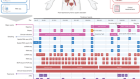
- NEWS AND VIEWS
Expanded ENCODE delivers invaluable genomic encyclopedia
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 583, 685-686 (2020)
doi: https://doi.org/10.1038/d41586-020-02139-1
References
International Human Genome Sequencing Consortium. Nature 431, 931–945 (2004).
Roadmap Epigenomics Consortium et al. Nature 518, 317–330 (2015).
Stunnenberg, H. G., International Human Epigenome Consortium & Hirst, M. Cell 167, 1145–1149 (2016).
The FANTOM Consortium and the RIKEN PMI and CLST (DGT). Nature 507, 462–470 (2014).
The ENCODE Project Consortium et al. Nature 583, 699–710 (2020).
Meuleman, W. et al. Nature https://doi.org/10.1038/s41586-020-2559-3 (2020).
Vierstra, J. et al. Nature 583, 729–736 (2020).
Partridge, E. C. et al. Nature 583, 720–728 (2020).
Grubert, F. et al. Nature 583, 737–743 (2020).
He, Y. et al. Nature 583, 752–759 (2020).
Gorkin, D. U. et al. Nature 583, 744–751 (2020).
He, P. et al. Nature 583, 760–767 (2020).
Van Nostrand, E. L. et al. Nature 583, 711–719 (2020).
The ENCODE Project Consortium. Nature 447, 799–816 (2007).
ENCODE Project Consortium. Nature 489, 57–74 (2012).
Yue, F. et al. Nature 515, 355–364 (2014).
The ENCODE Project Consortium et al. Nature 583, 693–698 (2020).
Yip, K. Y. et al. Genome Biol. 13, R48 (2012).
Regev, A. et al. eLife 6, 27041 (2017).
Dekker, J. et al. Nature 549, 219–226 (2017).

 Perspectives on ENCODE
Perspectives on ENCODE
 Read the paper: Expanded encyclopaedias of DNA elements in the human and mouse genomes
Read the paper: Expanded encyclopaedias of DNA elements in the human and mouse genomes
 Read the paper: Occupancy maps of 208 chromatin-associated proteins in one human cell type
Read the paper: Occupancy maps of 208 chromatin-associated proteins in one human cell type
 Read the paper: Landscape of cohesin-mediated chromatin loops in the human genome
Read the paper: Landscape of cohesin-mediated chromatin loops in the human genome
 Read the paper: A large-scale binding and functional map of human RNA-binding proteins
Read the paper: A large-scale binding and functional map of human RNA-binding proteins
 Read the paper: An atlas of dynamic chromatin landscapes in mouse fetal development
Read the paper: An atlas of dynamic chromatin landscapes in mouse fetal development
 Read the paper: Spatiotemporal DNA methylome dynamics of the developing mouse fetus
Read the paper: Spatiotemporal DNA methylome dynamics of the developing mouse fetus
 Read the paper: The changing mouse embryo transcriptome at whole tissue and single-cell resolution
Read the paper: The changing mouse embryo transcriptome at whole tissue and single-cell resolution
 Read the paper: Global reference mapping of human transcription factor footprints
Read the paper: Global reference mapping of human transcription factor footprints
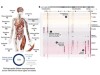 Read the paper: Index and biological spectrum of human DNase I hypersensitive sites
Read the paper: Index and biological spectrum of human DNase I hypersensitive sites
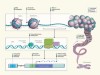 ENCODE explained
ENCODE explained
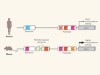 Mice in the ENCODE spotlight
Mice in the ENCODE spotlight