Abstract
Background:
Oesophageal adenocarcinoma or Barrett’s adenocarcinoma (EAC) is increasing in incidence and stratification of prognosis might improve disease management. Multi-colour Fluorescence in situ hybridisation (FISH) investigating ERBB2, MYC, CDKN2A and ZNF217 has recently shown promising results for the diagnosis of dysplasia and cancer using cytological samples.
Methods:
To identify markers of prognosis we targeted four selected gene loci using multi-colour FISH applied to a tissue microarray containing 130 EAC samples. Prognostic predictors (P1, P2, P3) based on genomic copy numbers of the four loci were statistically assessed to stratify patients according to overall survival in combination with clinical data.
Results:
The best stratification into favourable and unfavourable prognoses was shown by P1, percentage of cells with less than two ZNF217 signals; P2, percentage of cells with fewer ERBB2- than ZNF217 signals; and P3, overall ratio of ERBB2-/ZNF217 signals. Median survival times for P1 were 32 vs 73 months, 28 vs 73 months for P2; and 27 vs 65 months for P3. Regarding each tumour grade P2 subdivided patients into distinct prognostic groups independently within each grade, with different median survival times of at least 35 months.
Conclusions:
Cell signal number of the ERBB2 and ZNF217 loci showed independence from tumour stage and differentiation grade. The prognostic value of multi-colour FISH-assays is applicable to EAC and is superior to single markers.
Similar content being viewed by others
Main
Within the last three decades the incidence of oesophageal adenocarcinoma (EAC) has increased in Western countries and exceeded the levels of other cancers (Blot, 1994; Heitmiller and Sharma, 1994; Devesa et al, 1998; Vizcaino et al, 2002; Pohl and Welch, 2005; Brown et al, 2008; Umar and Fleischer, 2008). Otherwise, the 5-year survival of EAC has remained at less than 10% in the past two decades (Blot et al, 1991; Devesa et al, 1998). EAC is believed to develop via a stepwise process from Barrett’s metaplasia as a predisposing condition (Hamilton and Smith, 1987; Hameeteman et al, 1989; Miros et al, 1991; Reid, 1991; Haggitt, 1994; Spechler, 1994, 2002). Within a reflux-metaplasia-dysplasia-carcinoma sequence, chronic damage by gastro oesophageal reflux leads to development of metaplastic Barrett’s epithelium (BE). Further mutagenic events result in a low- to high-grade intraepithelial neoplasia-dysplasia-sequence and finally in invasive cancer (Spechler, 2002).
To detect EAC at an early, potentially curable stage, guidelines for gastro oesophageal reflux disease (GERD) outline that patients with GERD might benefit from an early endoscopic evaluation for Barrett’s oesophagus (BE), considering its conversion rate to EAC of 0.5–1% per year (Peitz and Malfertheiner, 2007; Thomas et al, 2007). The gold standard in estimating the prognosis of EAC remains to be clinical classifications like TNM and grading of EAC. However, these classifications are not suitable for every situation, for example, biopsy collection, and could be complemented by new biomarkers (Cestari et al, 2007; Huang, 2010). In EAC the detection of MDM2, VEGF and p53 have been suggested as markers of poor prognosis, but showed contradictory results across studies (Cavazzola et al, 2009; Prins et al, 2012; Renouf et al, 2013). A recent multi-centric study (OCCAM group) identified DCK, PAPSS2, SIRT2 and TRIM44 as prognostic biomarkers in EAC by extensive gene expression profiling and immunohistochemistry (Peters et al, 2010). New molecular methods like multiplex FISH analysis could be utilised for predictive stratification of patients with EAC to select individualised treatment.
Gain and loss of certain gene loci can contribute to the progression from BE to EAC, but less is known about their prognostic significance as biomarkers. As one example, CDKN2A (previously known as p16; 9p21) is involved in the clonal expansion of EAC progenitor cells (Barrett et al, 1999). Other gene loci such as ERBB2 (also known as Her2) (17q12), MYC (8q24) and ZNF217 (20q13.2) are also implicated in EAC development. All these genomic loci have shown promising results in detecting progression from BE to dysplasia and EAC in oesophageal brush cytology specimens (Brankley et al, 2006; Rygiel et al, 2007; Fritcher et al, 2008). Although it could be assumed that functionally important markers for progression might have an impact on prognosis in EAC, the Fluorescence In Situ Hybridisation (FISH) assay using multi-colour Locus-Specific Identifier (LSI) probes has never been applied to advanced EAC.
In brief, the single markers in this multi-colour FISH assay are characterised as follows:
ERBB2 is part of the human epidermal growth factor receptor family. It stimulates cell proliferation using the RAS-MAP-kinase pathway and blocks apoptosis using the mTOR pathway. ERBB2 was first evaluated as a target for trastuzumab therapy in invasive breast cancer, and later in gastric cancer and EAC (Huober et al, 2006).
ZNF217, also known as Zinc finger protein 217, encoded by ZNF217, blocks signals resulting from dysfunction and may promote neoplastic transformation and progression of malignancy (Huang et al, 2005).
ZNF217 has recently been implicated in ovarian cancer. There it could be linked to activated PIK3CA signalling, which puts it in a possible context to influence ERBB2 and MAPK signalling (Hou et al, 2014; Yoda et al, 2014).
Cyclin-dependent kinase inhibitor 2A (CDKN2A, p16Ink4A), also known as multiple tumour suppressor 1 (MTS-1), is a tumour suppressor encoded by CDKN2A (Nobori et al, 1994). Alterations such as loss of CDKN2A, usually by deletion, mutation or promoter hypermethylation, cause increased risk of developing malignant tumours such as adenocarcinoma, including EAC, because of its function in cell cycle regulation (Igaki et al, 1994).
MYC is a regulator gene that encodes a transcription factor and is located on chromosome 8. It is believed to regulate the expression of 15% of all genes through binding to enhancer box sequences. Mutations in MYC that cause its persistent expression have been found in many cancers (Cotterman et al, 2008). This leads to deregulated expression of genes involved in cell proliferation and results in cancers like Burkitt’s lymphoma (Hummel et al, 2006).
As a proof-of-principle, multiplex FISH analysis can be applied to formalin-fixed paraffin-embedded (FFPE) tissue, for example, for MYC and ZNF217 (Wiech et al, 2009).
