Abstract
Trimerotropis pallidipennis is an American grasshopper whose South American populations are polymorphic for pericentric inversions. Colonization of southern latitudes was by North American grasshoppers with basic chromosome arrangements, presumably along the Andean dry lands of South America. In Argentina, the frequencies of some of the rearrangements are correlated with geographical and climatic variables, following similar patterns among different ecological gradients, and are probably maintained by geographically variable coefficients of selection. Restriction site variation of mitochondrial DNA is used as a tool for determining the species history in relation to the formation of clines. Populations located along an altitudinal gradient, and others outside the cline, are analysed through phylogeographical studies. There is no strong geographical orientation in the unrooted tree connecting all 17 mitochondrial DNA haplotypes found. Many of them are present in most of the populations analysed, indicating high gene flow. The fact that there is no obvious differentiation in haplotype distribution between both extremes of the cline nor between chromosomally differentiated populations shows that the cline is not the result of a hybrid zone and reinforces the selection hypothesis. The estimation of the overall nucleotide divergence between the hypothetical ancestral haplotype and the other molecules shows that T. pallidipennis haplotypes started diverging from each other about 3 Myr ago. This result is in agreement with the time when the Isthmus of Panamá rose (2–3 Myr ago), probably favouring the migration of many species between both hemispheres.
Similar content being viewed by others
Introduction
The pattern of genetic variation in natural populations of species is a result of a complex interplay of natural selection and the species history. Whereas natural selection will differ in its effects on individual loci or gene clusters with epistatic effects, the influence of history should be more or less similar on all genetic markers (Hale & Singh, 1991). Sometimes the pattern of variation is seen as a clinal distribution of gene or chromosome frequencies along environmental gradients. Several models have been proposed to explain these patterns, and in some cases the clines are treated as hybrid zones.
South American populations of Trimerotropis pallidipennis (2n=23, X0 in males), polymorphic for three to seven pericentric inversions in four medium-sized chromosome pairs, exhibit a clinal distribution of inversion polymorphisms (Confalonieri & Colombo, 1989; Confalonieri, 1994; Matrajt et al., 1996). In fact, three inversions (4 AI, 7 SM2, 8 SM4) correlate simultaneously with altitude, latitude and minimum temperature, and two other inversions (6 M and 8 SM4) correlate with longitude and humidity in such a way that the frequency distribution of these chromosome rearrangements could be predicted for previously unstudied regions (Colombo & Confalonieri, 1996). According to White (1973) the colonization of southern latitudes by T. pallidipennis was probably by a North American grasshopper with basic chromosome arrangement, presumably along the Andean dry lands of South America. From here, it would have extended its distribution to lower altitudes (Vaio et al., 1979), where inversion sequences appear in polymorphic or fixed states. Chromosome clines observed in T. pallidipennis do not obviously fit any hybrid-zone model and are most probably maintained by geographically variable coefficients of selection (Matrajt et al., 1996).
The application of mitochondrial DNA (mtDNA) studies to population genetics has lead to a shift towards historical, phylogenetic perspectives on intraspecific population structure (Avise, 1994). Multiple alleles or haplotypes within a species can be ordered phylogenetically, yielding intraspecific phylogenies (gene genealogies), which can explain the types of population structures revealed (Avise et al., 1987).
Here we present the analysis of RFLP of mitochondrial DNA in T. pallidipennis as a tool for determining the species history in relation to the formation of current clines. For this purpose we studied the RFLPs of four populations belonging to an altitudinal cline, and of two other geographically isolated populations. We performed a phylogeographical study of the haplotypes found, and we discuss the observed genealogical lineages in the light of the different models for the origin of clines and the hypothetical migration routes proposed for this species.
Materials and methods
Population sampling
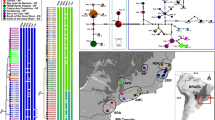
The locations of collection sites and sample sizes are shown in Fig. 1 and Tables 1 and 3, respectively. Four populations (Observatorio, Tunuyán, Uspallata and Puente del Inca) were collected along an altitudinal gradient which ranged from 827 m (Observatorio) to 2720 m above sea level (Puente del Inca). The first three belong to the biogeographical region known as Provincia del Monte belonging to the Chaco Dominion; they are sited at lower altitudes, with a prevalence of shrubs and a warm, dry climate. Puente del Inca is located in the Provincia Altoandina belonging to the Dominio Andino-Patagónico; it is at a higher altitude, with scarce vegetation consisting of shrubs and grasses and a moderate to cold, dry climate. The Southern sample of Bariloche (863 m) also belongs to the Dominio Andino-Patagónico, but to the Provincia Patagónica with a cold, dry climate and barren plains with shrubs. The northern sample of Quilmes (2150 m) belongs to the Provincia de la Prepuna from the Chaco Dominion. The climate is dry and warm and the vegetation consists mainly of low shrubs and several cactus species (Cabrera & Willink, 1973).
