Abstract
We examined expression of maspin and the epigenetic status of its gene in 40 primary hepato-biliary tract carcinomas and 11 cell lines originating from hepato-pancreatico-biliary tract carcinomas. Aberrant maspin expression was frequently observed immunohistochemically in biliary tract carcinomas (22/25, 88%) but not in hepatocellular carcinomas (HCCs) (0/15, 0%). Aberrant maspin expression by five pancreatico-biliary tract carcinoma cell lines was closely associated with demethylation at the maspin promoter. Five of six HCC cell lines were maspin-negative and exhibited extensive hypomethylation and hypoacetylation at the maspin promoter. Treatment with 5-aza-2’-deoxycytidine did not activate maspin expression in these five maspin-negative HCC cell lines, whereas treatment with Trichostatin A (TSA) activated maspin expression in two of them. Treatment with TSA increased histone acetylation in some HCC cell lines. These results suggest that aberrant maspin expression in biliary tract carcinomas is closely associated with demethylation at the promoter region, but that some HCC cell lines additionally require histone acetylation. In addition, the fact that maspin-negative HCC cell lines remain after treatment with TSA suggests the existence of other repressive factors controlling maspin expression.
Similar content being viewed by others
Main
Maspin is a member of the serine protease inhibitor family, which is related to the SERPIN family. The maspin gene was originally identified in normal mammary epithelial cells. A maspin-transfected mammary cancer cell line was found to have reduced capacity for tumorigenesis and metastasis in nude mice.1 Exogenous maspin protein can inhibit certain invasive and malignant characteristics of human breast carcinoma cell populations.1, 2, 3 Loss of maspin protein expression has frequently been observed, and is associated with poor prognosis, in breast, prostatic and oral cancers.4, 5, 6, 7, 8 However, it has been reported that maspin is overexpressed in pancreatic and ovarian cancers, whereas their normal tissues are maspin-negative.9, 10 Since maspin has a metastasis suppressor function in human breast cells, the gain of expression observed in these other tumor types is paradoxical.
Although the precise cellular and biochemical activities of maspin are currently unknown, we recently demonstrated that aberrant maspin expression was frequently observed in intestinal metaplasia of gastric epithelium and gastric cancers,11 and in undifferentiated carcinomas of the thyroid gland.12 This aberrant pattern of expression (where tumor cells are positive for maspin but their normal counterparts are negative) was closely associated with demethylation status at the 5′ regulatory region of the maspin gene.11, 12, 13 The concept that DNA methylation might play a role in the establishment and/or maintenance of tissue-specific gene expression was first proposed by Holliday14 and Riggs.15 Futscher et al16 demonstrated that this concept applies to cell-type-specific expression of the maspin gene in normal human tissues, supporting the hypothesis that disruption of cell-type-specific repression by demethylation at maspin CpG islands contributes to revealing metaplastic or dedifferentiated morphological phenotypes in normal or tumor cells.
To test our hypothesis, we initially used an immunohistochemical method to screen various human tumors and/or normal tissues for aberrant expression of the maspin gene in relation to metaplastic or dedifferentiated phenotypes. We found that aberrant maspin expression was frequently observed in biliary tract carcinomas, including cholangiocellular carcinomas (CCCs), common bile duct carcinomas (CBDCs) and gallbladder carcinomas (GBCs), whereas hepatocellular carcinomas (HCCs) were completely negative for maspin expression. In the series, biliary tract tumors demonstrated the expected relationship between DNA methylation and protein expression of the maspin gene, whereas the relationship was disrupted in HCCs. The present study describes in detail the epigenetic status and protein expression of the maspin gene in hepato-biliary tract carcinomas.