In this study we have applied four locus-specific probes (ERBB2 (17q12), MYC (8q24), CDKN2A (9p21), ZNF217 (20q13.2)) to FFPE EAC tissue specimens and assessed the prognostic value of these markers. A large, clinically well-defined EAC patient group with follow-up data was investigated on tissue microarrays (TMA).
Materials and methods
Patient samples, tumour classification and clinical data
One hundred and thirty EAC patient tissue samples were collected at the Institute of Pathology, Technical University of Munich (TUM) from 1991 to 2004 as previously described (Langer et al, 2006; Rauser et al, 2007). The presence of Barrett’s adenocarcinoma was confirmed histologically according to the tumour site and histological categories. None of the patients had received neoadjuvant chemotherapy or radiation. The average patients’ age was 63.4 years, ranging from 33 to 83 years. The median follow-up time was 43 months (see Table 1). All histological samples were FFPE. The tumours were classified according to the 2002 WHO classification of tumours from 2002 and the 6th edition of the TNM-classification for oesophageal tumours (Hamilton and Aaltonen, 2000). Invasion into blood vessels, lymphatic vessels or extension of tumour beyond submucosal gland tissue was documented separately. All cases were re-evaluated by an experienced pathologist (MS, TTR), including exact location of the tumour area on haematoxylin/eosin-stained (HE) slides before extracting tissue for the TMA. The clinical follow-up data were assembled separately from patient records. Twenty tissue samples of cardiac mucosa without metaplasia or dysplasia were used as representative control tissues. National and local ethical guidelines were followed.
TMA procedure
TMAs were constructed for each tumour as described previously (Bubendorf et al, 2001; Klopocki et al, 2004; Baumann et al, 2006). A representative tumour section was selected from a HE section of the paraffin block with the centre and representative region of tumour tissue identified (detailed in Supplementary Technical Notes).
Four-colour FISH
Analysis of multi-colour FISH on TMA slides is a suitable tool to identify specific genomic changes in tumours (Schwarz et al, 2008). FISH was performed on FFPE slides as previously described (Sassen et al, 2008). We used directly labelled LSI probes for ERBB2 (17q12), MYC (8q24), CDKN2A (9p21) and ZNF217 (20q13.2). The four-colour LSI FISH probe set was provided by Abbott Molecular (Des Plaines, IL, USA) and applied on TMA sections following the manufacturer’s protocols (detailed information in Supplementary Technical Notes).
Signal evaluation by epifluorescence microscopy
Slides were evaluated using a fluorescence microscope equipped with five bandpass filters specific for DAPI and each probe signal (ERBB2 (green), MYC (blue), CDKN2A (red) and ZNF217 (gold, Aristoplan from Leitz, Wetzlar, more details in Supplementary Technical Notes): Images were obtained with a Plan-Apochromatic lens (63 × , numerical aperture 1.4) and an Axio-Cam MRm camera (both from Zeiss, Jena, Germany). HE pictures were taken with a Leica DM 4000 B and DFC 320 camera (Leica AG, Wetzlar, Germany).
Enumeration and evaluation of FISH signals
To determine copy numbers at ERBB2 (17q12), MYC (8q24), CDKN2A (9p21) and ZNF217 (20q13.2) chromosomal loci, FISH signals for each of the four probes were enumerated in 130 EAC specimens with acceptable hybridisation quality. A conventional HE stain was performed to identify tumour areas for FISH analysis, as well as guarantee the representativeness of the TMA spots, and for reference histology.
For each patient specimen, a representative tumour area was selected for evaluation on one of the three replicate punch samples on the TMA, and 50 consecutive tumour cells were counted in the selected area. To assure the sampling method was representative, a 50-cell enumeration was conducted on all three punch samples of 10 individual, randomly selected EAC cases. The representation and homogeneity of the selected tumour areas during TMA generation was statistically confirmed for each of the four probes (ERBB2 (17q12), MYC (8q24), CDKN2A (9p21) and ZNF217 (20q13.2)) in the three resulting data sets by student’s t-test and interclass correlation (Cohen’s Kappa).
In the control group, 19 tissue specimens with acceptable hybridisation quality were enumerated. Nuclei in the cardiac glands were examined in the same way as the nuclei of tumour cells in the carcinoma group. For quality control, two reviewers experienced in FISH analysis evaluated at least 15% of all punches and comparison of their results showed high agreement.
Statistical analysis
In total, 130 EAC patient specimens were evaluated in this study. To select the final FISH parameter(s) that have significant impact on patients’ overall survival, the enumerated data for the 130 patients were split into two parts. A randomly selected subset of 64 patients was used as the training dataset, and the remaining subset of 66 patients was treated as the validation dataset.
Multiple (>30) FISH parameters derived from the copy numbers of the four FISH–LSI probes were first evaluated as possible prognostic indicators of overall survival within a training subset; the statistically significant parameters were further validated within the full 130-patient dataset. The FISH parameters were created from all possible combinations of the four probes used in the assay, and were derived either from percentages of cells with an abnormal copy number of one or more probes, or from the copy number ratios.
Correlations between FISH parameters and important clinical variables (age, gender, tumour grade and stage), were evaluated using the χ2-test. Frequency tables were generated to show the patient distribution within each level.
To select optimal cutoff values for each of the FISH parameters, patients were categorised into FISH positive (+) and FISH negative (−) groups based on each of the possible cutoff values. The log-rank test was used to compare overall survival between the FISH+ and FISH− groups. The hypothesis was that FISH+ patients have worse overall survival than FISH− patients. For each FISH parameter, the optimal cutoff value separating patients into favourable and unfavourable survival outcome groups was selected as the value that yields the lowest log-rank P-value. The parameters with log-rank P-values<0.05 at the optimal cutoff value were chosen for further analysis.
With the most promising FISH parameters selected and their optimal cutoffs derived from the log-rank test, a multivariate Cox proportional hazard model was used to select the final FISH parameters after adjusting for key clinical factors (age, gender, tumour grade and stage). This exercise was done using the training dataset. Once the final model was selected, it was confirmed with the validation dataset and applied to the full dataset. Furthermore, Kaplan–Meier method was used to obtain the overall survival curves, stratified by important FISH and clinical predictors, together with the corresponding median overall survival time. All analyses were performed using SAS (Version 8.2, SAS Institute Inc., Cary, NC, USA).