All insects were collected in the field and maintained alive; at the laboratory they were gutted, and in some cases dissected for chromosomal analysis, before being frozen in liquid nitrogen and stored at −80°C for RFLP studies.
mtDNA RFLP detection
Total DNA was prepared from whole gutted animals following Marchant (1988) and digested with restriction enzymes, electrophoresed through agarose, Southern blotted onto nylon membranes, and probed using standard methods. The probes used were three pUC18-derived plasmids, containing SacI fragments of the Caledia captiva mitochondrial genome and together representing the whole molecule (Marchant, 1988), kindly provided by A. Marchant. Probes were labelled with digoxigenin-dUTP by random priming and immunodetected, following the instructions of the manufacturer (Boehringer-Mannheim). Because of the heterologous nature of these probes, hybridization was carried out overnight at 42°C and posthybridization washes were performed in low stringency conditions (15 min in 2×SSC, 0.1% SDS; 15 min in 1×SSC, 0.1% SDS; and 15 min in 0.5×SSC, 0.1% SDS; all three washes were at 42°C).
Restriction enzyme analysis
The DNA from all animals studied was cut with the restriction enzymes AluI, PstI, BamHI, ScaI, XbaI, HaeIII and EcoRI. Sizes of restriction fragments were estimated as a function of their electrophoretic mobility following Maniatis et al. (1982) by comparison with various standard DNA markers: bacteriophage EcoRI–HindIII double digest (Promega) and φDNA–HaeIII digests (Promega). Most individuals could be characterized by four enzymes, XbaI, ScaI, HaeIII and EcoRI, which showed more than one restriction pattern to give a composite mtDNA genotype. These composite haplotypes were designated as a sequence of four uppercase letters, where each letter refers to a particular fragment pattern produced by one of the four restriction enzymes. For example, haplotype AAJX displayed pattern A of XbaI, pattern A of ScaI, etc. (see Table 2). Restriction maps were built by partial digestions which allowed inference of the relative position of restriction sites.
Data analysis
A data matrix consisting of presence/absence information for each restriction site in each mtDNA clone was used to compute the overall proportion of shared restriction sites between pairs of clones, and also to estimate the mtDNA sequence divergence for three categories: between-population and between-haplotype divergence, and within-population diversity. Pairwise measures of between-haplotype diversity were calculated using the maximum likelihood estimator of Nei (1987) (eqns 5.38, 5.50). Intrapopulational diversities (dx) were estimated according to eqn (10.19) of Nei (1987), which is simply an average of the haplotype distances of all possible pairwise comparisons of individuals in the population. Estimates of pairwise distances between populations (dxy) were calculated in a similar fashion (eqn 10.20 of Nei (1987)), except that pairwise comparisons were made between individuals from different populations.
The different clones were also interconnected in an unrooted phylogenetic network by means of the Wagner maximum parsimony, using the MIX program of the PHYLIP package (v. 3.5c) (Felsenstein, 1991), kindly provided by Joe Felsenstein. The CONSENSE program was employed to summarize all the most parsimonious topologies in a majority-rule consensus tree. A parsimony method was used because it was assumed that all branches had equal rates of change, given that they correspond to mitochondrial clones of the same species. This assumption was further confirmed with the consistent results obtained between maximum likelihood estimators of between-haplotype diversity and the consensus tree. The differentiation between populations was also measured by Nei's index (1987), using data on chromosome frequencies. These estimates of genetic distances were used to make an unweighted pair-grouped cluster analysis (UPGMA).