Materials and methods
Hepato-Biliary Tract Tumors and Cell Lines
Tumor specimens were obtained from 40 patients with hepato-biliary tract carcinomas, consisting of 15 HCCs, six CCCs, eight CBDCs and 11 GBCs, and were subjected to immunohistochemistry for maspin. All specimens were from autopsy cases at the Department of Pathology, Iwate Medical University School of Medicine (Morioka, Japan), between 1992 and 2002. The cases were 29 males and 11 females, in the age range 48–82 years (mean 68 years). To study maspin protein expression in normal tissues (hepatocytes, intrahepatic bile duct, common bile duct and gallbladder), 10 autopsied patients (mean age 68 years, age range 45–89?years) who had died of noncancerous disease were also examined. Additionally, specimens obtained from three autopsy cases (66, 68 and 75 years old) who died of noncancerous disease were subjected to genomic DNA extraction to determine the methylation status at the maspin promoter of the normal liver and epithelium of the common bile duct and gallbladder. Permission for this study was obtained from the Institutional Review Board (Iwate Medical University School of Medicine, Morioka, Japan).
Six human HCC cell lines (HepG2, Hep3B, HT17, HuH-7, Li-7 and PLC/RPF/5), three human CBDC cell lines (HuH-28, HuCCT1 and TFK-1), a human GBC cell line (OCUG-1) and a human PC cell line (PK-8) were obtained from the Cell Resource Center for Biomedical Research, Tohoku University (Sendai, Japan). These cells were maintained under the recommended conditions.
Immunohistochemistry
Four-micrometer slices were cut from formalin-fixed paraffin-embedded samples and were stained with hematoxylin and eosin. Serial sections were stained immunohistochemically. A microwave-based antigen-retrieval method with 10?mM citrate buffer (pH 6.0) for 15?min was used. After deparaffinization and antigen recovery, the sections were immersed in 0.3% hydrogen peroxide in methanol for 30?min to block endogenous peroxidase activity. After three washes with PBS for 5?min, the sections were incubated with 5% bovine serum albumin for 10?min to block nonspecific reactions. The sections were then incubated overnight at 4°C with anti-human maspin antibody (diluted 1:50; clone G167-70, Pharmingen, San Diego, CA, USA). Incubation with secondary antibody and peroxidase labeling were performed with a Simple Stain MAX-PO Kit (Nichirei, Tokyo, Japan). Color was produced with DAB substrate (Dako, Glostrup, Denmark), and counterstaining was performed with Mayer's hematoxylin. The extent of immunohistochemical reactivity for maspin was evaluated according to the incidence of maspin-positive cells and the relative density of maspin-positive cells. The incidence of maspin-positive cells was graded as follows: negative, no immunoreactive cells were seen; <20, 20–80 and >80%, the indicated proportion of immunoreactive cells. Relative density of maspin-positive cells was graded as follows: negative, no immunoreactivity; faint, moderate and strong, the indicated staining intensity.
Bisulfite-Modified Genomic DNA Sequencing
The methylation status of 19 CpG dinucleotides within the maspin gene promoter region was examined in bisulfite-modified genomic DNA, as previously described.17 Briefly, genomic DNA was digested with Pst I and then subjected to bisulfite modification as described elsewhere.16 The maspin gene promoter was amplified from bisulfite-modified DNA by using nested primers specific to the bisulfite-modified sequence of the maspin gene CpG islands. PCR primers and conditions were as previously described.16 PCR products were purified with a QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany), ligated to pGEM-T Easy Vectors (Promega Corp., Madison, WI, USA) and transformed into DH5α-competent cells (Toyobo, Tokyo, Japan). Ten subcloned colonies were chosen randomly. Plasmid DNA was purified with a PI-200 automatic DNA isolation system (Kurabo, Osaka, Japan). Cycle sequencing used a primer of the T7 promoter and a BigDye Terminator Cycle Sequencing FS Ready Reaction Kit (Applied Biosystems, ABI, Foster City, CA, USA). The product was analyzed with an ABI PRISM 3100 DNA Sequencer (ABI).
5-Aza-dC Treatment
Cells were seeded at a density of 5 × 105 cells/10?cm plate on day 1. After 24?h, 5-aza-2′-deoxycytidine (5-Aza-dC; Sigma, St Louis, MO, USA) was added to a final concentration of 10?μM. At 3 days (72?h) after 5-Aza-dC treatment, the cells were harvested for Western blotting and real-time quantitative-PCR (RQ-PCR) assay.
