Abstract
Variation in mitochondrial DNA (mtDNA) was surveyed, using restriction endonucleases, in the white-clawed crayfish, Austropotamobius pallipes lusitanicus, from 14 populations sampled in Spain. Four additional samples from France (1), Slovenia (1) and Italy (2) were also analysed. Among the 11 haplotypes listed, only one was detected from the 154 animals sampled from Spanish populations. This haplotype was also recorded in the Fosso di Ferfereta population (Italy). Estimates of nucleotide sequence divergence among haplotypes ranged from 0.45% to 17.4%. Interpopulational genetic relationships showed that Spanish populations were closely related to those of Fosso di Ferfereta with a small genetic distance (0.0003) found between them. AMOVA revealed that most of the genetic variance (71.97%) was attributed to variation between European regions. These results are in accordance with a drastic bottleneck event during the history of the Spanish populations. Four suggestions, based on human introduction, selection and recent or ancient historical events are discussed in relation to the lack of genetic variation in the Spanish crayfish stock.
Similar content being viewed by others
Introduction
The white-clawed crayfish, Austropotamobius pallipes, is a European native species of freshwater crayfish, which is mainly distributed throughout the western and southern regions of the continent. During the last few decades, the distribution of white-clawed crayfish populations has been greatly reduced due to the crayfish plague fungus (Diéguez-Uribeondo et al., 1997; Diéguez-Uribeondo & Söderhäll, 2000) and human alteration of the crayfish habitat due to pollution, and waterway management schemes such as channelisation (Holdich & Rogers, 1997). As a result, this species has been placed on the Red List of Threatened Animals of the International Union for the Conservation of Nature and Natural Resources (IUCN) as a vulnerable species (Baillie & Groombridge, 1996), and also in the Annexes II and IV of European Community Directives for the Conservation of Natural Habitats and Wild Flora and Fauna (92/43/EEC and 97/62/EU) as a species requiring special conservation measures. The decline of the white-clawed crayfish has been especially abrupt in the Iberian peninsula, where the current trend of disappearance ranges from 30% to 50% every five years, and it is considered to be at risk of extinction (Alonso et al., 2000). A plan for restoration of A. pallipes has been carried out (Diéguez-Uribeondo et al., 1997) and several conservation actions have been proposed in Spain and Portugal (Bernardo et al., 1997; Alonso et al., 2000, respectively). Thus, new interest has arisen in understanding the genetic diversity of this species. The success of any plan of restoration requires knowledge of the genetic make-up of the species. This is because, firstly, the potential to adapt to new or changing environments relies on genetic diversity. Secondly, an understanding of the genetic structure of wild populations will allow us to avoid genetic contamination of local populations during in situ management decisions, such as reintroductions, translocations or reinforcements of natural populations, and will also prevent the establishment of populations of non-autochthonous taxa. Thirdly, learning about the genetic diversity might also contribute to understanding of the distribution of the species in the past and the interpopulation relationships.
However, the taxonomical status of A. pallipes is currently unresolved (Grandjean et al., 1998). The first studies of its diversity were based on morphological and meristic characters, and its geographical distribution (Bott, 1950). Bott (1950) accepted the existence of at least three major groups or subspecies within A. pallipes: (1) A. pallipes pallipes, distributed in the British Isles, France, Switzerland, Northern Italy and Corsica; (2) A. p. italicus, occurring in Dalmatia, Italy, and Switzerland; and (3) A. p. lusitanicus, found in the Iberian peninsula. Nevertheless, there have been many discrepancies between studies of the taxonomical status of these groups (Karaman, 1963). Recent molecular studies by mtDNA and allozyme analyses appear to indicate that the A. pallipes taxon represents a highly structured species complex (Grandjean et al., 1997, 1998; Grandjean & Souty-Grosset, 2000a, b; Lörtscher et al., 1997; Santucci et al., 1997; Souty-Grosset et al., 1997; Largiader et al., 2000). Both types of marker showed a great genetic distance between the A. p. pallipes and A. p. italicus populations with a marked genetic heterogeneity within groups of subspecific status. This result was interpreted in the context of postglacial colonization from multiple refugia distributed around coastal Mediterranean areas.
