA was at least 6.4%. Moreover, in other cases, at least partial genotyping was possible.
For the first time in the literature, we report a successfully developed protocol of NIPT for the genotyping of compound heterozygous point mutations of AR monogenic diseases by coupling the dPCR technique with a sophisticated statistical analysis. This protocol is applicable to any AR monogenic diseases with various genotypes, including point mutations; it is not limited for a specific disease if the fraction of cffDNA is higher than a certain level. Improved techniques in obtaining distinctive clusters for each genotype would make this protocol popular. With the incorporation of our novel protocol, NIPT can become a popular tool for prenatal diagnosis, making it possible to prenatally diagnose a number of AR monogenic diseases with various genotypes. Through this prenatal diagnosis, timely management of diseases leading to better lives is feasible. This would pave the way for the establishment of a widely used prenatal diagnosis method in the near future.
Additional Information
How to cite this article: Chang, M. Y. et al. Development of novel noninvasive prenatal testing protocol for whole autosomal recessive disease using picodroplet digital PCR. Sci. Rep. 6, 37153; doi: 10.1038/srep37153 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Kong, C. W. et al. Risk factors for procedure-related fetal losses after mid-trimester genetic amniocentesis. Prenat Diagn 26, 925–930 (2006).
Lau, K. T. et al. [Outcome of 1,355 consecutive transabdominal chorionic villus samplings in 1,351 patients]. Chin Med J (Engl) 118, 1675–1681 (2005).
Tabor, A. et al. Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet 1, 1287–1293 (1986).
Barrett, A. N., McDonnell, T. C., Chan, K. C. & Chitty, L. S. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin Chem 58, 1026–1032 (2012).
Debrand, E., Lykoudi, A., Bradshaw, E. & Allen, S. K. A Non-Invasive Droplet Digital PCR (ddPCR) Assay to Detect Paternal CFTR Mutations in the Cell-Free Fetal DNA (cffDNA) of Three Pregnancies at Risk of Cystic Fibrosis via Compound Heterozygosity. PLoS One 10, e0142729 (2015).
Tsui, N. B. et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood 117, 3684–3691 (2011).
Bianchi, D. W. et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med 370, 799–808 (2014).
Gil, M. M., Quezada, M. S., Revello, R., Akolekar, R. & Nicolaides, K. H. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound in Obstetrics & Gynecology 45, 249–266 (2015).
Norton, M. E. et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med 372, 1589–1597 (2015).
Lo, Y. M. & Chiu, R. W. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet 8, 71–77 (2007).
Liao, G. J. et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem 57, 92–101 (2011).
Hindson, C. M. et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 10, 1003–1005 (2013).
Pekin, D. et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11, 2156–2166 (2011).
Taly, V. et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 59, 1722–1731 (2013).
Lohle, E., Holm, M. & Lehnhardt, E. Preconditions of language development in deaf children. Int J Pediatr Otorhinolaryngol 47, 171–175 (1999).
Lo, Y. M. et al. Presence of fetal DNA in maternal plasma and serum. Lancet 350, 485–487 (1997).
Faas, B. H., Beuling, E. A., Christiaens, G. C., von dem Borne, A. E. & van der Schoot, C. E. Detection of fetal RHD-specific sequences in maternal plasma. Lancet 352, 1196 (1998).
Lo, Y. M. et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med 339, 1734–1738 (1998).
Fan, H. C. & Quake, S. R. Detection of aneuploidy with digital polymerase chain reaction. Anal Chem 79, 7576–7579 (2007).
Lo, Y. M. et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci USA 104, 13116–13121 (2007).
Chiu, R. W. et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA 105, 20458–20463 (2008).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA 105, 16266–16271 (2008).
Chitty, L. S. et al. New aids for the non-invasive prenatal diagnosis of achondroplasia: dysmorphic features, charts of fetal size and molecular confirmation using cell-free fetal DNA in maternal plasma. Ultrasound Obstet Gynecol 37, 283–289 (2011).
Meaney, C. & Norbury, G. Noninvasive prenatal diagnosis of early onset primary dystonia I in maternal plasma. Prenat Diagn 29, 1218–1221 (2009).
Lam, K. W. et al. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to beta-thalassemia. Clin Chem 58, 1467–1475 (2012).
Lim, B. C. et al. Genetic diagnosis of Duchenne and Becker muscular dystrophy using next-generation sequencing technology: comprehensive mutational search in a single platform. J Med Genet 48, 731–736 (2011).
Meng, M. et al. Noninvasive prenatal testing for autosomal recessive conditions by maternal plasma sequencing in a case of congenital deafness. Genet Med 16, 972–976 (2014).
New, M. I. et al. Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab 99, E1022–E1030 (2014).
Yoo, S. K. et al. Noninvasive prenatal diagnosis of duchenne muscular dystrophy: comprehensive genetic diagnosis in carrier, proband, and fetus. Clin Chem 61, 829–837 (2015).
Lun, F. M. et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci USA 105, 19920–19925 (2008).
Norwitz, E. R. & Levy, B. Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol 6, 48–62 (2013).
Palomaki, G. E. et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med 13, 913–920 (2011).
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2013R1A1A2063237 to B.Y. Choi) and also by the Korean Health Technology R&D project, Ministry of Health & Welfare, Republic of Korea (HI15C1632 and HI14C1867) to B. Y. Choi).
Author information
Authors and Affiliations
Contributions
B.Y.C. and M.Y.C. designed the study and wrote the paper. A.R.K., M.Y.K., S.K. and J.Y. performed the experiments. B.Y.C., M.Y.C., C.K., S.A. and J.J.H. analysed the data. All authors discussed the results and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chang, M., Kim, A., Kim, M. et al. Development of novel noninvasive prenatal testing protocol for whole autosomal recessive disease using picodroplet digital PCR. Sci Rep 6, 37153 (2016). https://doi.org/10.1038/srep37153
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37153
This article is cited by
-
Nanostructures in non-invasive prenatal genetic screening
Biomedical Engineering Letters (2022)
-
One-step noninvasive prenatal testing (NIPT) for autosomal recessive homozygous point mutations using digital PCR
Scientific Reports (2018)
-
Using massively parallel shotgun sequencing of maternal plasmatic cell-free DNA for cytomegalovirus DNA detection during pregnancy: a proof of concept study
Scientific Reports (2018)