Abstract
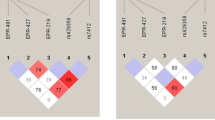
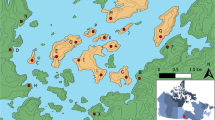
Communities with increased shared ancestry represent invaluable tools for genetic studies of complex traits. ‘1001 Dalmatians’ research program collects biomedical information for genetic epidemiological research from multiple small isolated populations (‘metapopulation’) in the islands of Dalmatia, Croatia. Random samples of 100 individuals from 10 small island settlements (n<2000 inhabitants) were collected in 2002 and 2003. These island communities were carefully chosen to represent a wide range of distinct and well-documented demographic histories. Here, we analysed their genetic make-up using 26 short tandem repeat (STR) markers, at least 5 cM apart. We found a very high level of differentiation between most of these island communities based on Wright's fixation indexes, even within the same island. The model-based clustering algorithm, implemented in STRUCTURE, defined six clusters with very distinct genetic signatures, four of which corresponded to single villages. The extent of background LD, assessed with eight linked markers on Xq13-21, paralleled the extent of differentiation and was also very high in most of the populations under study. For each population, demographic history was characterised and 12 ‘demographic history’ variables were tentatively defined. Following stepwise regression, the demographic history variable that most significantly predicted the extent of LD was the proportion of locally born grandparents. Strong isolation and endogamy are likely to be the main forces maintaining this highly structured overall population.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Peltonen L, Palotie A, Lange K : Use of population isolates for mapping complex traits. Nat Rev Genet 2000; 1: 182–190.
Wright AF, Carothers AD, Pirastu M : Population choice in mapping genes for complex diseases. Nat Genet 1999; 23: 397–404.
Heutink P, Oostra BA : Gene finding in genetically isolated populations. Hum Mol Genet 2002; 11: 2507–2515.
Varilo T, Peltonen L : Isolates and their potential use in complex gene mapping efforts. Curr Opin Genet Dev 2004; 14: 316–323.
Fraumene C, Petretto E, Angius A, Pirastu M : Striking differentiation of sub-populations within a genetically homogeneous isolate (Ogliastra) in Sardinia as revealed by mtDNA analysis. Hum Genet 2003; 114: 1–10.
Bulayeva K, Jorde LB, Ostler C, Watkins S, Bulayev O, Harpending H : Genetics and population history of Caucasus populations. Hum Biol 2003; 75: 837–853.
Marjanovic D, Kapur L, Drobnic K, Budowle B, Pojskic N, Hadziselimovic R : Comparative study of genetic variation at 15 STR loci in three isolated populations of the Bosnian mountain area. Hum Biol 2004; 76: 15–31.
Rudan P, Simic D, Smolej-Narancic N et al: Isolation by distance in Middle Dalmatia-Yugoslavia. Am J Phys Anthropol 1987; 74: 417–426.
Waddle DM, Sokal RR, Rudan P : Factors affecting population variation in eastern Adriatic isolates (Croatia). Hum Biol 1998; 70: 845–864.
Rudan I, Campbell H, Rudan P : Genetic epidemiological studies of eastern Adriatic Island isolates, Croatia: objective and strategies. Coll Antropol 1999; 23: 531–546.
Roberts DF, Noor ZM, Papiha SS, Rudan P : Genetic variation in Brac, Croatia. Ann Hum Biol 1992; 19: 539–557.
Martinovic I, Mastana S, Janicijevic B et al: VNTR DNA variation in the population of the island of Hvar, Croatia. Ann Hum Biol 1998; 25: 489–499.
Tolk HV, Pericic M, Barac L et al: MtDNA haplogroups in the populations of Croatian Adriatic Islands. Coll Antropol 2000; 24: 267–280.
Barac L, Pericic M, Klaric IM et al: Y chromosomal heritage of Croatian population and its island isolates. Eur J Hum Genet 2003; 11: 535–542.
Forenbaher S : The earliest islanders of the eastern Adriatic. Coll Antropol 1999; 23: 521–530.
Rudan I, Stevanovic R, Vitart V et al: Lost in transition –the Island of Susak (1951–2001). Coll Antropol 2004; 28: 403–421.
Laan M, Paabo S : Demographic history and linkage disequilibrium in human populations. Nat Genet 1997; 17: 435–438.
Nei M, Roychoudhury AK : Sampling variances of heterozygosity and genetic distance. Genetics 1974; 76: 379–390.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Cavalli-Sforza LL, Edwards AW : Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet 1967; 19 (Suppl 19): 233.
Pritchard JK, Stephens M, Donnelly P : Inference of population structure using multilocus genotype data. Genetics 2000; 155: 945–959.
Li N, Stephens M : Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 2003; 165: 2213–2233.
Hedrick PW : Gametic disequilibrium measures: proceed with caution. Genetics 1987; 117: 331–341.
Aulchenko YS, Axenovich TI, Mackay I, van Duijn CM : miLD and booLD programs for calculation and analysis of corrected linkage disequilibrium. Ann Hum Genet 2003; 67: 372–375.