For the Spanish populations, Santucci et al. (1997) reported an absence of genetic variability between and within two Spanish populations studied, which appear to be closely related to populations sampled from the north Apennines. These molecular data are concordant with the recent morphological studies that show that the criterion used by Bott (1950) to distinguish italicus from lusitanicus (i.e. presence of hairs on the upper border of the endopod of the second male gonopod in lusitanicus) have a very limited taxonomic value because a high percentage, ≈90%, of Spanish males have no hairs on the gonopod (Almaça, 1987; Grandjean et al., 1998). Thus, these genetic studies suggest that only a rapidly evolving genetic marker will have sufficient variation to distinguish populations of Spanish origin, i.e. A. p. lusitanicus populations.
Mitochondrial DNA provides a useful marker system (Avise, 1989). It has a lower effective population size than nuclear genes, approximately equal to half of the effective number of females, and is therefore more subject to drift than are nuclear genes (Avise, 1994). So mtDNA is a powerful instrument to detect genetic variation within and between populations. In the white-clawed crayfish, previous studies from other European samples suggest that analysis of the mtDNA would be valuable in order to characterize the genetic structure of Spanish populations.
The purpose of this study was to use the mitochondrial DNA (mtDNA) analysed by RFLP to survey the extent and the pattern of genetic variation across the range of the theoretical A. p. lusitanicus group or cluster. Some additional samples located in Italy, Slovenia and France are also being analysed to clarify our understanding of the genetic structure observed in Spanish populations.
Materials and methods
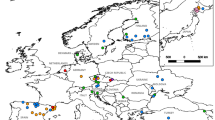
Specimens of A. pallipes were collected from 14 locations across the geographical range described by Bott (1950) for A. p. lusitanicus in Spain (Fig. 1). Populations originated from four main geographical areas: (1) three from the Cantabric sea region located at Guipuzkoa, Santesteban and Baztan; (2) nine from the Mediterranean sea region at different points of the river Ebro catchment, i.e. Aoiz, Etxarri, Izaga, Estella, Mendillorri, Pamplona, Salazar, and Tafalla, Puertos de Beseitle; (3) one from the Mediterranean sea region in Granada, which represents the southern limit of A. pallipes distribution; and (4) one population from the Duero catchment at Leon. Four additional populations were collected from Serremijanes (France) from the Rhône catchment, Pragatto, Fosso di Ferfereta (Italy) from the Arno and Po catchments and Rizana (Slovenia) from the Danube catchment (Fig. 1).
Location map of sampled white-clawed crayfish (Austropotamobius pallipes) populations. Numbers preceding location names refer to positions on the map and those in parentheses refer to the sample size. 1 Baztan (14), 2 Santesteban (8), 3 Etxarri (12), 4 Mendillorri (8), 5 Pamplona (5), 6 Aoiz (8), 7 Guiperezkoa (16), 8 Izaga (8), 9 Salazar (8), 10 Taffala (10), 11 Estella (15), 12 Puertos de Beseitle (12), 13 Granada (13), 14 Leon (15), 15 Serremijanes (12) (France), 16 Fosso di Ferferita (16) (Italy), 17 Pragatto (14) (Italy), 18 Rizana (20) (Slovenia).
The method described in Grandjean & Souty-Grosset (1996) was used to extract total mtDNA from samples. MtDNA samples were digested with six restriction endonucleases, four 4-base cutters (HaeIII, AccII, TaqI and HpaII) and two 6-base cutters (HindIII and EcoRI), according to the instructions of the manufacturer (Gibco BRL, Life Technologies S.A.R.L., Cergy Pontoise, France). The resulting restriction fragments were separated in 1.5% agarose gels in Tris-EDTA buffer (30 mM; 60 mM) for 15 h at 30 V. Gels were stained with SYBR Green I (FMC Bioproducts, Tebu S.A., 78610 Le Perray en Yvelines, France) and visualized with a UV light transilluminator. The restriction fragment pattern from each endonuclease was given a letter, and each individual was characterized by a haplotype composed of six letters. All individuals sharing a common composite phenotype were regarded as belonging to the same mtDNA matrilineal clone.
Because small mtDNA fragments less than 400 base pairs could not been scored, estimates of nucleotide sequence divergence (p) among mtDNA haplotypes were calculated by the method of Nei & Li (1979) from the total proportion of shared mtDNA fragments between two individuals. Measures of diversity within populations were estimated using the DA program in the REAP 4.0 package (McElroy et al., 1992). Genetic heterogeneity within populations was estimated by haplotype diversity (h) and nucleotide diversity (p) within populations, and nucleotide divergence (dxy) among populations (Nei & Tajima, 1981). The distance matrix of net nucleotide divergence among populations was used as the input for a principal coordinate analysis (PCO).