Trichostatin A Treatment
Treatment with a histone deacetylase (HDAC) inhibitor (Trichostatin A (TSA); Sigma, St Louis, MO, USA) was performed according to the method used by Maass et al18 to examine maspin expression in breast cancer cell lines. Cells were seeded at a density of 1 × 106 cells/10?cm plate on day 1. After 24?h, TSA was added to a final concentration of 1?μM. At 2 days (48?h) after TSA treatment, the cells were subjected to chromatin immunoprecipitation (ChIP) assay, Western blotting and RQ-PCR assay.
Western Blotting
The cell pellet was dissolved in 1.0%. NP-40 lysis buffer [50?mM HEPES (pH 7.5)/1?mM EDTA/150?mM NaCl/2.5?mM EGTA/1.0% NP-40] and rotated at 4°C for 30?min. Insoluble material was spun down (20?min, 14?000?rpm) and the clear supernatants were collected. The protein concentration of the lysates was measured with a Bio-Rad DC Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA, USA). Cell lysates containing equal amounts of protein were mixed with 6 × concentrated loading dye, heated for 4?min at 95°C and subjected to SDS-PAGE on a 10% polyacrylamide gel (Bio-Rad). The proteins were then transferred to a PVDF membrane (Amersham Biosciences, Buckinghamshire, UK) by electroblotting. The same primary monoclonal antibody for maspin used in immunohistochemistry was diluted 1:500 in PBST. The membrane was incubated for 1?h at room temperature and washed. For the secondary antibody, anti-mouse IgG (Amersham Biosciences) was diluted 1:10000 in blocking buffer. The membrane was incubated for 45?min at room temperature and washed. Maspin protein was detected with ECL Plus (Amersham Biosciences). Equal loading was confirmed by incubation with an antiactin antibody (C-2; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
RQ-PCR for Maspin mRNA
Total RNA was isolated with TRIZOL reagent (Invitrogen, Carlsbad, CA, USA). mRNA was reverse-transcribed with a ThermoScript™ RT-PCR system and oligo(dT) (Invitrogen) to produce cDNA. For the RQ-PCR assay, primers and a fluorogenic probe were designed with Primer Express software (ABI): maspin F (5′-CGA CCA GAC CAA AAT CCT TG-3′), maspin R (5′-GAA CGT GGC CTC CAT GTT C-3′), probe (5′-FAM-CAA CAA GAC AGA CAC CAA ACC AGT GCA G-TAMURA-3′). An ABI PRISM 7700 Sequence Detector (ABI) was used for RQ-PCR. The reaction mix contained 50?ng of cDNA, 200?nmol/l of each primer, 5?μM of probe and 25?μl of TaqMan Universal PCR Master Mix (ABI) in a final volume of 50?μl. The cDNA was subjected to 50 cycles of a two-step PCR consisting of 15?s denaturation at 95°C and 1?min combined annealing/extension at 60°C. Plasmids were diluted in a precise series, ranging from 50?pg to 0.005 fg (2 × 107 to two copies). For normalization of each target in the samples, the copy number of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The normalized values of maspin mRNA were expressed as the ratio of maspin copy number per 104 copy number of GAPDH, as previously described.11
ChIP Assay
The ChIP assay with acetyl-histone H3 and H4, and dimethyl histone H3 (Lys9) was performed according to the manufacturer's instructions (Upstate Biotech, Lake Placid, NY, USA) with slight modifications. Cells were rinsed in 1 × HBSS with 0.1% EDTA and treated with 1% formaldehyde for 10?min at 37°C to form DNA–protein crosslinks. The cells were rinsed in ice-cold 1 x HBSS containing 0.1% EDTA and protease inhibitors (1?mM PMSF/1?μg/ml aprotinin/1?