Results
Morphological homogeneity in different TMA punches from the tumour centre correlates with FISH results
Initially a series of 10 tumour specimens were analysed. In HE staining, the punch biopsies from tumour centres showed the same morphology, cytology and tumour grading. To exclude molecular heterogeneity in the tumour punches taken during TMA generation, three separate sets of 50 consecutive cells were counted for each of our individual probes. Student’s t-tests showed no significant differences between means and each counted subset; further on, interclass correlation coefficients were calculated and showed significant homogeneity between the chosen tumour punch biopsies in the TMA (details in Supplementary Statistical Notes).
The effect of nuclear truncation is comparable in EAC tissue and normal tissue
In this study, FISH analysis was performed on histological specimens. In comparison to cytological specimens, FISH on histologic tissues showed a loss of up to 30% owing to nuclear truncation, which depends on slide thickness (Nielsen et al, 2012). The effect was equal in tumour and normal tissues. There was no effect of the greater diameter of tumour cell nuclei vs those of normal tissue (data not shown). Therefore, these FISH-stained 3 μm-thick TMA sections were deemed appropriate for FISH analysis. In normal tissue the mean values of signals/cell for CDKN2A, ERBB2, ZNF217 and MYC were determined to be 1.68, 1.78, 1.77 and 1.74, respectively.
Fluorescence in situ hybridisation parameters distinguish between normal and malignant tissue
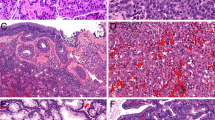
The multi-colour probe set utilised in this study (ERBB2, MYC, CDKN2A and ZNF217, Figure 1) was originally selected to aid in diagnosing dysplasia and EAC in patients with Barrett’s metaplasia (Brankley et al, 2006). We observed that EAC was associated with gains in the ZNF217 and ERBB2 signals. As expected, MYC also showed a slight gain in signal, while CDKN2A showed a signal loss in EAC. MYC and CDKN2A indicated malignancy, but revealed no further prognostic potential when analysed in multivariate analysis. ERBB2 as a single marker correlated with tumour grade.
(A Oesophageal adenocarcinoma with HE staining (TMA): magnification 400 × , for verifying carcinoma cells against epithelium, low- or high-grade dysplasia and normal soft tissue in between tumour cells. (B) Oesophageal adenocarcinoma with HE staining (TMA): magnification 500 × , for counting and verifying carcinoma tissue and epithelium, low- or high-grade dysplasia and normal soft tissue. (C) Representative sample of EAC (TMA): FISH showing gain in ERBB2 and ZNF217, loss of CDKN2A and unspecific signals for MYC. Fluorescence in situ hybridisation–LSI probes for ERBB2 (green), MYC (blue), CDKN2A (red) and ZNF217 (gold). Note the gain in strong ERBB2 spots in comparison to the low number of ZNF217, indicating an unfavourable EAC with regard to our predictors P1–P3. (D) Tissue from normal cardiac mucosa with the FISH–LSI probes for ERBB2 (green), MYC (blue), CDKN2A (red) and ZNF217 (gold) as the control group and standard values.
Table 2 shows the performance of selected FISH parameters with cutoffs (percentage of positive cells or ratio of two probe signals) for normal tissue and EAC. As is evident from the table, we confirmed that the probes used in this study were highly sensitive and specific for EAC when compared to normal tissue, with the highest sensitivity and specificity achieved with ERBB2 and ZNF217, or composite parameters based on these loci.
Selected FISH parameters predict overall survival in EAC patients
Multiple parameters derived from the copy numbers of ERBB2, MYC, CDKN2A, and ZNF217 were screened by univariate log-rank analysis of overall survival at multiple cutoff values within a randomly selected training subset, as described in the Materials and Methods section.
For each of the parameters, the optimal cutoffs were selected that minimised log-rank P-values. Those parameters with optimal cutoffs that were statistically significant at the 0.05 level were selected as potential prognostic indicators of overall survival. The selected parameters were further evaluated for significance in univariate analysis with a validation data subset, as well as with the complete 130-specimen dataset. With the selected cutoffs, parameters P1, P2 and P3 listed below were shown to have statistically significant effects on survival in randomly selected training and validation subsets of the data, as well as in the full dataset, discriminating patients with longer median survival from those with shorter survival times. Therefore, parameters P1, P2 and P3 were shown to dichotomise between favourable and unfavourable outcomes with respect to overall survival:
-
P1: percentage of cells with less than two signals for the ZNF217 locus, with favourable outcome below a cutoff of 16%.
-
P2: percentage of cells with fewer ERBB2 than ZNF217 signals (ERBB2/ZNF217<1), with favourable outcome observed at or above a cutoff of 28%.
-
P3: the overall ratio of ERBB2 to ZNF217 signals, with favourable outcome observed below a cutoff of 1.5.
The results of the log-rank test for each of the selected predictors in the combined full dataset of 130 patients are shown in Table 3.
Analysis of covariates and proportional hazards model to predict overall survival
As shown above, FISH parameters were found to be potential predictors of survival. These parameters were further evaluated in a covariate analysis with clinico-pathological variables to understand the relative importance of molecular and clinical parameters that might be associated with disease outcome. Correlation between several clinico-pathological variables, as well as correlation of these variables with FISH, was assessed in univariate and multivariate (Cox proportional hazards) analyses, testing the effect of these covariates on patient survival. The clinico-pathological variables included anatomical stage (I, IIA, IIB, III and IV), blood vessel invasion, lymphangiosis carcinomatosa, depth of tissue infiltration (T-stage), lymph node status (N-stage) and tumour grade. In a univariate analysis these were the best predictors of survival. However, when entered in a multivariate model, it was observed that the clinical parameters, including lymph node status, were correlated with the anatomical stage. Therefore, out of the clinical parameters tested, two were selected for further analysis: tumour grade and anatomical stage.
Tumour grade and anatomical stage were also found to be correlated with FISH parameters P1, P2 and P3. Therefore, a stratified analysis was performed to assess the prognostic value of FISH in the presence of these important clinico-pathological covariates.
Multivariate model analysis of stratified data
The greatest predictive power in stratifying the patient population in this study was observed when FISH was combined with grade or stage in the multivariate analysis (Figures 2, 3, 4, 5 and Table 4). In the multivariate model analysis carried out using SAS, the PHRED procedure, statistical significance was achieved with both the training and validation sets for all three FISH parameters (P1–P3, log-rank P<0.0001). The model with the highest statistical scores of all models possible for all given covariates is shown in Table 4. This model is based on the entire patient group (complete dataset) and reports survival times and hazard ratios for each FISH parameter.