Results
Chromosome analysis
Chromosome characteristics are given in Table 1. Because testes were not available from the population of Quilmes, data from Ampimpa are given instead, a locality which is only 25 km from Quilmes and almost at the same altitude and latitude. All four populations along the altitudinal cline (Puente del Inca, Uspallata, Tunuyán and Observatorio) showed chromosome frequencies that correlate very significantly with altitude (Matrajt et al., 1996). Previous macrogeographical studies have revealed that the altitude at which monomorphism for standard sequences is reached, decreases with increasing latitude as a consequence of minimum temperature dependence (Colombo & Confalonieri, 1996). Bariloche (Table 1) shows the frequencies predicted, according to Colombo & Confalonieri (1996), by its southern latitudinal and low altitudinal situation (41° Lat. S, 836 m, respectively): monomorphism for 4 A and 8 A and a very high incidence of 6 A and 7 A standard sequences. An unweighted pair-grouped (UPGMA) method applied to Nei's genetic distances (Nei, 1987) obtained from chromosomal frequencies confirms these observations (Fig. 2a), because it shows that Bariloche is chromosomally more similar to Puente del Inca than to the other samples of the cline. It also shows that the other samples are grouped in agreement with their altitudes, the most differentiated being those of both extremes of the cline: Puente del Inca and the group formed by Observatorio.
Mitochondrial DNA analysis
All enzymes produced detectable bands totalling in size ≈16±0.5 kb. Fragment patterns and sizes for each enzyme are in Table 2. A total of 19 different morphs were observed for all seven restriction enzymes. Each revealed more than one restriction pattern (Table 2).
Seventeen different mtDNA clones were resolved for all six populations. The frequencies of these composite haplotypes in each sample are indicated in Table 3. The fragment variation could be explained by the gain or loss of particular restriction sites. The presence/absence data matrix for each restriction site and all haplotypes is given in Table 4. A total of 18 sites were studied for the polymorphic restriction enzymes of which 14 were variable and nine were informative for the parsimony analysis.
The haplotypes could be connected in a most parsimonious unrooted tree. The cladistic analysis produced 19 minimal trees of 23 steps each which were combined in a majority rule consensus tree resulting in a cladogram depicted in Fig. 3. This phylogenetic network is formed by a principal clade with haplotype BIJX occupying a central position. This suggests that BIJX is more ancestral, an idea supported also by its frequency distribution (Table 3). Most of the other haplotypes are endemic in the population where they were detected.
Unrooted phylogenetic tree of the 17 haplotypes of Trimerotropis pallidipennis mtDNA. This topology corresponds to the majority rule consensus of the 19 most parsimonious trees of length 23. The numbers on each branch indicate which site changed (from 0 to 1 or vice versa) following the nomenclature of Table 4.
If the geographical origins of mtDNA clones are superimposed on this cladogram, it can be seen that endemic haplotypes from the most geographically distant samples (Bariloche and Quilmes) are the most distantly connected in the tree. Haplotypes belonging to the altitudinal cline are distributed in the phylogenetic network without an obvious pattern, but with clones from Uspallata tending to be near those from Quilmes.
The estimates of the number of nucleotide differences per site between pairs of haplotypes (dij) were obtained by the maximum likelihood estimation method (Nei, 1987) which gave a mean value for sequence divergence of 4.8%, ranging from 0.6–16%. If this estimate is obtained from the mean divergence of sequences from the hypothetical ancestral mtDNA clone (BIJX), then the value diminishes to 3.6%.
The extent of interpopulation variation (dxy) (corrected for within-sample variation (dx) shown in Table 3) was used to apply the unweighted pair-group method, UPGMA of cluster analysis (Fig. 2b). The resulting phenetic tree shows clustering of populations that is mostly in agreement with the evolutionary relationships of clones shown by the cladogram and with geographical distances. It is not congruent with the chromosomal differentiation observed among samples (Fig. 2a). Compared with mitochondrial markers, population differentiation for chromosomal rearrangements reflects more the climatic conditions of sample locations rather than their geographical proximities.
Discussion
Several models have been proposed to explain clinal distribution of genetic markers, of which the main ones are ecological selection for broad clines and hybrid zones for narrow ones. Fundamentally, a hybrid zone is a cline or set of clines between two parapatric hybridizing taxa for genes and the distinguishing characters they determine (Hewitt, 1988). These clines may originate in two ways: primarily or secondarily. In the former case, the differences evolve in a continuous distribution, for example an environmental gradient that favours different alleles on either side; natural selection progressively sharpens the incipient cline until it becomes a narrow hybrid zone between two internally coadapted genotypes (Endler, 1986). In a secondary zone, the differences evolve while the two populations are geographically isolated, so that when their ranges alter and meet, a steep cline is formed as they hybridize (Hewitt, 1988). In this latter case, deterministic forces may not be involved in the formation of clines, but are necessary for a long continuation of this pattern of variation.
