Abstract
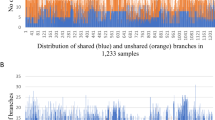
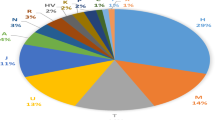
The initial belief that haplotype block boundaries and haplotypes were largely shared across populations was a foundation for constructing a haplotype map of the human genome using common SNP markers. The HapMap data document the generality of a block-like pattern of linkage disequilibrium (LD) with regions of low and high haplotype diversity but differences among the populations. Studies of many additional populations demonstrate that LD patterns can be highly variable among populations both across and within geographic regions. Because of this variation, emphasis has shifted to the generalizability of tagSNPs, those SNPs that capture the bulk of variation in a region. We have examined the LD and tagSNP patterns based upon over 2000 individual samples in 38 populations and 134 SNPs in 10 genetically independent loci for a total of 517 kb with an average density of 1 SNP/5 kb. Four different ‘block’ definitions and the pairwise LD tagSNP selection algorithm have been applied. Our results not only confirm large variation in block partition among populations from different regions (agreeing with previous studies including the HapMap) but also show that significant variation can occur among populations within geographic regions. None of the block-defining algorithms produces a consistent pattern within or across all geographic groups. In contrast, tagSNP transferability is much greater than the similarity of LD patterns and, although not perfect, some generalizations of transferability are possible. The analyses show an asymmetric pattern of tagSNP transferability coinciding with the subsetting of variation attributed to the spread of modern humans around the world.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Risch N, Merikangas K : The future of genetic studies of complex human diseases. Science 1996; 273: 1516–1517.
Kidd KK, Morar B, Castiglione CM et al: A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Hum Genet 1998; 103: 211–227.
Kidd JR, Pakstis AJ, Zhao H et al: Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 2000; 66: 1882–1899.
Reich DE, Cargill M, Bolk S et al: Linkage disequilibrium in the human genome. Nature 2001; 411: 199–204.
Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES : High-resolution haplotype structure in the human genome. Nat Genet 2001; 29: 229–232.
Patil N, Berno AJ, Hinds DA et al: Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 2001; 294: 1719–1723.
Gabriel SB, Schaffner SF, Nguyen H et al: The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
International HapMap Consortium: International HapMap project. Nature 2003; 426: 789–796.
International HapMap Consortium: A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Clark AG, Weiss KM, Nickerson DA et al: Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 1998; 63: 595–612.
Templeton AR, Clark AG, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF : Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet 2000; 66: 69–83.
Wang N, Akey JM, Zhang K, Chakraborty R, Jin L : Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am J Hum Genet 2002; 71: 1227–1234.
González-Neira A, Calafell F, Navarro A et al: Geographic stratification of linkage disequilibrium: a worldwide population study in a region of chromosome 22. Hum Genomics 2004; 1: 399–409.
González-Neira A, Ke X, Lao O et al: The portability of tagSNPs across populations: a worldwide survey. Genome Res 2006; 16: 323–330.
Sawyer SL, Mukherjee N, Pakstis AJ et al: Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet 2005; 13: 677–686.
Liu N, Sawyer SL, Mukherjee N et al: Haplotype block structures show significant variation among populations. Genet Epidemiol 2004; 27: 385–400.
Tishkoff SA, Kidd KK : Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet 2004; 36 (Suppl 11): S21–S27.
Osier MV, Cheung KH, Kidd JR, Pakstis AJ, Miller PL, Kidd KK : ALFRED: an allele frequency database for diverse populations and DNA polymorphisms – an update. Nucleic Acids Res 2001; 29: 317–319.
Osier MV, Cheung KH, Kidd JR, Pakstis AJ, Miller PL, Kidd KK : ALFRED: an allele frequency database for anthropology. Am J Phys Anthropol 2002; 119: 77–83.
Wright S : Evolution and the Genetics of Populations. Vol 2: The Theory of Gene Frequencies. University of Chicago Press: Chicago IL, 1969.