Predictor 1 (P1): P1 discriminates patient survival the best in low-stage groups. Kaplan–Meier survival analysis of P1: percentage of cells with less than two signals for ZNF217. Best dichotomization of data for favourable outcome below a cutoff of 16%. Results grouped by clinical stage. Median survival in months for each category is shown in parentheses (log-rank test, P<0.0001).
Predictor 2 (P2): P2 is able to separate patients with low-stage tumours into groups with good and poor prognosis. Kaplan–Meier survival analysis of P2: percentage of cells with fewer ERBB2 than ZNF217 signals (ERBB2/ZNF217<1), with favourable outcome observed at or above a cutoff of 28%, grouped by clinical stage. Median survival in months for each category is shown in parentheses (log-rank test, P<0.0001).
Predictor 3 (P3): P3 is able to separate patients with low-stage tumours into groups with good and poor prognosis. Kaplan–Meier survival analysis of P3: the overall ratio of ERBB2 to ZNF217 signals, with favourable outcome observed below a cutoff ratio of 1.5, grouped by clinical stage. The late crossing of the curves in high-stage groups is owing to the small number of patients with these tumour stages. Median survival in months for each category is shown in parentheses (log-rank test, P<0.0001).
A combination of predictors is able to identify individuals with a poorer prognosis in low- and high-tumour stages (A), and by grade in the standard tripartite grading system (B). Prognosis grouped by histological grade or stage and FISH results, favourable if one of the three FISH predictors P1–P3=P is favourable in Kaplan–Meier survival analysis. Median survival in months for P for each category is shown in tables under the plots. Regarding stage (A), FISH results separate low stages IA and IIA significantly (log-rank test P<0.0001) but not higher stages (log-rank test, P=0.8060). Regarding grade (B), FISH results tend to separate G1 EAC (log-rank test, P=0.0586), G2 EAC not significantly (log-rank test, P=0.5596), but especially G3 EAC (log-rank test P=0.0007).
Each of the predictors P1, P2 and P3 adds additional prognostic information to histological grade
As is evident from Table 4, each of the three FISH predictors, P1, P2 and P3 added prognostic value to that of each histological grade in the common tripartite grading system. Furthermore, defining the FISH+ group of patients as those with poorer prognosis for each of the selected FISH parameters, the same trend for a combination of all three parameters combined (P positive defined as either P1, P2, or P3 positive) was shown in the survival analysis (Figure 5A). For each grade, FISH (P) stratified patients into FISH positive and negative groups based on the difference in the overall survival. For example, in well differentiated tumours patients’ survival could be separated in longer than study duration in comparison to 59.67 months as well as in poor differentiated tumours with a distinction of 47.0 vs 11.00 months, as is evident from Figure 5.
Fluorescence in situ hybridisation parameters and tumour stages
In contrast, the contributions of P1, P2 and P3, as well as the three parameters in combination, were more significant at low stages (I and IIA), where FISH discriminated between patients with favourable prognosis and those with survival similar to that of higher-stage patients (IIB, III and IV) (Figures 2, 3, 4 and 5B). Specifically, the effect was most significant for Stage IIA patients (data not shown). As stage IIA and IIB differ by the status of lymph node metastasis, it is possible that FISH allows detection of a subset of patients with worse outcome, possibly owing to occult metastasis, who may need to be re-evaluated for treatment.
ZNF217/cell ratio and ERBB2/cell ratio influence
Fluorescence in situ hybridisation parameters P1, P2 and P3, derived from the copy number of ZNF217 loci per cell, have high-prognostic value. Thus, we examined this FISH parameter in more detail. ZNF217/cell did not show statistical significance in the initial analysis; however, it was significant when combined with stage or grade (Figure 6). Interestingly, we found evidence that a relatively low level of ZNF217/cell (<2.8) was associated with a poorer prognosis, similar to the trend observed with P1 and P2 (Figure 6), while higher ratios were associated with a more favourable outcome. This trend was consistent with the effect of ZNF217 copy number on P1, P2 and P3.
ZNF217 /cell serves as a predictor and subdivides individuals into stage groups and within each grade according to prognosis. Kaplan–Meier survival analysis of the FISH parameter ZNF217/cell; definition and cutoff: ZNF217/cell with a cutoff of 2.8 and a favourable outcome above the cutoff, grouped by clinical stage (A), and histological grade (B). ZNF217/cell and stages I and IIA, III and IV (log-rank test, P<0.0001); ZNF217/ cell and grades 1–3 (log-rank test, P=0.0009). Median survival in months for each category is shown in tables under the plots.
In contrast, the ERBB2/cell number was directly correlated with poorer prognosis. We found a significant correlation between ERBB2 gain and poorer survival. Kaplan–Meier survival analysis indicated a median survival time greater than the study duration for patients with low ERBB2 copy number (<43% cells with ERBB2 >2 signals/cell) and 43 months for patients with higher copy number (Supplementary Figure 1A). This trend is consistent with the effect of ERBB2 copy number on P2 and P3. So besides the three predictors P1–P3, the percentage of cells with ERBB2 >2 signals/cell may serve as a predictor.
Combining ERBB2 and ZNF217 into a ratio to yield a single predictor was fulfilled with our predictor P3, which could be used as a separate prognostic marker for the whole study collective (Supplementary Figure 1B).
Discussion
The clinical management of EAC is still limited in stratifying patients into prognostic groups (Esofago, 1991) and a search for comprehensive markers to improve disease management is warranted (Ong et al, 2010; Moyes and Going, 2011).
Biomarkers in EAC are being investigated with a plethora of molecular techniques. In addition to classical histology, immunohistochemistry, methylation patterns or genetic aberrations like LOH of a certain chromosomal region (Albrecht et al, 2004; Ong et al, 2010), as well as single nucleotide polymorphism microarray (SNP-chip), are being utilised as research tools, but still lack clinical significance (Akagi et al, 2009; Wiech et al, 2009).
Regarding prognosis, MDM2, VEGF and p53 evolved as additional single markers in EAC but don’t seem to be robust across studies (Cavazzola et al, 2009; Prins et al, 2012; Renouf et al, 2013). Hence, extending the analysis to multiple targets and evaluating its feasibility for routine protocols is required. Fluorescence in situ hybridisation meets these criteria.