In T. pallidipennis, selection of some sort is very likely to be acting, because of the repetition of patterns of variation of inversion polymorphisms over a wide geographical area and their stability over time (Confalonieri & Colombo, 1989; Confalonieri, 1994; Colombo & Confalonieri, 1996; Matrajt et al., 1996). If the current clines were primary hybrid zones, selection could be acting against recombinant genotypes based on inversion types. However, as the populations are all polymorphic except for some at the very extremes of the distribution of the species in arid and semiarid regions of Argentina (Go¸ni et al., 1985; Confalonieri, 1994), the width of the hybrid zone seems to be too extensive, exceeding in some cases more than 1000 km and involving almost the entire distribution of populations from southern latitudes. Therefore, we proposed the hypothesis of geographically variable selection maintaining the clines (Matrajt et al., 1996).
The analysis of mtDNA site variation reported here provides new evidence favouring this hypothesis. If the clines originated as a consequence of secondary contact, then the populations sited at either side of the hybrid zone would possibly show clear genetic differentiation for mtDNA, either because gene flow would be limited by the lower fitness of recombinants or because of historical isolation (secondary zones) between both extremes of the cline. Examples of this have been observed across hybrid zones between chromosomally or morphologically differentiated taxa in other grasshopper species (Shaw et al., 1980, 1987; Marchant, 1988; Marchant & Shaw, 1993; King et al., 1996). However, in the case of T. pallidipennis there is no obvious pattern of distribution of mitochondrial haplotypes parallel to the chromosomal differentiation of populations. Furthermore, the net percentage of nucleotide differentiation between populations at the extremes of the altitudinal cline is lower than between other samples geographically distant but with more similar chromosomal characteristics.
In many studies involving phylogeographical analysis of mtDNA, one or several haplotype groups that correspond to geographical areas of the species distribution are frequently observed. These groups are associated as clades in the analyses of haplotype differentiation (Avise, 1986, 1994), and generally have the most frequent genotype in a central position connected with other genotypes that are not so widely distributed — or are even endemic — and that differ from the central haplotype in a small number of restriction sites. Both subspecies of Caledia captiva (Marchant & Shaw, 1993) and populations of Drosophila subobscura (Latorre et al. 1992), D. melanogaster (Hale & Singh, 1991) and Chelonia mydas (Encalada et al. 1996), among many other examples (Avise, 1994), conform to this typical pattern.
In T. pallidipennis, genetically related clones tend to be geographically contiguous or overlapping. However, although populations from the BIJX clade tend to belong to central–southern latitudes, and those from the CAMX–CAJX clade are restricted to central–northern latitudes, a strong geographical orientation is not evident. Differences in mobility of individuals and in environmental fragmentation appear to exert important influences on patterns of mtDNA phylogeographical structure (Avise, 1994). Therefore, the lack of geographical differentiation confirms the existence of high levels of gene flow previously inferred from allozyme data (Matrajt et al., 1996), and reinforces the selection hypothesis.
Migration routes
Most of the species of the genus Trimerotropis inhabit arid regions in western North America. Trimerotropis pallidipennis is one of the few trimerotropines to have successfully extended its distribution to Andean South America (White, 1973) where it shows a greater ecological amplitude, along a wider altitudinal range, than populations from North America (John, 1983).
Geological and palaeontological findings provide evidence that during most of the Cretaceous (Raven & Axelrod, 1975) and even at the end of the Jurassic (Harrington, 1962), South America was separated from North America and Central America by a Pacific–Atlantic marine water barrier. In the late Cretaceous an arc of volcanic islands apparently allowed migration between the two areas. The contemporary Central American land bridge arose in the late Pliocene to early Pleistocene, about 2–3 Myr ago, and since then a direct faunal and floral exchange has occurred. In the case of vertebrates, there was an important migration from North to South America, after the direct connection between these lands was established (Simpson, 1950).
By contrast, the most important insect migrations occurred in the opposite direction (from South to North America) during two time periods (Halffter, 1964, 1974): the first started in the upper Cretaceous or Palaeocene, and lasted until the Eocene; and the second would have occurred between the Pliocene and early Pleistocene — after the Panamá isthmus was established. During this latter migration, many Nearctic and Holarctic elements also invaded the Neotropical Region by following the mountain systems. This is called the Nearctic dispersal pattern, that involves only species restricted to highlands (Halffter, 1974).