Kaessmann H, Zollner S, Gustafsson AC et al: Extensive linkage disequilibrium in small human populations in Eurasia. Am J Hum Genet 2002; 70: 673–685.
Katoh T, Mano S, Ikuta T et al: Genetic isolates in East Asia: a study of linkage disequilibrium in the X chromosome. Am J Hum Genet 2002; 71: 395–400.
Angius A, Bebbere D, Petretto E et al: Not all isolates are equal: linkage disequilibrium analysis on Xq13.3 reveals different patterns in Sardinian sub-populations. Hum Genet 2002; 111: 9–15.
Latini V, Sole G, Doratiotto S et al: Genetic isolates in Corsica (France): linkage disequilibrium extension analysis on the Xq13 region. Eur J Hum Genet 2004; 12: 613–619.
Vitart V, Carothers AD, Hayward C et al: Increased level of linkage disequilibrium in rural compared with urban communities: a factor to consider in association-study design. Am J Hum Genet 2005; 76: 763–772.
Budowle B, Shea B, Niezgoda S, Chakraborty R : CODIS STR loci data from 41 sample populations. J Forensic Sci 2001; 46: 453–489.
Martinovic Klaric I, Jin L, Chakraborty R et al: Inter- and intra-island genetic diversity in Adriatic populations of Croatia: implications for studying complex diseases in isolated populations. Am J Hum Genet 2001; 68: 1237M.
Workman PL, Lucarelli P, Agostino R et al: Genetic differentiation among Sardinian villages. Am J Phys Anthropol 1975; 43: 165–176.
Harding RM, McVean G : A structured ancestral population for the evolution of modern humans. Curr Opin Genet Dev 2004; 14: 667–674.
Wakeley J : Metapopulation models for historical inference. Mol Ecol 2004; 13: 865–875.
Eller E, Hawks J, Relethford JH : Local extinction and recolonization, species effective population size, and modern human origins. Hum Biol 2004; 76: 689–709.
Rosenberg NA, Pritchard JK, Weber JL et al: Genetic structure of human populations. Science 2002; 298: 2381–2385.
Laan M, Wiebe V, Khusnutdinova E, Remm M, Paabo S : X-chromosome as a marker for population history: linkage disequilibrium and haplotype study in Eurasian populations. Eur J Hum Genet 2005; 13: 452–462.
Fischer J, Bouadjar B, Heilig R et al: Mutations in the gene encoding SLURP-1 in Mal de Meleda. Hum Mol Genet 2001; 10: 875–880.
Bohacek N : Tristan da Cunha and Susak. Lijec Vjesn 1964; 86: 1412–1416.
Falchi M, Forabosco P, Mocci E et al: A genomewide search using an original pairwise sampling approach for large genealogies identifies a new locus for total and low-density lipoprotein cholesterol in two genetically differentiated isolates of Sardinia. Am J Hum Genet 2004; 75: 1015–1031.
Williams-Blangero S, Correa-Oliveira R, Vandeberg JL et al: Genetic influences on plasma cytokine variation in a parasitized population. Hum Biol 2004; 76: 515–525.
Blangero J : Localization and identification of human quantitative trait loci: king harvest has surely come. Curr Opinon Genet Dev 2004; 14: 233–240.
Acknowledgements
The authors are indebted to the study participants and to many students of the faculty of Medicine, University of Zagreb, local general practitioners and nurses, employees of the University of Rijeka and Split, the Croatian Institute of Public Health and the Institutes of Public Health in Split and Dubrovnik, for their help in the field work. We thank HGU MRC staff, Susan Fitzsimmons and Isla Campbell for help in genotyping, and Douglas Stuart for figure reproduction. We also thank Jim Wilson for critical reading of the manuscript. VV holds an MRC Special Training Fellowship in Bioinformatics. This work was supported by the British Scholarship Trust (IR and ZB), the University of Zagreb (‘Miroslav Cackoviç’ fellowship, ZB), Overseas Research Scheme (IR), the University of Edinburgh (scholarships, IR and OP), the Croatian Ministry of Science (Grant no 0108330 to IR), the British Council, the Wellcome Trust, the Royal Society (HC and IR), and the MRC.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Vitart, V., Biloglav, Z., Hayward, C. et al. 3000 years of solitude: extreme differentiation in the island isolates of Dalmatia, Croatia. Eur J Hum Genet 14, 478–487 (2006). https://doi.org/10.1038/sj.ejhg.5201589
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201589
Keywords
This article is cited by
-
The ethnobotany and biogeography of wild vegetables in the Adriatic islands
Journal of Ethnobiology and Ethnomedicine (2019)
-
Socioeconomic inequalities show remarkably poor association with health and disease in Southern Croatia
International Journal of Public Health (2015)
-
Inference of identity by descent in population isolates and optimal sequencing studies
European Journal of Human Genetics (2013)
-
Does inbreeding affect N-glycosylation of human plasma proteins?
Molecular Genetics and Genomics (2011)
-
Comparison of participant information and informed consent forms of five European studies in genetic isolated populations
European Journal of Human Genetics (2010)