Multi-colour FISH used on cytological oesophageal brushing specimens has shown potential to serve as a diagnostic tool to determine the progression from BE to high-grade dysplasia and EAC with high sensitivity and specificity (Fritcher et al, 2008). This probe set consisting of ERBB2, CDKN2A, ZNF217 and MYC was transferred to manifested EAC to address a possible prognostic impact. To our knowledge this is the first study in EAC combining FISH biomarkers with outstanding clinical follow-up data on 130 individuals over 160 months.
We defined three essential predictors of overall survival: P1, P2 and P3, and compared them to the clinico-pathological parameters in univariate and multivariate analysis (Cox proportional hazards) to analyse the effect of these covariates on patient survival. We determined that each of the FISH parameters added additional predictive information independent of histological grade. Thus, FISH may be able to detect individuals with high-risk EAC, adding information to the classical histopathological staging (Figures 2, 3, 4).
In patient follow-up of over 160 months, all three FISH parameters P1, P2 and P3 predicted significant survival differences of about 40 months. This allowed the identification of patients with at least 25% prolonged lifetime. We further developed a subgroup analysis in low- and high-grade stages (I and IIA vs IIB, III and IV). Fluorescence in situ hybridisation predictors were able to better distinguish favourable and unfavourable outcomes in low tumour stages. Hence, the useful multi-colour FISH approach for the analysis of progression from BE to EAC in brush cytology sends further light on EAC’s prognosis (Brankley et al, 2012; Yao et al, 2008). In detail, the biomarkers ZNF217 and ERBB2 added more prognostic significance than CDKN2A or MYC. The latter still provided diagnostic power for detection of EAC. As seen from Table 2, all four gene loci are increased in EAC at high sensitivity and specificity, but our data support a particular role of ZNF217 and ERBB2 in EAC.
Taking a closer look at the effect of the ZNF217 locus on survival, ZNF217 showed a relatively low-level gain that was associated with a poorer prognosis, while higher copy number was linked to better outcome. Low-level gain in ZNF217, in contrast to high-copy number, appears to be unfavourable for survival in this analysis. This interesting finding may be related to the notion that initial genomic instability is followed by the selection of more stable tumour cells with low-level to moderately amplified ZNF217 in aggressive cancer cells, similar to ovarian clear cell carcinoma (Rahman et al, 2012). Our finding that EAC patients possessing higher copy number ZNF217 have better outcomes contrasts to literature reports, where Huang et al (2005) showed that a higher copy number of ZNF217 was associated with a poorer prognosis in breast cancer. Littlepage et al (2012) showed that ZNF217 as a candidate oncogene could be a target of treatment in breast cancer and is associated with poor prognosis. Those findings regarding ZNF217 seem not to be a universal phenomenon and actually are still under debate. Our data on ZNF217 are so far the first correlated to prognosis in EAC. It seems that ZNF217 has a different role in EAC prognosis as compared with breast cancer. In our study the clinical associations of the three predictors (P1–P3) have in common the finding that higher levels of ZNF217 are associated with better outcome.
ERBB2 represents the second essential part of our predictors (P1–P3). Knowledge about Her2/ERBB2 has grown substantially within the last years since the introduction of trastuzumab as a targeted therapy for gastric cancer and EAC as well (Ross and McKenna, 2001; Bookman, 2003; Gravalos and Jimeno, 2008; Aitken et al, 2010; Bang et al, 2010; Rüschoff et al, 2010). It is known that up to 25% of EACs overexpress Her2/ERBB2 in immunohistochemistry or FISH analysis. These patients might therefore be eligible for targeted therapy and benefit from a therapy-associated better outcome. Assessment of ERBB2 by means of a theranostic marker evoked the problem of tumour heterogeneity in EAC. It is strongly recommended to use whole tumour sections for diagnostic purposes. Additionally, immunohistochemistry was judged superior to FISH analysis as it maintains an overview of a possibly heterogeneous cancer sample (Rüschoff et al, 2010, 2012). In a study setting, TMA techniques are generally very useful for the simultaneous analysis of large sample numbers, as in our case. TMA generation itself is targeted on homogeneity in collected core biopsies. Possibly this is the reason behind our more homogenous ERBB2 results. The perspectives of whole slide imaging and analysis by automated digital pathology will be applicable for Multi-colour FISH analysis in the near future (Dobson et al, 2010; Braun et al, 2013; Furrer et al, 2013). This will enable a concrete mapping of tumour heterogeneity with FISH comparable to immunohistochemistry. Beyond theranostic aspects, the role of ERBB2 as a prognostic marker is still under debate (Bang et al, 2010). We found a significant correlation between low levels of ERBB2 and better outcome and an association with tumour grade and stage. Specifically, low copy number of ERBB2, which in this context is statistically defined as less than 43% of cells with ERBB2 >2/cell, was significantly associated with improved survival (Supplementary Figure 1A). This is in line with a recent meta-analysis showing 7 months of gain in survival in patients without Her2/ERBB2 expression. However, Gowryshankar et al (2014) also referred to the heterogeneous results of the prognostic impact of ERBB2 in EAC among different studies (Tanner et al, 2005; Gravalos and Jimeno, 2008; Yoon et al, 2012a, 2012b).
To obtain more objective detection methods and classifications of Her2/ERBB2, positivity should be standardized for study purposes, for example, with the envisioned partially automated sample scoring system (Dobson et al, 2010).
This is particularly mandatory as multiplex approaches complicate the number of possible combinations—we calculated n>30 mathematical terms—and might imply confusing thresholds. This accounts for our markers P1 and P2, which achieved the best discriminative P-values, but might be regarded as counterintuitive with respect to their definitions and cutoffs.
Fluorescence in situ hybridisation parameters defined as ratios are more convenient. Thus, P3 is described as the overall ratio of the ERBB2 to ZNF217 signals. Its cutoff is set at 1.5 and maintains high significance. That makes P3 the most valuable for current routine diagnostics. Additionally, ratio-based FISH predictors are advantaged as they are less dependent on the thickness of slides or nuclear truncation (see additional Supplementary Technical Notes). However, the combination of all three predictors (P1–P3) added the utmost prognostic value in our study.