The geographical distribution of haplotypes was quantified by an analysis of variance approach adapted for molecular data (Excoffier et al., 1992) as implemented in the computer program ARLEQUIN. This program calculates a correlation statistic, referred to as ΦST, that reflects the percentage of genetic variance explained by the geographical stratification of the sample. The statistical significance of the Φ-values is tested by generating a null distribution of values from 1000 random permutations of the data matrix. The degree of geographical heterogeneity of mtDNA haplotype distributions was assessed using a (χ2) statistic as described by Roff & Bentzen (1989). The significance level was obtained by 10 000 Monte Carlo randomizations using the MONTE program from the REAP 4.0 package (McElroy et al., 1992).
Results
Among the 11 haplotypes recorded in this study (Table 1), one was found in 154 animals sampled from 14 Spanish populations. The other 10 haplotypes were found in the four populations sampled in other parts of Europe, four in Pragatto and two in Fosso di Ferfereta, respectively (Italy), three in Serremijanes (France) and two in Rizana (Slovenia) (Table 1). Most of the haplotypes found in this study were specific for each population except the Spanish haplotype, which also occurred in Fosso di Ferfereta. Statistically significant differences in haplotype frequencies were observed among all populations (χ2=658.91, P < 0.0001). Significant differences were found between Rizana, Serremijanes and Pragatto with all the other populations. Chi-squared tests of haplotype frequencies revealed homogeneity among all Spanish populations and with that of Fosso di Ferfereta (χ 2=29.39, P=0.028).
Estimates of nucleotide sequence divergence (p) among haplotypes ranged from 0.45% to 17.4%. Haplotype diversity and nucleotide divergence ranged from 0 to 0.773 and from 0 to 0.091, respectively (Table 2). No genetic variation was found within or among the Spanish populations. Genetic distance among populations ranged from 0.0003 to 0.164 (Table 3). Spanish populations appear to be closely related to those of Fosso di Ferfereta (Italy), with a small genetic distance (0.0003) between them (Fig. 2).
Estimates of variance components within populations, between populations within region and between regions, calculated using AMOVA, revealed that most of the total variance (71.97%) could be attributed to variation between regions. However, significant amounts were also attributable to variation among populations within regions (4.79%) and within populations (23.24%).
The PCO graphical representation based on Nei’s genetic distance values among populations (Fig. 2) showed three groups well separated according the first two principal components which explain 66% and 33% of total variance, respectively. Cluster (A) includes the 14 Spanish populations and the two Italian ones, cluster (B) contains the population sampled in Slovenia (Rizana) and cluster (C) includes the French population (Serremijanes) (Fig. 2).
Discussion
The lack of genetic variability found among and within Spanish populations using mtDNA markers is particularly surprising when compared to the samples of other European origins included in this study, which all showed some degree of intraspecific variation. Moreover, previous data obtained from samples of the clusters A. p. italicus (Santucci et al., 1997) and A. p. pallipes (Grandjean et al., 1997; Grandjean & Souty-Grosset, 2000a, b) also revealed a high intraspecific variation between populations of the same cluster, with a strong genetic structuring (FST = 0.8 for A. p. italicus from allozymes and ΦST = 0.74 for A. p. pallipes from mtDNA).
Although more sensitive techniques could be applied to detect some degree of genetic variability, i.e. microsatellites, RAPD-PCR, on their own or in combination with allozyme studies, the low level of genetic variation detected among Spanish populations does not appear to be due to any methodological bias. The enzymes used in this study have revealed haplotype variation within the other sampled European populations (Table 1). Moreover, for example, these enzymatic markers have also shown mitochondrial genetic variation ranging from 0.56% to 1.43% in A. p. pallipes populations that are closely geographically related (Grandjean & Souty-Grosset, 2000a). In addition, the lack of genetic variation cannot be blamed on sampling since 14 populations have been sampled constituting a total sample size of 154 animals from four different hydrographic drainages. This represents a more widespread sample than previous studies made from mtDNA in other part of its range.