μg/ml pepstatin A), scraped and collected by centrifugation at 4°C. Cells were then resuspended in PIPES buffer (5?mM PIPES, pH 8.0/85?mM KCl/0.5% NP40) containing protease inhibitors and incubated for 10?min on ice. Cells were then collected by centrifugation and resuspended in SDS lysis buffer containing protease inhibitors and incubated on ice for 10?min. The DNA–protein complexes were sonicated to lengths between 200 and 1000?bp as determined by gel electrophoresis. Samples were centrifuged at 14?000?rpm at 4°C to spin out cell debris, and then the supernatant was diluted 10-fold with ChIPs dilution buffer containing protease inhibitors. One-tenth of the sample was set aside for input control, and the remaining sample was then precleared with Salmon Sperm DNA/Protein A Agarose (Upstate Biotech, Lake Placid, NY, USA). Following preclearing, the samples were split into thirds, with two of the three samples being treated with each antibody, while the third was left as a minus antibody (−Ab) control. All samples were rotated overnight at 4°C. The chromatin–antibody complexes were collected with Salmon Sperm DNA/Protein A Agarose and then sequentially washed with the manufacturer's low salt high salt and LiCl buffers, then twice with TE buffer (10?mM Tris-HCl, 1?mM EDTA). The chromatin–antibody complexes were eluted and the DNA–protein crosslinks were reversed with 400?mM NaCl at 65°C for 6?h for all samples, including the input DNA control. All samples were treated with proteinase K, and each antibody-enriched fraction of genomic DNA was recovered by phenol/chloroform extraction and ethanol precipitations.
The RQ-PCR assay was used to analyze ChIP DNA with the ABI Prism 7700 sequence detector (ABI). Primers and fluorogenic probes for the maspin and GAPDH promoters were designed with Primer Express software (ABI): maspin CF (5′-CCA CCA ACG TGT CTG AGA AAT T-3′), maspin CR (5′-TCT TGG CAG CTT GTC CAC A-3′), maspin Cprobe (5′-FAM-CAC AGC CCC TTC CTG CCC GAA C-TAMURA-3′), GAPDH CF (5′-CTA GTG TCC TGC TGC CCA C-3′), GAPDH CR (5′-AGG TCT TGA GGC CTG AGC TAC-3′), GAPDH Cprobe (5′-FAM-AGC GGT TTT ACG GGC-TAMURA-3′). PCR conditions and reaction mixtures were the same as for the quantification of maspin mRNA. A standard curve method was used for the quantitative evaluation of ChIP DNAs. Plasmids into which maspin or GAPDH PCR products had been inserted were diluted in a precise series, ranging from 5?pg to 0.005?fg (2 × 106 to two copies). For normalization of each target in the samples, the copy number of the input control was used. The normalized values of maspin ChIPs DNA were expressed as the ratio of maspin copy numbers of the acetyl-histone H3-, acetyl-histone H4- or dimethyl histone H3 (Lys9)-enriched fractions to each input.
Results
Immunohistochemistry for Maspin in Primary Hepato-Biliary Tract Carcinomas
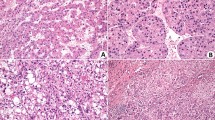
We immunohistochemically screened maspin expression in 40 hepato-biliary tract carcinomas. Immunoreactivity for maspin was completely negative in all HCCs (0/15, 0%) and their corresponding background hepatocytes, whereas maspin expression was highly positive in biliary tract carcinomas (Figure1 and Table 1). No immunoreactivity for maspin was also observed in normal hepatocytes, intrahepatic bile duct common bile duct and gallbladder from autopsy cases of noncancerous disease (Figure 1). Immunoreactivity and the incidence of maspin-positive cells are summarized in Table 1. The high incidence of maspin overexpression in biliary tract carcinomas led us to speculate that cell-type-specific maspin repression was frequently disrupted in these carcinomas, whereas the repressive character was preserved in HCCs.
Immunohistochemistry for maspin in primary hepato-biliary tract carcinomas; (a) hepatocellular; (b) cholangiocellular; (c) common bile duct; (d) gallbladder carcinomas; (e) normal liver; (f) normal gallbladder; (all scale bars=200?μm). Immunoreactivity for maspin is observed in the cholangiocellular, common bile duct and gallbladder carcinomas, but not in the hepatocellular carcinoma. Normal hepatocytes, biliary ducts (arrows), and gallbladder epithelium were completely negative for maspin.