In conclusion, we see great advantage in analysing several biomarkers in a single assay. This can be demonstrated by multi-colour FISH in comparison to single marker approaches, for example, investigation of p53, MDM2 and VEGF in EAC (Reid et al, 2001; Cavazzola et al, 2009; Prins et al, 2012; Renouf et al, 2013). An extensive whole genome analysis from the OCCAM group detected DCK, PAPSS2, SIRT2 and TRIM44 as potential prognostic biomarkers on the transcriptional and immunohistochemical level (Peters et al, 2010). Taking into account that tumour genetics and gene expressions interfere in a complex manner, FISH is an ancillary approach investigating prognosis in EAC. Our multi-colour-FISH assay is easy to use, ready for clinical application on FFPE-material, and combines the sensitivity, specificity and diagnostic value of CDKN2A, ZNF217, ERBB2 and MYC and the prognostic value of ZNF217 and ERBB2 into predictors like P1, P2 and P3. In particular the predictor P3 seems to be appropriate for diagnostic feasibility (Supplementary Figure 1B). In addition, it is important to note that our predictors P1–P3 were not planned a priori and, therefore, the definitions and their results might require validation in an independent study. All predictors might provide an additive opportunity for early intervention and improved outcome in patients with EAC (Reid et al, 2001; Quaroni et al, 2009).
Change history
10 June 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Aitken SJ, Thomas JS, Langdon SP, Harrison DJ, Faratian D (2010) Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann Oncol 21: 1254–1261.
Akagi T, Ito T, Kato M, Jin Z, Cheng Y, Kan T, Yamamoto G, Olaru A, Kawamata N, Boult J, Soukiasian HJ, Miller CW, Ogawa S, Meltzer SJ, Koeffler HP (2009) Chromosomal abnormalities and novel disease-related regions in progression from Barrett’s esophagus to oesophageal adenocarcinoma. Int J Cancer 125: 2349–2359.
Albrecht B, Hausmann M, Zitzelsberger H, Stein H, Siewert JR, Hopt U, Langer R, Höfler H, Werner M, Walch A (2004) Array-based comparative genomic hybridisation for the detection of DNA sequence copy number changes in Barrett’s adenocarcinoma. J Pathol 203: 780–788.
Bang Y-J, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Rüschoff J, Kang Y-K (2010) Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 376: 687–697.
Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Patricia C, Paulson TG, Rabinovitch PS, Reid BJ (1999) Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 22: 106–109.
Baumann S, Keller G, Pühringer F, Napieralski R, Feith M, Langer R, Höfler H, Stein HJ, Sarbia M (2006) The prognostic impact of O6-Methylguanine-DNA Methyltransferase (MGMT) promotor hypermethylation in oesophageal adenocarcinoma. Int J Cancer 119: 264–268.
Blot WJ (1994) OEsophageal cancer trends and risk factors. Semin Oncol 21: 403–410.
Blot WJ, Devesa SS, Kneller RW, Fraumeni JF (1991) Rising incidence of adenocarcinoma of the esophagus and gastric cardia. JAMA 265: 1287–1289.
Bookman Ma (2003) Evaluation of monoclonal humanized anti-HER2 antibody, trastuzumab, in patients with recurrent or refractory ovarian or primary peritoneal carcinoma with overexpression of HER2: a phase ii trial of the gynecologic oncology group. J Clin Oncol 21: 283–290.
Brankley SM, Fritcher EGB, Smyrk TC, Keeney ME, Campion MB, Voss JS, Clayton AC, Wang KK, Lutzke LS, Kipp BR, Halling KC (2012) Fluorescence in situ hybridisation mapping of esophagectomy specimens from patients with Barrett’s esophagus with high-grade dysplasia or adenocarcinoma. Hum Pathol 43: 172–179.
Brankley SM, Wang KK, Harwood AR, Miller DV, Legator MS, Lutzke LS, Kipp BR, Morrison LE, Halling KC (2006) The development of a fluorescence in situ hybridisation assay for the detection of dysplasia and adenocarcinoma in Barrett’s esophagus. J Mol Diagn 8: 260–267.
Braun M, Kirsten R, Rupp N, Moch H (2013) Quantification of protein expression in cells and cellular subcompartments on immunohistochemical sections using a computer supported image analysis system. Histol Histopathol. 28: 605–610.
Brown LM, Devesa SS, Chow W-H (2008) Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst 100: 1184–1187.
Bubendorf L, Nocito A, Moch H, Sauter G (2001) Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol 195: 72–79.
Cavazzola LT, Rosa ARP, da, Schirmer CC, Gurski RR, Telles JPB, Mielke F, Meurer L, Edelweiss MIA, Kruel CDP (2009) Immunohistochemical evaluation for P53 and VEGF (vascular endothelial growth factor) is not prognostic for long term survival in end stage oesophageal adenocarcinoma. Rev Col Bras Cir 36: 24–34.
Cestari R, Villanacci V, Rossi E, Della Casa D, Missale G, Conio M, Grigolato P, Bassotti G (2007) Fluorescence in situ hybridisation to evaluate dysplasia in Barrett’s esophagus: a pilot study. Cancer Lett 251: 278–287.
Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, Knoepfler PS (2008) N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res 68: 9654–9662.
Devesa SS, Blot WJ, Fraumeni JF (1998) Changing patterns in the incidence of oesophageal and gastric carcinoma in the United States. Cancer 83: 2049–2053.
Dobson L, Conway C, Hanley A, Johnson A, Costello S, O’Grady A, Connolly Y, Magee H, O’Shea D, Jeffers M, Kay E (2010) Image analysis as an adjunct to manual HER-2 immunohistochemical review: a diagnostic tool to standardize interpretation. Histopathology 57: 27–38.
Esofago D (1991) BARRETT ’ S ESOPHAGUS: epidemiological and clinical results of a multicentric survey. Int J Cancer 48: 364–368.
Fritcher EB, Brankley S, Kipp B, Voss J (2008) A comparison of conventional cytology, DNA ploidy analysis, and fluorescence in situ hybridisation for the detection of dysplasia and adenocarcinoma in patients with. Hum Pathol 39: 1128–1135.
Furrer D, Jacob S, Caron C, Sanschagrin F, Provencher L, Diorio C (2013) Validation of a new classifier for the automated analysis of the human epidermal growth factor receptor 2 (HER2) gene amplification in breast cancer specimens. Diagn Pathol 8: 17.
Gowryshankar A, Nagaraja V, Eslick GD (2014) HER2 status in Barrett’s esophagus & oesophageal cancer: a meta analysis. J Gastrointest Oncol 5: 25–35.
Gravalos C, Jimeno A (2008) HER2 in gastric cancer: a new prognostic factor and a novel therapeutic target. Ann Oncol 19: 1523–1529.