A tentative hypothesis for explaining this lack of variability in Spanish A. pallipes would be that it arises from a founder effect due to an anthropogenic origin of crayfish in Spain. Given the close genetic relationship between Spanish and Italian populations, particularly those of Fosso di Ferfereta, we could assume a translocation of crayfish from Italy to Spain. This assumption is in accordance with Laurent (1988) who speculated about the possible colonization of the Iberian peninsula by A. pallipes due to a recent introduction of crayfish from Italian populations. Human translocations of crayfish are considered to be a frequent event in Europe (Grandjean et al., 1997; Lörtscher et al., 1997; Santucci et al., 1997). According to Laurent & Suscillon (1962), these practices have been performed on both the local and larger scales across Europe. For example, they reported that restocking attempts were performed using freshwater crayfish from Italy and Spain in the 19th century, reconstituting a part of the ‘French stock’, decimated by pathogens introduced by exotic crayfish. There is a lack of historical documents describing the past distribution of A. pallipes in the Iberian peninsula. The first account of crayfish distribution was that of Madoz (1850–1853). This has been taken as proof of an anthropogenic origin of Spanish stocks (Albrecht, 1982). Certainly, more sensitive techniques could be applied to detect some degree of repetitive genetic variability, i.e. microsatellites, RAPD-PCR, or a combination with allozyme studies, to confirm or rule out the present hypothesis.
A second hypothesis would include historical events linked to population fragmentation, which has become particularly acute in Spain in the last decades. Thus the number and size of the populations has been drastically reduced, the species now being confined to headwater river systems (Alonso et al., 2000). In most organisms, the consequence of small population size is that the rate of allelic loss increases and the average individual heterozygosity, relative to the overall population, decreases, due to a combination of genetic drift and inbreeding (Saccheri et al., 1998). However some authors have shown that bottlenecks are expected to strongly depress neutral variability within populations but to increase differentiation among populations (Tajima, 1989; Barton & Whitlock, 1995). Thus, we would expect to observe a genetic structuring of Spanish populations, if there had been recent population bottlenecks, as recorded within the northern group A. p. pallipes, and the southern group, A. p. italicus. Perhaps a further analysis of Iberian populations of A. pallipes from other geographical origins could add more information.
A third hypothesis would postulate more ancient population bottlenecks, perhaps related to palaeogeographic events during the Pleistocene. It is believed that such ancient population bottlenecks have had a great influence on the current genetic structure of the European species (Hewitt, 1996). Several authors have reported that most of European species show three main haplotype lineages related to the existence of three different glacial refugia: (1) the Balkans, (2) Iberia and (3) the Italian peninsula (Hewitt, 1996; Taberlet et al., 1998). Thus the lack of genetic variability between and within Spanish populations could be the consequence of a drastic bottleneck during the Pleistocene. Thus, our results are consistent with a single refugium for this subspecies from which all extant populations have been derived. However, the lack of genetic divergence observed among Spanish populations contrasts with the strong genetic heterogeneity found within A. p. pallipes and A. p. italicus clusters by Grandjean et al. (1997), Santucci et al. (1997) and Grandjean & Souty-Grosset, 2000a, b). According to Santucci et al. (1997) these results could be related to range fragmentations and subsequent recolonizations from multiple French and Italian refugia during the last glacial events. However, it seems hardly conceivable that the Iberian refugia area could have been smaller than each of the other two (Dawson, 1992), inducing a more drastic bottleneck. Another objection to this hypothesis is that significant genetic structuring has generally been detected among major Pleistocene isolates of palaearctic species (Hewitt, 1996), which we did not find in this study of the Italian and Spanish samples.
A fourth hypothesis would implicate the selection due to the impact of the present disease, the crayfish plague, affecting the species, which caused an abrupt fall of the populations of A. pallipes in the last 20 years over its range. The incidence of infections could be greater in Spain than that in neighbouring regions (Italy, France and the Balkans) involving a selection and a restriction of the A. pallipes genetic diversity to the present genotypes. Thus, the current populations could be more adapted to diseases and/or the present habitat of this species in the Iberian peninsula, i.e. the headwater systems.
