Abstract
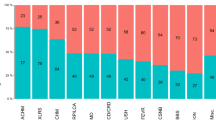
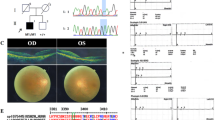
Inherited retinal degeneration (IRD) is a common cause of visual impairment (prevalence ∼1/3500). There is considerable phenotype and genotype heterogeneity, making a specific diagnosis very difficult without molecular testing. We investigated targeted capture combined with next-generation sequencing using Nimblegen 12plex arrays and the Roche 454 sequencing platform to explore its potential for clinical diagnostics in two common types of IRD, retinitis pigmentosa and cone-rod dystrophy. 50 patients (36 unknowns and 14 positive controls) were screened, and pathogenic mutations were identified in 25% of patients in the unknown, with 53% in the early-onset cases. All patients with new mutations detected had an age of onset <21 years and 44% had a family history. Thirty-one percent of mutations detected were novel. A de novo mutation in rhodopsin was identified in one early-onset case without a family history. Bioinformatic pipelines were developed to identify likely pathogenic mutations and stringent criteria were used for assignment of pathogenicity. Analysis of sequencing metrics revealed significant variability in capture efficiency and depth of coverage. We conclude that targeted capture and next-generation sequencing are likely to be very useful in a diagnostic setting, but patients with earlier onset of disease are more likely to benefit from using this strategy. The mutation-detection rate suggests that many patients are likely to have mutations in novel genes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Jaakson K, Zernant J, Külm M et al: Genotyping microarray (gene chip) for the ABCR (ABCA4) gene. Hum Mutat 2003; 22: 395–403.
Klevering BJ, Yzer S, Rohrschneider K et al: Microarray-based mutation analysis of the ABCA4 (ABCR) gene in autosomal recessive cone-rod dystrophy and retinitis pigmentosa. Eur J Hum Genet 2004; 12: 1024–1032.
Audo I, Bujakowska K, Leveillard T et al: Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis 2012; 7: 8.
Neveling K, Collin RW, Gilissen C et al: Next generation genetic testing for retinitis pigmentosa. Hum Mutat 2012; 33: 963–967.
World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. J Postgrad Med 2002; 48: 206–208.
Downes SM, Packham ER, Cranston T, Clouston P, Seller A, Nemeth AH : Detection rate of pathogenic mutations in ABCA4 using direct sequencing: clinical and research implications. Arch Ophthalmol, (in press).
den Hollander AI, Koenekoop RK, Yzer S et al: Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006; 79: 556–561.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Ng PC, Henikoff S : Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863–874.
Li B, Krishnan V, Mort ME et al: Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009; 25: 2744–2750.
Yeo G, Burge CB : Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 2004; 11: 377–394.
Reese MG, Eeckman FH, Kulp D, Haussler D : Improved splice site detection in Genie. J Comput Biol 1997; 4: 311–323.
Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C : Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67.
Davies WIL, Downes SM, Fu JK et al: Next Generation Sequencing (NGS) in healthcare delivery: lessons from the functional analysis of rhodopsin. Genet Med 2012; e-pub ahead of print 12 July 2012; doi:10.1038/gim.2012.73.
Margulies M, Egholm M, Altman WE et al: Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437: 376–380.
Inglehearn CF, Keen TJ, Bashir R et al: A completed screen for mutations of the rhodopsin gene in a panel of patients with autosomal dominant retinitis pigmentosa. Hum Mol Genet 1992; 1: 41–45.
Maugeri A, Klevering BJ, Rohrschneider K et al: Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am J Hum Genet 2000; 67: 960–966.
Shroyer NF, Lewis RA, Lupski JR : Complex inheritance of ABCR mutations in Stargardt disease: linkage disequilibrium, complex alleles, and pseudodominance. Hum Genet 2000; 106: 244–248.
September AV, Vorseter AA, Ramesar RS, Greenberg LJ : Mutation spectrum and founder chromosomes for the ABCA4 gene in South African patients with Stargardt disease. Invest Ophthalmol Vis Sci 2004; 45: 1705–1711.
Zernant J, Schubert C, Im KM et al: Analysis of the ABCA4 gene by next-generation sequencing. Invest Ophthalmol Vis Sci 2011; 52: 8479–8487.
McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP : Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA 1995; 92: 3249–3253.
Weigell-Weber M, Fokstuen S, Török B, Niemeyer G, Schinzel A, Hergersberg M : Codons 837 and 838 in the retinal guanylate cyclase gene on chromosome 17p: hot spots for mutations in autosomal dominant cone-rod dystrophy? Arch Ophthalmol 2000; 118: 300.
McLaughlin ME, Sandberg MA, Berson EL, Dryja TP : Recessive mutations in the gene encoding the −subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat Genet 1993; 4: 130–134.
den Hollander AI, ten Brink JB, de Kok YJ et al: Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat Genet 1999; 23: 217–221.
den Hollander AI, Davies J, van der Velde-Visser SD et al: CRB1 mutation spectrum in inherited retinal dystrophies. Hum Mutat 2004; 24: 355–369.
Brancati F, Barrano G, Silhavy JL et al: CEP290 mutations are frequently identified in the oculo-renal form of Joubert syndrome-related disorders. Am J Hum Genet 81: 104–113.
Hoppman-Chaney N, Peterson LM, Klee EW, Middha S, Courteau LK, Ferber MJ : Evaluation of oligonucleotide sequence capture arrays and comparison of next-generation sequencing platforms for use in molecular diagnostics. Clin Chem 2010; 56: 1297–1306.
Acknowledgements
This work was supported by the Oxford Partnership Comprehensive Biomedical Research Centre with funding from the Department of Health’s NIHR Biomedical Research Centre Programme. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health. This work was also supported in part by Roche Diagnostics, AK was funded by Ataxia UK, and JB and JR were funded by The Wellcome Trust (grant 075491/Z/04). WILD was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) grant to MWH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Rights and permissions
About this article
Cite this article
Shanks, M., Downes, S., Copley, R. et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur J Hum Genet 21, 274–280 (2013). https://doi.org/10.1038/ejhg.2012.172
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2012.172
Keywords
This article is cited by
-
Genetic characteristics of 234 Italian patients with macular and cone/cone-rod dystrophy
Scientific Reports (2022)
-
Targeted next generation sequencing and family survey enable correct genetic diagnosis in CRX associated macular dystrophy – a case report
BMC Ophthalmology (2021)
-
Genetic characteristics and epidemiology of inherited retinal degeneration in Taiwan
npj Genomic Medicine (2021)
-
The mutation spectrum in familial versus sporadic congenital cataract based on next-generation sequencing
BMC Ophthalmology (2020)
-
The psychosocial and service delivery impact of genomic testing for inherited retinal dystrophies
Journal of Community Genetics (2019)