Haggitt RC (1994) Barrett’s esophagus, dysplasia, and adenocarcinoma. Hum Pathol 25: 982–993.
Hameeteman W, Tytgat GN, Houthoff HJ, van den Tweel JG (1989) Barrett’s esophagus: development of dysplasia and adenocarcinoma. Gastroenterology 96: 1249–1256.
Hamilton SR, Aaltonen LA (2000) Pathology and Genetics. Tumours of the Digestive System—WHO—OMS—. WHO: Lyon, France.
Hamilton SR, Smith RR (1987) The relationship between columnar epithelial dysplasia and invasive adenocarcinoma arising in Barrett’s esophagus. Am J Clin Pathol 87: 301–312.
Heitmiller RF, Sharma RR (1994) Comparison of prevalence and resection rates in patients with oesophageal squamous cell carcinoma and adenocarcinoma. J Thorac Cardiovasc Surg 112: 130–136.
Hou J, Jiang D, Zhang J, Gavine PR, Xu S, Liu Y, Xu C, Huang J, Tan Y, Wang H, Lu Y, Zheng L, Hou Y, Tan L (2014) Frequency, characterization, and prognostic analysis of PIK3CA gene mutations in Chinese oesophageal squamous cell carcinoma. Hum Pathol 45: 352–358.
Huang Q (2010) Biomarkers in Barrett’s oesophagus. Biochem Soc Trans 2: 343–347.
Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, Feuerstein B, Mills GB, Stokoe D, Yaswen P, Collins C (2005) ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet 14: 3219–3225.
Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TFE, Bernd H-W, Cogliatti SB, Dierlamm J, Feller AC, Hansmann M-L, Haralambieva E, Harder L, Hasenclever D, Kühn M, Lenze D, Lichter P, Martin-Subero JI, Möller P, Müller-Hermelink H-K, Ott G, Parwaresch RM, Pott C, Rosenwald A, Rosolowski M, Schwaenen C, Stürzenhofecker B, Szczepanowski M, Trautmann H, Wacker H-H, Spang R, Loeffler M, Trümper L, Stein H, Siebert R (2006) A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med 354: 2419–2430.
Huober J, Jackisch C, Untch M, Möbus V, Wallwiener D, Kaufmann M, Minkwitz G (2006) Adjuvant therapy with trastuzumab (Herceptin) in primary breast cancer. Zentralbl Gynakol 128: 30–37.
Igaki H, Sasaki H, Kishi T, Sakamoto H, Tachimori Y, Kato H, Watanabe H, Sugimura T, Terada M (1994) Highly frequent homozygous deletion of the p16 gene in oesophageal cancer cell lines. Biochem Biophys Res Commun 203: 1090–1095.
Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, Singer G, Stöhr R, Simon R, Sauter G, Leibiger H, Essers L, Weber B, Hermann K, Rosenthal A, Hartmann A, Dahl E (2004) Loss of SFRP1 is associated with breast cancer progression and poor prognosis in early stage tumors. Int J Oncol 25: 641–649.
Langer R, Von Rahden BHA, Nahrig J, Von Weyhern C, Reiter R, Feith M, Stein HJ, Siewert JR, Höfler H, Sarbia M (2006) Prognostic significance of expression patterns of c-erbB-2, p53, p16INK4A, p27KIP1, cyclin D1 and epidermal growth factor receptor in oesophageal adenocarcinoma: a tissue microarray study. J Clin Pathol 59: 631–634.
Littlepage LE, Adler AS, Kouros-Mehr H, Huang G, Chou J, Krig SR, Griffith OL, Korkola JE, Qu K, Lawson DA, Xue Q, Sternlicht MD, Dijkgraaf GJP, Yaswen P, Rugo HS, Sweeney CA, Collins CC, Gray JW, Chang HY, Werb Z (2012) The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov 2: 638–651.
Miros M, Kerlin P, Walker N (1991) Only patients with dysplasia progress to adenocarcinoma in Barrett ’s oesophagus. Gut 32: 1441–1446.
Moyes LH, Going JJ (2011) Still waiting for predictive biomarkers in Barrett’s oesophagus. J Clin Pathol 64: 742–750.
Nielsen KV, Ejlertsen B, Møller S, Jensen M-B, Balslev E, Müller S, Knoop A, Mouridsen HT (2012) Lack of independent prognostic and predictive value of centromere 17 copy number changes in breast cancer patients with known HER2 and TOP2A status. Mol Oncol 6: 88–97.
Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA (1994) Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368: 753–756.
Ong C-AJ, Lao-Sirieix P, Fitzgerald RC (2010) Biomarkers in Barrett’s esophagus and oesophageal adenocarcinoma: predictors of progression and prognosis. World J Gastroenterol 16: 5669–5681.
Peitz U, Malfertheiner P (2007) Barrett carcinoma—diagnosis, screening, surveillance, endoscopic treatment, prevention. Z Gastroenterol 45: 1264–1272.
Peters CJ, Rees JRE, Hardwick RH, Hardwick JS, Vowler SL, Ong CJ, Zhang C, Save V, O’Donovan M, Rassl D, Alderson D, Caldas C, Fitzgerald RC (2010) A 4-gene signature predicts survival of patients with resected adenocarcinoma of the esophagus, junction, and gastric cardia. Gastroenterology 139: 1995–2004.e15.
Pohl H, Welch HG (2005) The role of overdiagnosis and reclassification in the marked increase of oesophageal adenocarcinoma incidence. J Natl Cancer Inst 97: 142–146.
Prins MJD, Verhage RJJ, ten Kate FJW, van Hillegersberg R (2012) Cyclooxygenase isoenzyme-2 and vascular endothelial growth factor are associated with poor prognosis in oesophageal adenocarcinoma. J Gastrointest Surg 16: 956–966.
Quaroni L, Zhao R, Casson AG (2009) Shining light on Barrett’s esophagus. Expert Rev Gastroenterol Hepatol 3: 577–580.
Rahman MT, Nakayama K, Rahman M, Nakayama N, Ishikawa M, Katagiri A, Iida K, Nakayama S, Otsuki Y, Shih I-M, Miyazaki K (2012) Prognostic and therapeutic impact of the chromosome 20q13.2 ZNF217 locus amplification in ovarian clear cell carcinoma. Cancer 118: 2846–2857.