Gu S, Pakstis AJ, Kidd KK : HAPLOT: a graphical comparison of haplotype blocks, tagSNP sets and SNP variation for multiple populations. Bioinformatics 2005; 21: 3938–3939.
Wang N, Deng M, Chen T, Waterman MS, Sun F : A dynamic programming algorithm for haplotype partitioning. Proc Natl Acad Sci USA 2002; 99: 7335–7339.
Barrett JC, Fry B, Maller J, Daly MJ : Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
Clayton D : http://www.nature.com/ng/journal/v29/n2/extref/ng1001-233-S10.pdf, 2001.
Johnson GCL, Esposito L, Barratt BJ et al: Haplotype tagging for the identification of common disease genes. Nat Genet 2001; 29: 233–237.
Nothnagel M, Furst R, Rhode K : Entropy as a measure for linkage disequilibrium over multilocus haplotype blocks. Hum Hered 2003; 54: 186–198.
de Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D : Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217–1223.
de Bakker PIW, Graham RR, Alshuler D, Henderson BE, Haiman CA : Transferability of Tag SNPs to capture common genetic variation in DNA repair genes across multiple populations. Pacific Symp Biocomput 2006; 11: 478–486.
Abecasis GR, Cookson WO : GOLD – graphical overview of linkage disequilibrium. Bioinformatics 2000; 16: 182–183.
Wall JD, Pritchard JK : Assessing the performance of the haplotype block model of linkage disequilibrium. Am J Hum Genet 2003; 73: 502–515.
Kidd KK, Pakstis AJ, Speed WC, Kidd JR : Understanding human DNA sequence variation. J Hered 2004; 95: 406–420.
Hey J : On the number of New World founders: a population genetic portrait of the peopling of the Americas. PLoS Biol 2005; 3: e193.
Fallin D, Schork NJ : Accuracy of haplotype frequency estimation for biallelic loci, via the Expectation-Maximization Algorithm for unphased diploid genotype data. Am J Hum Genet 2000; 67: 947–959.
Shifman S, Darvasi A : The value of isolated populations. Nat Genet 2001; 28: 309–310.
Terwilliger JD, Hiekkalinna T : An utter refutation of the ‘Fundamental Theorem of the HapMap’. Eur J Hum Genet 2006; 14: 426–437.
Acknowledgements
We thank F Black, B Bonne-Tamir, L Cavalli-Sforza, K Dumars, J Friedlaender, D Goldman, E Grigorenko, SLB Kajuna, NJ Karoma, KS Kendler, WC Knowler, S Kungulilo, D Lawrence, R-B Lu, A Odunsi, F Okonofua, F Oronsaye, J Parnas, L Peltonen, LO Schulz, D Upson, D Wallace, KM Weiss, S Williams, OV Zhukova for helping assemble the diverse population collection used in this study. Some cell lines were made available by the Coriell Institute for Medical Research and by the National Laboratory for the Genetics of Israeli Populations. Special thanks are given to the many hundreds of individuals who volunteered to give blood samples for studies. This work was supported in part by NIH GM57672.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Gu, S., Pakstis, A., Li, H. et al. Significant variation in haplotype block structure but conservation in tagSNP patterns among global populations. Eur J Hum Genet 15, 302–312 (2007). https://doi.org/10.1038/sj.ejhg.5201751
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201751
Keywords
This article is cited by
-
Genetic analysis of novel phenotypes for farm animal resilience to weather variability
BMC Genetics (2019)
-
Application of six IrisPlex SNPs and comparison of two eye color prediction systems in diverse Eurasia populations
International Journal of Legal Medicine (2014)
-
Association of Hepatocyte Nuclear Factor 4 Alpha Polymorphisms with Type 2 Diabetes With or Without Metabolic Syndrome in Malaysia
Biochemical Genetics (2012)
-
The complex global pattern of genetic variation and linkage disequilibrium at catechol-O-methyltransferase
Molecular Psychiatry (2010)
-
Evaluation of genetic tests for susceptibility to common complex diseases: why, when and how?
Human Genetics (2010)