Maspin mRNA and Protein Expression in Hepato-Biliary Tract Carcinoma Cell Lines
Overexpression of maspin protein was observed in two (HuCCT1 and TFK-1) of three CBDC cell lines, the GBC cell line (OCUG-1) and the PC cell line (PK-8) (Figure 2a). Only one (HT17) of the six HCC cell lines exhibited a maspin signal, and this was relatively weak (Figure 2b). One of the CBDC cell lines (HuH-28) and 5 of the HCC cell lines (HepG2, Hep3B, HuH-7, Li-7 and PLC/RPF/5) were negative (Figure 2). Maspin mRNA expression correlated well with the Western blotting results (Table 2). Thus, maspin mRNA and protein expression by human hepato-biliary tract carcinoma cell lines was compatible with the immunostaining results in the primary tumors.
Western blotting analyses of maspin expression in pancreatico-biliary tract (a) and hepatocellular (b) carcinoma cell lines. Equal loading was confirmed by incubation with an anti-actin antibody (43 kD, C-2; Santa Cruz Biotechnology). (a) Maspin protein expression was observed frequently in pancreatico-biliary tract carcinoma cell lines, and was only undetectable in the HuH-28 common bile duct carcinoma cell line. (b) A relatively weaker signal for maspin was observed in only one HCC cell line (HT17); the other HCC cell lines were negative.
Methylation Status at the Maspin Promoter in Hepato-Biliary Tract Carcinoma Cell Lines
The maspin promoter of the maspin-negative CBDC cell line (HuH-28) was relatively hypermethylated, but those of the maspin-positive cell lines originating from pancreatico-biliary tract carcinomas were hypomethylated (Figure 3a). Surprisingly, all HCC cell lines exhibited extensive hypomethylation at the maspin promoter (Figure 3b). Further, bisulfite-modified genomic DNA sequencing showed extensive hypermethylation in normal tissues of the liver and of the epithelia of the gallbladder and the common bile duct (Figure 4). In previous studies, demethylation status at the maspin promoter and aberrant maspin gene expression were well correlated, whereas in the HCC cell lines the sequencing results are inconsistent with mRNA/protein expression status.
Summary of 5-methylcytosine levels obtained by bisulfite-modified genomic DNA sequencing of the maspin promoter in pancreatico-biliary tract (a) and hepatocellular (b) carcinoma cell lines. In all, 10 cloned PCR products were sequenced to determine the percentage methylation of the 19 CpG sites in the region analyzed. The y-axis represents percentage cytosine methylation and the x-axis shows the nucleotide position relative to the transcription start site. Each row of circles represents the methylation pattern obtained from individual clones of the maspin gene promoter, with the number of the clone at the right. Each circle represents a CpG dinucleotide site. The filled circles are methylation-positive and the open circles are methylation-negative. The number above the circle indicates the position of each CpG site from the major transcription start site. The allele status of each clone at the SNP site (−156 from the major transcription start site) is indicated on the right.
Summary of 5-methylcytosine levels obtained by bisulfite-modified genomic DNA sequencing of the maspin promoter in the normal liver and epithelia of the gallbladder and common bile duct. Ten cloned PCR products were sequenced for each autopsy case, and the percentage methylation of the 19 CpG sites in the region was analyzed. The percentage shown is the average of the three autopsy cases.
Treatment with TSA and 5-Aza-dC, and ChIP Assay
To test whether cytosine methylation or histone acetylation in the maspin promoter is functionally linked to its silencing in HCC cell lines, we treated the cells with 5-Aza-dC, a demethylating drug, and TSA, a histone acetylating drug.
After treatment with 5-Aza-dC, gain of maspin expression was observed only in one cell line (HT17), and the others did not exhibit maspin expression even after 5-Aza-dC treatment (Figure 5b). The response of each cell line was compatible with the result of bisulfite-modified genomic DNA sequencing. We also examined the effects of 5-Aza-dC treatment at 2 and 50?μM, but the responses at 2?μM were almost identical and those at 50?μM were not evaluable because of strong cytotoxicity (data not shown).