The mean value of the estimated divergence time among South American mitochondrial haplotypes of T. pallidipennis can provide a crude estimate of the time when colonization of South America by these grasshoppers from northern latitudes might have occurred. The evolutionary rate of Drosophila mtDNA has been shown to be 60% of that of mammalian mtDNA (Tamura, 1992). According to this finding, if we apply a calibration of 1.2% sequence divergence per million years (60% of the value given by Wilson et al. (1985) for mammalian mtDNA), haplotypes of T. pallidipennis would have diverged from the hypothetical most ancestral one (BIJX) at about 3 Myr ago, during the late Pliocene. Therefore, this result agrees with the time when highland species of insects invaded South America — Halffter's (1974) Nearctic dispersal pattern — and with the hypothetical migration route proposed for T. pallidipennis. Furthermore, it supports with biological data (or mitochondrial RFLP characters) other geological and palaeontological findings that infer the time when the Panamá isthmus arose.
References
Avise, J. C. (1986). Mitochondrial DNA and the evolutionary genetics of higher animals. Phil Trans R Soc B, 312: 325–342.
Avise, J. C. (1994). Molecular Markers, Natural History and Evolution. Chapman and Hall, New York.
Avise, J. C., Arnold, J., Ball, R. M., Bermingham, E., Lamb, J. E., Neigel, J. E. et al. (1987). Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Ann Rev Ecol Syst, 18: 489–522.
Cabrera, A. L. and Willink, A. (1973). Biogeografía de América Latina. Organización de Estados Americanos, Washington, DC.
Colombo, P. C. and Confalonieri, V. A. (1996). An adaptive pattern of inversion polymorphisms in Trimerotropis pallidipennis (Orthoptera). Correlation with environmental variables: an overall view. Hereditas, 125: 289–296.
Confalonieri, V. A. (1994). Inversion polymorphisms and natural selection in Trimerotropis pallidipennis (Orthoptera): correlations with geographical gradients. Hereditas, 121: 79–86.
Confalonieri, V. A. and Colombo, P. C. (1989). Inversion polymorphisms in Trimerotropis pallidipennis (Orthoptera): clinal variation along an altitudinal gradient. Heredity, 62: 107–112.
Encalada, S. E., Lahanas, P. N., Bjornadal, K. A., Bolten, A. B., Miyamoto, M. M. and Bowen, B. W. (1996). Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: a mitochondrial DNA control region sequence assessment. Mol Ecol, 5: 473–483.
Endler, J. A. (1986). Natural Selection in the Wild. Princeton University Press, Princeton, NJ.
Felsenstein, J. F. (1991). PHYLIP. Phylogeny inference package. Version 3.5. University of Washington, Seattle.
Goñi, B., Vaio, E., Beltrami, M., Leira, M., Crivel, M., Panzera, F. et al. (1985). Geographic patterns of chromosomal variation in populations of grasshopper (Trimerotropis pallidipennis) from southern Argentina. Can J Genet Cytol, 27: 259–271.
Hale, L. R. and Singh, R. S. (1991). A comprehensive study of genic variation in natural populations of Drosophila melanogaster IV. Mitochondrial DNA variation and the role of history vs selection in the genetic structure of geographic populations. Genetics, 129: 102–117.
Halffter, G. (1964). La entomofauna americana, ideas acerca de su orígen y distribución. Folia Entomol Mex, 6: 1–108.
Halffter, G. (1974). Elements anciens de l'entomofaune neotropicale: ses implications biogeographiques. Quaest Entomol, 10: 223–262.
Harrington, H. J. (1962). Paleogeographic development of South America. Bull Am Ass Petrol Geol, 46: 1773–1814.
Hewitt, G. M. (1988). Hybrid zones: natural laboratories for evolutionary studies. Trends Ecol Evol, 3: 158–167.
John, B. (1983). The role of chromosome change in the evolution of orthopteroid insects. In: Sherma, A. K. and Sherma, A. N. (eds) Chromosomes in the Evolution of Eukaryotic Groups, vol. I, pp. 1–114. C.R.C. Press, Boca Raton, FL.
