Abstract
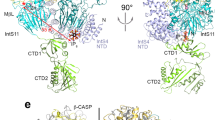
The cleavage factor Im (CF Im), consists of a 25 kDa subunit (CF Im25) and one of three larger subunits (CF Im59, CF Im68, CF Im72), and is an essential protein complex for pre-mRNA 3′-end cleavage and polyadenylation. It recognizes the upstream sequence of the poly(A) site in a sequence-dependent manner. Here we report the crystal structure of human CF Im, comprising CF Im25 and the RNA recognition motif domain of CF Im68 (CF Im68RRM), and the crystal structure of the CF Im-RNA complex. These structures show that two CF Im68RRM molecules bind to the CF Im25 dimer via a novel RRM-protein interaction mode forming a heterotetramer. The RNA-bound structure shows that two UGUAA RNA sequences, with anti-parallel orientation, bind to one CF Im25-CF Im68RRM heterotetramer, providing structural basis for the mechanism by which CF Im binds two UGUAA elements within one molecule of pre-mRNA simultaneously. Point mutation and kinetic analyses demonstrate that CF Im68RRM can bind the immediately flanking upstream region of the UGUAA element, and CF Im68RRM binding significantly increases the RNA-binding affinity of the complex, suggesting that CF Im68 makes an essential contribution to pre-mRNA binding.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Zhao J, Hyman L, Moore C . Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 1999; 63:405–445.
Takagaki Y, Manley JL . Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 2000; 20:1515–1525.
Gilmartin GM, Nevins JR . An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev 1989; 3:2180–2190.
Brown KM, Gilmartin GM . A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 2003; 12:1467–1476.
Rüegsegger U, Blank D, Keller W . Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1998; 1:243–253.
Rüegsegger U, Beyer K, Keller W . Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 1996; 271:6107–6113.
Venkataraman K, Brown KM, Gilmartin GM . Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 2005; 19:1315–1327.
Bienroth S, Keller W, Wahle E . Assembly of a processive messenger RNA polyadenylation complex. EMBO J 1993; 12:585–594.
Kubo T, Wada T, Yamaguchi Y, Shimizu A, Handa H . Knock-down of 25 kDa subunit of cleavage factor Im in Hela cells alters alternative polyadenylation within 3′-UTRs. Nucleic Acids Res 2006; 34:6264–6271.
Bessman MJ, Frick DN, O'Handley SF . The MutT proteins or "Nudix" hydrolases, a family of versatile, widely distributed, "housecleaning" enzymes. J Biol Chem 1996; 271:25059–25062.
Dreyfuss G, Swanson MS, Pinol-Roma S . Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem Sci 1988; 13:86–91.
Swanson MS, Nakagawa TY, LeVan K, Dreyfuss G . Primary structure of human nuclear ribonucleoprotein particle C proteins: conservation of sequence and domain structures in heterogeneous nuclear RNA, mRNA, and pre-rRNA-binding proteins. Mol Cell Biol 1987; 7:1731–1739.
Adam SA, Nakagawa T, Swanson MS, Woodruff TK, Dreyfuss G . mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol Cell Biol 1986; 6:2932–2943.
Smith CW, Valcarcel J . Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 2000; 25:381–388.
Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM . Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem 2004; 279:35788–35797.
Coseno M, Martin G, Berger C, et al. Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res 2008; 36:3474–3483.
Yang Q, Gilmartin GM, Doublié S . Structural basis of UGUA recognition by the Nudix protein CFIm25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci USA 2010; 107:10062–10067.
Calero G, Wilson KF, Ly T, et al. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat Struct Biol 2002; 9:912–917.
Mazza C, Ohno M, Segref A, Mattaj IW, Cusack S . Crystal structure of the human nuclear cap binding complex. Mol Cell 2001; 8:383–396.
Handa N, Nureki O, Kurimoto K, et al. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature 1999; 398:579–585.
Holm L, Sander C . Mapping the protein universe. Science 1996; 273:595–603.
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 50(Pt 5):760–763.
Clery A, Blatter M, Allain FH . RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 2008; 18:290–298.
Maris C, Dominguez C, Allain FH . The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 2005; 272:2118–2131.
Kadlec J, Izaurralde E, Cusack S . The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 2004; 11:330–337.