An alternative hypothesis can be drawn by combining all four hypotheses. It could assumed that the present range of A. p. italicus and A. p. lusitanicus, is a relict of a broader one, including France. The disjunction of its range could have been caused by the spread of A. p. pallipes to the south during the Miocene, as suggested by Karaman (1963). A. p. pallipes might have been a more effective competitor than A. p. italicus. In support of this hypothesis, Santucci et al. (1997) cited the example of the Chamois Rupinaica originating from Eastern Europe, which excluded R. pyrenaica from most of its previous range, including the Alps and the Pyrenees. Both Italian and Spanish populations could be the relict of an extinct intermediate population located in France, which extended their range, either by stream captures after glaciation periods, as mentioned by Largiader et al. (2000) in the case of white fish, Coregonus sp. and brown trout, Salmon trutta (Largiader et al., 1996), or by translocations of crayfish by humans (Laurent, 1988), or both.
The explanation of the genetic structure of the Iberian A. pallipes populations remains an intriguing question with great interest palaeogeographically and for the management of this endangered species. New studies should be carried out by using more sensitive techniques for detecting genetic diversity, i.e. microsatellites and RAPD, as well as research into Pleistocene fossil records and also other literature sources about the presence of A. pallipes in the Iberian peninsula.
References
Albrecht, H. (1982). Das system der europäischen Flusskrebse (Decapoda, Astacidae): Vorschlag und Begrundung. Mitt Hamburg Zool Mus Inst, 79: 187–210.
Almaça, C. (1987). On the Portuguese populations of Austropotamobius pallipes (Lereboullet, 1858). Investigacion Pesquera, 51: 403–411.
Alonso, F., Temino, C. and Diéguez-Uribeondo, J. (2000). Status of the white clawed crayfish, Austropotamobius pallipes (Lereboullet, 1858), in Spain: Distribution and legislation. Bull Fr Pêche Pisc, 356: 31–54.
Avise, J. C. (1989). Gene trees and organismal histories: a phylogenetic approach to population biology. Evolution, 43: 1192–1208.
Avise, J. C. (1994). Molecular Markers, Natural History and Evolution. New York, Chapman & Hall.
Baillie, J. and Groombridge, B. (1996). IUCN red list of threatened animals. IUCN, Gland, Switzerland and Cambridge, U.K.
Barton, N. H. and Whitlock, M. (1995) The Evolution of Metapopulations. Metapopulation Dynamics, (Ed. by L. Hanski and M. Gilpin.). Academic Press, New York.
Bernardo, J. M., Ilheu, M. and Costa, A. M. (1997). Distribution, population structure and conservation of Austropotamobius pallipes in Portugal. Bull Fr Pêche Pisc, 347: 753–763.
Bott, R. (1950). Die Flusskrebse Europas (Decapoda, Astacidae). Proc Senck Nat Soc, 483: 1–36.
Dawson, A. G. (1992) Ice Age Earth. Routledge, London.
Diéguez-Uribeondo, J. and Söderhäll, K. (2000). RAPD evidence for the origin of an outbreak of crayfish plague in Spain. Freshwater Crayfish, 12: 313–318.
Diéguez-Uribeondo, J., Temino, C. and Muzquiz, J. L. (1997). The crayfish plague fungus (Aphanomyces astaci) in Spain. Bull Fr Pêche Pisc, 347: 753–763.
Excoffier, L., Smouse, E. and Quattro, J. M. (1992). Analyses of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479–491.
Grandjean, F. and Souty-Grosset, C. (1996). Isolation and characterisation of mtDNA from the endangered white-clawed crayfish Austropotamobius pallipes pallipes (Lereboullet, 1858). Bull Fr Pêche Pisc, 343: 175–182.
Grandjean, F. and Souty-Grosset, C. (2000a). Genetic and morphological variation in the endangered crayfish species, Austropotamobius pallipes (Lereboullet) (Crustacea, Astacidae) from the Poitou–Charentes region (France). Aquat Sci, 62: 1–19.
Grandjean, F. and Souty-Grosset, C. (2000b). Mitochondrial DNA variation and population genetic structure of the white-clawed crayfish, Austropotamobius pallipes pallipes. Cons. Genet., in press:.
Grandjean, F., Souty-Grosset, C. and Holdich, D. M. (1997). Geographical variation of mitochondrial DNA between European populations of the white-clawed crayfish Austropotamobius pallipes. Freshwater Biol, 37: 493–501.
Grandjean, F., Gouin, N., Frelon, M. and Souty-Grosset, C. (1998). Genetic and morphological systematic studies on the crayfish Austropotamobius pallipes. J Crust Biol, 18: 549–555.
Hewitt, G. M. (1996). Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc, 58: 247–276.
