Abstract
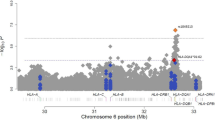
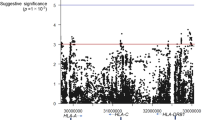
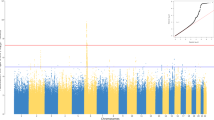
Peanut allergy (PA) is a common and serious food allergy and its prevalence has increased in the past decade. Although there is strong evidence of inheritance, the genetic causes of this disease are not well understood. Previously, a large-scale genome-wide association study described an association between human leukocyte antigen (HLA)-DQB1 and asthma; the aim of this study was to evaluate the association between HLA-DQB1 and PA. Genotypic and allelic profiles were established for 311 Caucasian members of a well-described Canadian group of children with PA and 226 Caucasian controls. Firth’s logistic regression analyses showed associations between HLA-DQB1 alleles and PA for DQB1*02 (P=1.1 × 10−8, odds ratio (OR)=0.09 (CI=0.03–0.23)) and DQB1*06:03P alleles (P=2.1 × 10−2, OR=2.82 (CI=1.48–5.45)). This study of HLA in PA demonstrates specific association between two allelic groups of the HLA-DQB1 gene (DQB1*02 and DQB1*06:03P) and PA, highlighting its possible role in the development of this disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hourihane JO : Peanut allergy. Pediatr Clin North Am 2011; 58: 445–458, xi.
Ben-Shoshan M, Harrington DW, Soller L et al: A population-based study on peanut, tree nut, fish, shellfish, and sesame allergy prevalence in Canada. J Allergy Clin Immunol 2010; 125: 1327–1335.
Moffatt MF, Gut IG, Demenais F et al: A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 2010; 363: 1211–1221.
Simpson AB, Yousef E, Hossain J : Association between peanut allergy and asthma morbidity. J Pediatr 2010; 156: 781. e771.
Shreffler WG, Charlop-Powers Z, Sicherer SH : Lack of association of HLA class II alleles with peanut allergy. Ann Allergy Asthma Immunol 2006; 96: 865–869.
Dreskin SC, Tripputi MT, Aubrey MT et al: Peanut-allergic subjects and their peanut-tolerant siblings have large differences in peanut-specific IgG that are independent of HLA class II. Clin Immunol 2010; 137: 366–373.
Howell WM, Turner SJ, Hourihane JO, Dean TP, Warner JO : HLA class II DRB1, DQB1 and DPB1 genotypic associations with peanut allergy: evidence from a family-based and case-control study. Clin Exp Allergy 1998; 28: 156–162.
Nguyen-Luu NU, Ben-Shoshan M, Alizadehfar R et al: Inadvertent exposures in children with peanut allergy. Pediatr Allergy Immunol 2012; 23: 133–139.
Chan-Yeung M, Manfreda J, Dimich-Ward H, Ferguson A, Watson W, Becker A : A randomized controlled study on the effectiveness of a multifaceted intervention program in the primary prevention of asthma in high-risk infants. Arch Pediatr Adolesc Med 2000; 154: 657–663.
Liem JJ, Huq S, Kozyrskyj AL, Becker AB : Should younger siblings of peanut-allergic children be assessed by an allergist before being fed peanut? Allergy Asthma Clin Immunol 2008; 4: 144–149.
Robinson J, Halliwell JA, McWilliam H, Lopez R, Parham P, Marsh SGE : The IMGT/HLA Database. Nucleic Acids Research 2013; 41: D1222–D1227.
Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE : IMGT/HLA - a sequence database for the human major histocompatibility complex. Tissue Antigens 2000; 55: 280–287.
Hollenbach JA, Mack SJ, Gourraud PA et al: A community standard for immunogenomic data reporting and analysis: proposal for a STrengthening the REporting of Immunogenomic Studies statement. Tissue Antigens 2011; 78: 333–344.
HLA nomenclature: nomenclature for factors of the HLA system; 2012; Accessed on September 2012. http://hla.alleles.org/alleles/p_groups.html.
Heinze G, Schemper M : A solution to the problem of separation in logistic regression. Stat Med 2002; 21: 2409–2419.
Miretti MM, Walsh EC, Ke X et al: A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 2005; 76: 634–646.
Ramagopalan SV, Ebers GC : Multiple sclerosis: major histocompatibility complexity and antigen presentation. Genome Med 2009; 1: 105.
Acknowledgements
We thank all the individuals involved in this study for their participation and the funding organizations for their support. This study was supported by the AllerGen NCE (the Allergy, Genes and Environment Network of Centers of Excellence). VT Vaillancourt is an AllerGen NCE trainee and is supported by the Corporation de recherche et d’action sur les maladies héréditaires (CORAMH) and the Campagne de développement de l’Université du Québec à Chicoutimi. Y Asai is supported by a Canadian Institutes for Health Research (CIHR) Research fellowship, an AllerGen CAIDATI training award and the Canadian Dermatology Foundation (CDF). M Ben-Shoshan is an Emerging Clinician-Scientist of the AllerGen NCE. A Kozyrskyj is the WCHRI Research Chair in Maternal-Child Health and the Environment. D Daley has received grant support from CIHR and is a Michael Smith Foundation for Health Research Career Scholar and she holds a Tier II Canadian Research Chair (Genetics of Common Complex Diseases). A Clarke is a National Research Scholar of the Fonds de recherche du Québec–santé (FRQS). P Hull is supported by the CDF and the University of Saskatchewan’s Department of Medicine Research Fund. C Laprise holds the Canada Research Chair on Genetic Determinants in Asthma and is the director of the Inflammation and Remodeling Strategic Group of the Respiratory Health Network of the FRQS. A Kozyrskyj, A Becker, D Daley, A Sandford and C Laprise are AllerGen NCE members. The Canadian studies (SAGE and CAPPS studies) were supported by the AllerGen NCE. The McGill PA case group is supported by the Foundations of the McGill University Health Center and the Montreal Children’s Hospital as well as by grants from the Canadian Allergy, Asthma and Immunology Foundation and the AllerGen NCE.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Madore, AM., Vaillancourt, V., Asai, Y. et al. HLA-DQB1*02 and DQB1*06:03P are associated with peanut allergy. Eur J Hum Genet 21, 1181–1184 (2013). https://doi.org/10.1038/ejhg.2013.13
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.13
Keywords
This article is cited by
-
Association of HLA-DQ and IL13 gene variants with challenge-proven shrimp allergy in West Bengal, India
Immunogenetics (2020)
-
Genome-wide association study of self-reported food reactions in Japanese identifies shrimp and peach specific loci in the HLA-DR/DQ gene region
Scientific Reports (2018)
-
Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy
Nature Communications (2017)
-
Genome-wide association study identifies peanut allergy-specific loci and evidence of epigenetic mediation in US children
Nature Communications (2015)