Bono F, Ebert J, Unterholzner L, et al. Molecular insights into the interaction of PYM with the Mago-Y14 core of the exon junction complex. EMBO Rep 2004; 5:304–310.
Lau CK, Diem MD, Dreyfuss G, Van Duyne GD . Structure of the Y14-Magoh core of the exon junction complex. Curr Biol 2003; 13:933–941.
Fribourg S, Gatfield D, Izaurralde E, Conti E . A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 2003; 10:433–439.
Selenko P, Gregorovic G, Sprangers R, et al. Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell 2003; 11:965–976.
Kielkopf CL, Rodionova NA, Green MR, Burley SK . A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 2001; 106:595–605.
Price SR, Evans PR, Nagai K . Crystal structure of the spliceosomal U2B"-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature 1998; 394:645–650.
Deo RC, Bonanno JB, Sonenberg N, Burley SK . Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 1999; 98:835–845.
Markham NR, Zuker M . UNAFold: software for nucleic acid folding and hybriziation. Methods Mol Biol 2008; 453:3–31.
Hiller M, Zhang Z, Backofen R, Stamm S . Pre-mRNA secondary structures influence exon recognition. PLoS Genet 2007; 3:e204.
Tian B, Hu J, Zhang H, Lutz CS . A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 2005; 33:201–212.
Otwinowski Z, Minor W . Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol 1997; 276:307–326.
Vagin A, Teplyakov A . MOLREP: an automated program for molecular replacement. J Appl Cryst 1997; 30:1022–1025.
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ . Phaser crystallographic software. J Appl Cryst 2007; 40:658–674.
Emsley P, Cowtan K . Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60(Pt 12 Pt 1):2126–2132.
Murshudov GN, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 1997; 53(Pt 3):240–255.
Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998; 54(Pt 5):905–921.
Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 2007; 35:W375–W383.
Acknowledgements
We thank the staff at BSRF beamline 3W1A and SSRF beamline BL17U for assistance with synchrotron data collection and Dr S Barabino (University of Milano-Bicocca) for plasmids, and Dr C Zhao and Dr J Zang (University of Science and Technology of China) for their valuable advice on experiments and manuscript. Financial support for this project was provided by research grants from the National Natural Science Foundation of China (30025012, 30900224 and 10979039), the Chinese Ministry of Science and Technology (2006CB806500, 2006CB910200 and 2006AA02A318), the Chinese Academy of Sciences (KSCX2-YW-R-60), the Chinese Ministry of Education (20070358025) and the Natural Science Foundation of Anhui Province (090413081). Heng Li was supported by the Graduate Innovation Foundation of USTC (KD2007034).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
CF Im 25 interacts with the RRM domain of CF Im68. (PDF 122 kb)
Supplementary information, Figure S2
Size exclusion chromatography assay. (PDF 128 kb)
Supplementary information, Figure S3
The L10 loop of CF Im25 interacts with CF Im68RRM. (PDF 223 kb)
Supplementary information, Figure S4
RNA recognition by CF Im25. (PDF 197 kb)
Supplementary information, Figure S5
Sensorgrams of CF Im68RRM, CF Im25, the CF 8 Im25-CF Im68RRM complex and the mutants binding to immobilized RNAs. (PDF 150 kb)
Supplementary information, Figure S6
Structure of the CF Im25-CF Im59RRM complex. (PDF 89 kb)
Supplementary information, Figure S7
(A) Surface and surface charge of the CF Im25-CF Im68RRM complex. (PDF 378 kb)
Rights and permissions
About this article
Cite this article
Li, H., Tong, S., Li, X. et al. Structural basis of pre-mRNA recognition by the human cleavage factor Im complex. Cell Res 21, 1039–1051 (2011). https://doi.org/10.1038/cr.2011.67
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2011.67
Keywords
This article is cited by
-
Targeting the polyadenylation factor EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica
Scientific Reports (2018)
-
Importance of amino acids Leu135 and Tyr236 for the interaction between EhCFIm25 and RNA: a molecular dynamics simulation study
Journal of Molecular Modeling (2018)
-
Plant polyadenylation factors: conservation and variety in the polyadenylation complex in plants
BMC Genomics (2012)