Western blotting analyses of maspin expression before (a) and after treatment with 5-Aza-dC (b) or TSA (c) in hepatocellular carcinoma cell lines. Equal loading was confirmed by incubation with an antiactin antibody (C-2, Santa Cruz Biotechnology). (a) Maspin protein expression was observed in only one cell line (HT17). (b) A relative signal increase was observed in HT17 after treatment with 5-Aza-dC. (c) Gain of maspin expression was observed in three HCC cell lines (Hep3B, HT17, and PLC/RPF/5) after treatment with TSA.
In three (Hep3B, HT17 and PLC/RPF/5) of the six HCC cell lines, maspin expression was activated by treatment with TSA (Figure 5c). The RQ-PCR assay for maspin mRNA showed identical results (Table 2). To assess whether the chromatin structure at the maspin promoter participates in the regulation of this gene, we performed ChIP experiments with antibodies directed against acetylated histones H3 and H4 or dimethyl histone H3 (Lys9), and amplified the maspin promoter of the hepato-biliary carcinoma cell lines (Figure 6). Three independent experiments were performed in each cell line. The acetylation status of these nucleosome constituents was tightly correlated with cytosine methylation status and gene expression in biliary tract carcinoma cell lines (Table 3). On the other hand, maspin-negative HCC cell lines exhibiting a hypomethylated promoter showed a chromatin structure with hypoacetylated histones (Table 3). After treatment with TSA, increased histone acetylation was observed in three (Hep3B, HT17 and PLC/RPF/5) of the six HCC cell lines, although the extent of histone acetylation in the HCC cell lines was less than that in the pancreatico-biliary tract carcinomas. We failed to demonstrate any differences in the degree of histone methylation (dimethyl H3K9) between maspin-negative cell lines in comparison with positive cell lines, using the same primer for the ChIP assay with acetyl-histone H3 and H4 antibodies (Table 3). Table 4 gives a summary of maspin mRNA/protein expression, DNA methylation and histone acetylation status in the hepatocellular carcinoma cell lines.
Quantitative chromatin immunoprecipitation assay for maspin promoter in a common bile duct carcinoma cell line (HuCCT1) (a and b) and an HCC cell line (Hep3B) (c and d) with acetyl-histone H3 (a and f) and H4 (b, d and g) antibodies. Gene promoter-specific RQ-PCR was carried out on DNA from the immunoprecipitated chromatin (a, b, d, f and g) as well as input DNA (c, e and h). (a) Representative amplification plots of HuCCT1 (a, b and c). Fluorescent signals of H3 (a) or H4 (b) are observed at earlier cycles than that of input. The degree of histone acetylation of a common bile duct carcinoma cell line (HuCCT1) is markedly higher than that in a hepatocellular carcinoma cell line (Hep3B) (see Figure 1c). As it has been confirmed that the acetylation level of HuCCT1 is higher, we did not examine TSA treatment. (c) Representative amplification plots of Hep3B before (d and e) and after (f, g and h) treatment with TSA. No amplification plot was observed for H3 before treatment with TSA. We judged that the histone H3 acetylation status of the hepatocellular carcinoma cell line, Hep3B, was absent or extremely low. Increasing histone acetylation was observed in H3 (f) and H4 (d–g) before and after treatment with TSA. (b and d), Standard curve for the RQ-PCR.
Discussion
The high incidence of aberrant maspin expression in biliary tract carcinomas suggests that they share the same genetic pathway with pancreatic carcinomas (PCs).19, 20 In previous reports, gain of maspin expression was correlated well with demethylation at the maspin promoter region.11, 12, 13, 16, 19, 21 Although the methylation status of biliary tract carcinoma cell lines was consistent with this pattern, most maspin-negative HCC cell lines surprisingly exhibited not only hypomethylated but also hypoacetylated CpG islands. In other types of tumors, hypomethylation at the maspin promoter is usually concomitant with hyperacetylation status.22 The unexpected epigenetic status of HCC cell lines suggested the existence of other repressive factors controlling maspin expression.