Rauser S, Weis R, Braselmann H, Feith M, Stein HJ, Langer R, Hutzler P, Hausmann M, Lassmann S, Siewert JR, Höfler H, Werner M, Walch A (2007) Significance of HER2 low-level copy gain in Barrett’s cancer: implications for fluorescence in situ hybridisation testing in tissues. Clin Cancer Res 13: 5115–5123.
Reid BJ (1991) Barrett’s esophagus and oesophageal adenocarcinoma. Gastroenterol Clin North A 20: 817–834.
Reid BJ, Prevo LJ, Galipeau PC, Sanchez CA, Longton G, Levine DS, Blount PL, Rabinovitch PS (2001) Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol 96: 2839–2848.
Renouf DJ, Zhai R, Sun B, Xu W, Cheung WY, Heist RS, Kulke MH, Cescon D, Asomaning K, Marshall AL, Li S, Christiani DC, Liu G (2013) Association of MDM2 T309G and p53 Arg72Pro polymorphisms and gastroesophageal reflux disease with survival in oesophageal adenocarcinoma. J Gastroenterol Hepatol 28: 1482–1488.
Ross JS, McKenna BJ (2001) The HER-2/neu oncogene in tumors of the gastrointestinal tract. Cancer Investig 19: 554–568.
Rüschoff J, Dietel M, Baretton G, Arbogast S, Walch A, Monges G, Chenard M-P, Penault-Llorca F, Nagelmeier I, Schlake W, Höfler H, Kreipe HH (2010) HER2 diagnostics in gastric cancer-guideline validation and development of standardized immunohistochemical testing. Virchows Arch 457: 299–307.
Rüschoff J, Hanna W, Bilous M, Hofmann M, Osamura RY, Penault-Llorca F, van de Vijver M, Viale G (2012) HER2 testing in gastric cancer: a practical approach. Mod Pathol 25 (5): 637–650.
Rygiel AM, van Baal JWPM, Milano F, Wang KK, ten Kate FJ, Fockens P, Rosmolen WD, Bergman JJGHM, Peppelenbosch MP, Krishnadath KK (2007) Efficient automated assessment of genetic abnormalities detected by fluorescence in situ hybridisation on brush cytology in a Barrett esophagus surveillance population. Cancer 109: 1980–1988.
Sassen A, Rochon J, Wild P, Hartmann A, Hofstaedter F, Schwarz S, Brockhoff G (2008) Cytogenetic analysis of HER1/EGFR, HER2, HER3 and HER4 in 278 breast cancer patients. Breast Cancer Res 10: R2.
Schwarz S, Rechenmacher M, Filbeck T, Knuechel R, Blaszyk H, Hartmann a, Brockhoff G (2008) Value of multicolour fluorescence in situ hybridisation (UroVysion) in the differential diagnosis of flat urothelial lesions. J Clin Pathol 61: 272–277.
Spechler SJ (1994) Barrett’s esophagus. Gastroenterologist 2: 273–284.
Spechler SJ (2002) Clinical Practice. Barrett’s esophagus. N Engl J Med 346: 836–842.
Tanner M, Hollmén M, Junttila TT, Kapanen AI, Tommola S, Soini Y, Helin H, Salo J, Joensuu H, Sihvo E, Elenius K, Isola J (2005) Amplification of HER-2 in gastric carcinoma: association with Topoisomerase IIalpha gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Oncol 16: 273–278.
Thomas T, Abrams KR, De Caestecker JS, Robinson RJ (2007) Meta analysis: cancer risk in Barrett’s oesophagus. Aliment Pharmacol Ther 26: 1465–1477.
Umar SB, Fleischer DE (2008) OEsophageal cancer: epidemiology, pathogenesis and prevention. Nat Clin Pract Gastroenterol Hepatol 5: 517–526.
Vizcaino AP, Moreno V, Lambert R, Parkin DM (2002) Time trends incidence of both major histologic types of oesophageal carcinomas in selected countries, 1973-1995. Int J Cancer 99: 860–868.
Wiech T, Nikolopoulos E, Weis R, Langer R, Bartholomé K, Timmer J, Walch AK, Höfler H, Werner M (2009) Genome-wide analysis of genetic alterations in Barrett’s adenocarcinoma using single nucleotide polymorphism arrays. Lab Invest 89: 385–397.
Yao H-Q, He S, Wu Y-P, Wang X-C, Han Y-L, Xu X, Cai Y, Wang G-Q, Wang M-R (2008) Application of multicolor fluorescence in situ hybridisation to early diagnosis of oesophageal squamous cell carcinoma. Ai Zheng 27: 1137–1143.
Yoda Y, Takeshima H, Niwa T, Kim JG, Ando T, Kushima R, Sugiyama T, Katai H, Noshiro H, Ushijima T (2014) Integrated analysis of cancer-related pathways affected by genetic and epigenetic alterations in gastric cancer. Gastric Cancer e-pub ahead or print 9 February 2014 doi:10.1007/s10120-014-0348-0.
Yoon HH, Shi Q, Sukov WR, Lewis Ma, Sattler Ca, Wiktor AE, Wu T-T, Diasio RB, Jenkins RB, Sinicrope Fa (2012a) Adverse Prognostic Impact of Intratumor Heterogeneous HER2 Gene Amplification in Patients With OEsophageal Adenocarcinoma. J Clin Oncol 30 (32): 3932–3938.
Yoon HH, Shi Q, Sukov WR, Wiktor AE, Khan M, Sattler Ca, Grothey A, Wu T-T, Diasio RB, Jenkins RB, Sinicrope Fa (2012b) Association of HER2/ErbB2 expression and gene amplification with pathologic features and prognosis in oesophageal adenocarcinomas. Clin Cancer Res 18: 546–554.
Acknowledgements
We thank Rudolf Jung for excellent technical assistance, especially in staining FISH, the TMA procedure and the team and laboratory of the Institute of Pathology, University Regensburg. We would also like to thank Annika Geppert for excellent help during writing the manuscript. The present work was performed in fulfilment of the requirements for obtaining the degree ‘Dr med’ at the Friedrich-Alexander-University Erlangen-Nuremberg (FAU).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Geppert, CI., Rümmele, P., Sarbia, M. et al. Multi-colour FISH in oesophageal adenocarcinoma—predictors of prognosis independent of stage and grade. Br J Cancer 110, 2985–2995 (2014). https://doi.org/10.1038/bjc.2014.238
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/bjc.2014.238
Keywords
This article is cited by
-
Pathogenetische Aspekte bei Vorläuferläsionen gastrointestinaler Tumoren
Der Pathologe (2016)