Holdich, D. M. and Rogers, W. D. (1997). The white-clawed crayfish, Austropotamobius pallipes, in Great Britain and Ireland with particular reference to its conservation in Great Britain. Bull Fr Pêche Pisc, 347: 597–616.
Karaman, M. S. (1963). Ein Beitrag sur Systematik der Astacidae. Crustaceana, 3: 174–191.
Largiader, C. R., Scholl, A. and Guyomard, R. (1996). The role of natural and artificial propagation in gene diversity of brown trout (Salmo trutta L.) of the upper Rhone drainage. In: Kirchhofer, A. and Hefti, D. (eds) Conservation of Endangered Freshwater Fishes in Europe, pp. 181–197. Birkhaüser-Verlag, Basel.
Largiader, C. R., Herger, F., Lortscher, M. and Scholl, A. (2000). Assessment of natural and artificial propagation of the white-clawed crayfish (Austropotamobius pallipes species complex) in the Alpine region with nuclear and mitochondrial markers. Mol Ecol, 9: 25–37, 10.1046/j.1365-294x.2000.00830.x.
Laurent, P. (1988). Austropotamobius pallipes and A. torrentium, with observations on their interactions with others species in Europe. In: Holdich, D. M. and Lowery, R. S. (eds) Freshwater Crayfish: Biology, Management and Exploitation, pp. 341–364.
Laurent, P. and Suscillon, M. (1962). Les écrevisses en France. Ann Stat Centr Hydro, 9: 336–395.
Lörstcher, M., Stucki, T. P., Clalüna, M. and Scholl, A. (1997). Phylogeographic structure of Austropotamobius pallipes populations in Switzerland. Bull Fr Pêche Pisc, 347: 649–661.
Madoz, P. (1850–1853). Diccionario geografico-estadistico-historico de Espana y sus posesiones de Ultramar. Madrid.
Mcelroy, D., Moran, P., Bermingham, E. and Kornfield, I. (1992). REAP: an integrated environment for the manipulation and phylogenetic analysis of restriction data. J Hered, 83: 157–158.
Nei, M. and Li, W. H. (1979). Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA, 76: 5269–5273.
Nei, M. and Tajima, F. (1981). DNA polymorphism detectable by restriction endonucleases. Genetics, 97: 145–163.
Roff, D. A. and Bentzen, P. (1989). The statistical analysis of mitochondrial DNA polymorphism: c2 and the problem of small samples. Mol Biol Evol, 6: 539–545.
Saccheri, I., Kuussaari, M., Kankare, M., Vikman, P. et al (1998). Inbreeding and extinction in a butterfly metapopulation. Nature, 392: 491–494, 10.1038/33136.
Santucci, F., Iaconelli, M., Andreani, P., Clanchi, R. et al (1997). Allozyme diversity of European freshwater crayfish of the genus Austropotamobius. Bull Fr Pêche Pisc, 347: 663–676.
Souty-Grosset, C., Grandjean, F., Raimond, R., Frelon, M. et al (1997). Conservation genetics of the white-clawed crayfish, Austropotamobius pallipes: the usefulness of the mitochondrial DNA marker. Bull Fr Pêche Pisc, 347: 677–692.
Taberlet, P., Fumagalli, L., Wust-Saucy, A. -G. and Cosson, J. -F. (1998). Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol, 7: 453–464.
Tajima, F. (1989). The effect of change in population size on DNA polymorphism. Genetics, 123: 597–601.
Acknowledgements
This work was supported by Servicio de Medio Ambiente del Gobierno de Navarra. Samples were provided by Dr Jose Maria Gil, Javier Alba, Inigo Mendiola and Francesca Gherardi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Grandjean, F., Gouin, N., Souty-Grosset, C. et al. Drastic bottlenecks in the endangered crayfish species Austropotamobius pallipes in Spain and implications for its colonization history. Heredity 86, 431–438 (2001). https://doi.org/10.1046/j.1365-2540.2001.00849.x
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1046/j.1365-2540.2001.00849.x
Keywords
This article is cited by
-
Freshwater biodiversity in the rivers of the Mediterranean Basin
Hydrobiologia (2013)
-
The Decline of the Endangered Populations of the Native Freshwater Crayfish (Austropotamobius pallipes) in Southern Spain: It is Possible to Avoid Extinction?
Hydrobiologia (2006)
-
Origin and colonization history of the white-clawed crayfish, Austropotamobius pallipes, in Ireland
Heredity (2003)