Zou et al23 identified two closely spaced p53 consensus binding sites in the maspin promoter region, and found that wild-type p53 is a positive transcription factor in human prostatic cancer cell lines. Several immunohistochemical studies have suggested inverse correlations between mutant p53 and maspin expression.7, 24, 25 Recently, Oshiro et al19 demonstrated that mutations of the p53 gene and aberrant cytosine methylation contribute to silencing of maspin expression in breast cancer cell lines. We screened p53 gene mutations in our series of cell lines. Four of the six HCC cell lines and three of the five pancreatico-biliary tract carcinoma cell lines had p53 gene mutations (data not shown). There was no correlation between the presence of p53 gene mutations and maspin expression. If mutant p53 negatively controls maspin expression in primary biliary tract carcinomas, the incidence of aberrant maspin expression detected by immunohistochemistry would be lower, since the incidence of p53 gene mutations is over 50% and equal to that of HCCs.26, 27 Thus, loss of p53 function as a result of mutation might be insufficient to repress aberrant maspin expression in pancreatico-biliary tract carcinomas, whereas it contributes to silencing of maspin expression in tumors where maspin acts as a tumor suppressor, such as breast cancers.21
The question remains as to why some HCC cell lines with hypomethylation at the maspin promoter do not gain aberrant maspin expression after treatment with TSA. Histone H3 lysine 9 (H3K9) methylation is another epigenetic factor for silencing gene expression.28, 29 Histone methylation does not always correlate with DNA methylation. We failed to demonstrate the gain of H3K9 methylation in HCC cell lines in comparison with maspin-positive cell lines in experiments using the same primer for ChIP assay with acetyl-histone H3 and H4 antibodies. A clear explanation for maspin repression in HCC has not yet been provided, since in this study we did not systematically examine both methylation and acetylation status at the maspin promoter. Although gain of maspin expression in some HCC cell lines may be caused by histone acetylation, further studies are needed to define the mechanism of repression of the maspin gene in HCC cell lines.
References
Zou Z, Anisowicz A, Hendrix MJ, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science 1994;263:526–529.
Odero-Marah VA, Khalkhali-Ellis Z, Chunthapong J, et al. Maspin regulates different signaling pathways for motility and adhesion in aggressive breast cancer cells. Cancer Biol Ther 2003;2:398–403.
Pemberton PA, Wong DT, Gibson HL, et al. The tumor suppressor maspin does not undergo the stressed to relaxed transition or inhibit trypsin-like serine proteases. Evidence that maspin is not a protease inhibitory serpin. J Biol Chem 1995;270:15832–15837.
Umekita Y, Ohi Y, Sagara Y, et al. Expression of maspin predicts poor prognosis in breast-cancer patients. Int J Cancer 2002;100:452–455.
Maass N, Hojo T, Rosel F, et al. Down regulation of the tumor suppressor gene maspin in breast carcinoma is associated with a higher risk of distant metastasis. Clin Biochem 2001;34:303–307.
Maass N, Teffner M, Rosel F, et al. Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol 2001;195:321–326.
Machtens S, Serth J, Bokemeyer C, et al. Expression of the p53 and Maspin protein in primary prostate cancer: correlation with clinical features. Int J Cancer 2001;95:337–342.
Xia W, Lau YK, Hu MC, et al. High tumoral maspin expression is associated with improved survival of patients with oral squamous cell carcinoma. Oncogene 2000;19:2398–2403.
Sood AK, Fletcher MS, Gruman LM, et al. The paradoxical expression of maspin in ovarian carcinoma. Clin Cancer Res 2002;8:2924–2932.
Maass N, Hojo T, Ueding M, et al. Expression of the tumor suppressor gene Maspin in human pancreatic cancers. Clin Cancer Res 2001;7:812–817.