King, T. M., Wallis, G. P., Hamilton, S. A. and Fraser, J. R. (1996). Identification of a hybrid zone between distinctive colour variants of the alpine weta Hemideina maori (Orthoptera: Stenopelmatidae) on the rock and pillar range, southern New Zealand. Mol Ecol, 5: 583–587.
Latorre, A., Hernández, C., Martínez, D., Castro, J. A., Ramón, M. and Moya, A. (1992). Population structure and mitochondrial DNA gene flow in Old World populations of Drosophila subobscura. Heredity, 68: 15–24.
Maniatis, T., Fritsch, E. F. and Sambrook, J. (1982). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor Publications, Cold Spring Harbor, NY.
Marchant, A. D. (1988). Apparent introgression of mitochondrial DNA across a narrow hybrid zone in the Caledia captiva species-complex. Heredity, 60: 39–46.
Marchant, A. D. and Shaw, D. D. (1993). Contrasting patterns of geographic variation shown by mtDNA and karyotype organization in two subspecies of Caledia captiva (Orthoptera). Mol Biol Evol, 10: 855–872.
Matrajt, M., Confalonieri, V. A. and Vilardi, J. C. (1996). Parallel adaptive patterns of allozyme and inversion polymorphisms on an ecological gradient. Heredity, 76: 346–354.
Nei, M. (1987). Molecular Evolutionary Biology. Columbia University Press, New York.
Raven, P. H. and Axelrod, D. I. (1975). History of the flora and fauna of Latin America. The theory of plate tectonics provides a basis for reinterpreting the origins and distributions of the biota. Am Sci, 63: 420–429.
Shaw, D. D., Coates, D. J. and Arnold, M. (1987). Complex patterns of chromosomal variation along a latitudinal cline in the grasshopper Caledia captiva. Genome, 30: 108–117.
Shaw, D. D., Moran, C. and Wilkinson, P. (1980). Chromosomal reorganization, geographic differentiation and the mechanism of speciation in the genus Caledia. In: Blackman, R. L., Hewitt, G. M. and Ashburner, M. (eds) Insect Cytogenetics, pp. 171–194. Blackwell Scientific Publications, Oxford.
Simpson, G. G. (1950). History of the fauna of Latin America. Am Sci, 38: 361–389.
Tamura, K. (1992). The rate of nucleotide substitution in Drosophila mitochondrial DNA. Mol Biol Evol, 9: 814–825.
Vaio, E., Goñi, B. and Rey, C. (1979). Chromosome polymorphism in populations of the grasshopper Trimerotropis pallidipennis from Southern Argentina. Chomosoma, 71: 371–386.
White, M. J. D. (1973). Animal Cytology and Evolution. 3rd edn. Cambridge University Press, London.
Wilson, A. C., Cann, R. L., Carr, S. L., George, M. J. R., Gyllenster, U. B., Helm-Bychowski, K. et al. (1985). Mitochondrial DNA and two perspectives on evolutionary genetics. Biol J Linn Soc, 26: 375–400.
Acknowledgements
We acknowledge Dr Adam Marchant, who kindly mailed the probes of Caledia captiva and gave us many other suggestions. We are very grateful to Dr Mariana Matrajt for some technical assistance and to Dr Analía Lanteri for helpful discussions. We are also indebted to Dr P. C. Colombo for critical reading of the manuscript. Financial support was from the Universidad de Buenos Aires (Argentina).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Confalonieri, V., Sequeira, A., Todaro, L. et al. Mitochondrial DNA and phylogeography of the grasshopper Trimerotropis pallidipennis in relation to clinal distribution of chromosome polymorphisms. Heredity 81, 444–452 (1998). https://doi.org/10.1046/j.1365-2540.1998.00405.x
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00405.x
Keywords
This article is cited by
-
Evolution of chromosome number in grasshoppers (Orthoptera: Caelifera: Acrididae)
Organisms Diversity & Evolution (2022)
-
Highly divergent karyotypes and barcoding of the East African genus Gonatoxia Karsch (Orthoptera: Phaneropterinae)
Scientific Reports (2021)
-
Adaptive genetic diversity and population structure of the “algarrobo” [Prosopis chilensis (Molina) Stuntz] analysed by RAPD and isozyme markers
European Journal of Forest Research (2010)
-
Genetic structure of Partamona helleri (Apidae, Meliponini) from Neotropical Atlantic rainforest
Insectes Sociaux (2010)