Akiyama Y, Maesawa C, Ogasawara S, et al. Cell-type-specific repression of the maspin gene is disrupted frequently by demethylation at the promoter region in gastric intestinal metaplasia and cancer cells. Am J Pathol 2003;163:1911–1919.
Ogasawara S, Maesawa C, Yamamoto M, et al. Disruption of cell-type-specific methylation at the maspin gene promoter is frequently involved in undifferentiated thyroid cancers. Oncogene 2004;23:1117–1124.
Wada K, Maesawa C, Akasaka T, et al. Aberrant expression of the maspin gene associated with epigenetic modification in melanoma cells. J Invest Dermatol 2004;122:805–811.
Holliday R, Pugh JE . DNA modification mechanisms and gene activity during development. Science 1975;187:226–232.
Riggs AD . X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 1975;14:9–25.
Futscher BW, Oshiro MM, Wozniak RJ, et al. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet 2002;31:175–179.
Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res 1994;22:2990–2997.
Maass N, Biallek M, Rosel F, et al. Hypermethylation and histone deacetylation lead to silencing of the maspin gene in human breast cancer. Biochem Biophys Res Commun 2002;297:125–128.
Fitzgerald M, Oshiro M, Holtan N, et al. Human pancreatic carcinoma cells activate maspin expression through loss of epigenetic control. Neoplasia 2003;5:427–436.
Ohike N, Maass N, Mundhenke C, et al. Clinicopathological significance and molecular regulation of maspin expression in ductal adenocarcinoma of the pancreas. Cancer Lett 2003;199:193–200.
Domann FE, Rice JC, Hendrix MJ, et al. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer 2000;85:805–810.
Oshiro MM, Watts GS, Wozniak RJ, et al. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene 2003;22:3624–3634.
Zou Z, Gao C, Nagaich AK, et al. p53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem 2000;275:6051–6054.
Zhang W, Zhang M . Tissue microarray analysis of maspin expression and its reverse correlation with mutant p53 in various tumors. Int J Oncol 2002;20:1145–1150.
Song SY, Lee SK, Kim DH, et al. Expression of maspin in colon cancers: its relationship with p53 expression and microvessel density. Dig Dis Sci 2002;47:1831–1835.
Dong M, Nio Y, Yamasawa K, et al. p53 alteration is not an independent prognostic indicator, but affects the efficacy of adjuvant chemotherapy in human pancreatic cancer. J Surg Oncol 2003;82:111–120.
Weihrauch M, Lehnert G, Kockerling F, et al. p53 mutation pattern in hepatocellular carcinoma in workers exposed to vinyl chloride. Cancer 2000;88:1030–1036.
Rougeulle C, Chaumeil J, Sarma K, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol 2004;24:5475–5484.
Soejima H, Nakagawachi T, Zhao W, et al. Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene 2004;23:4380–4388.
Acknowledgements
This work was supported, in part, by the Open Translational Research Project, Advanced Medical Science Center, Iwate Medical University and Grants-in-Aid 16590290 from the Ministry of Education, Science, Sports and Culture of Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fujisawa, K., Maesawa, C., Sato, R. et al. Epigenetic status and aberrant expression of the maspin gene in human hepato-biliary tract carcinomas. Lab Invest 85, 214–224 (2005). https://doi.org/10.1038/labinvest.3700214
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/labinvest.3700214
Keywords
This article is cited by
-
Cytologic predictors of malignancy in bile duct brushings: a multi-reviewer analysis of 60 cases
Modern Pathology (2017)
-
An immunohistochemical marker panel including claudin-18, maspin, and p53 improves diagnostic accuracy of bile duct neoplasms in surgical and presurgical biopsy specimens
Virchows Archiv (2015)
-
Effect of epigenetic modification of maspin on extravillous trophoblastic function
Journal of Huazhong University of Science and Technology [Medical Sciences] (2012)
-
Maspin: molecular mechanisms and therapeutic implications
Cancer and Metastasis Reviews (2012)
-
DNA methylation profiles of protease nexin 1 (SERPINE2) gene in human cell lines
Chinese Journal of Cancer Research (2011)